Abstract
The structure of RNase P protein from the hyperthermophilic bacterium Thermotoga maritima was determined at 1.2-Å resolution by using x-ray crystallography. This protein structure is from an ancestral-type RNase P and bears remarkable similarity to the recently determined structures of RNase P proteins from bacteria that have the distinct, Bacillus type of RNase P. These two types of protein span the extent of bacterial RNase P diversity, so the results generalize the structure of the bacterial RNase P protein. The broad phylogenetic conservation of structure and distribution of potential RNA-binding elements in the RNase P proteins indicate that all of these homologous proteins bind to their cognate RNAs primarily by interaction with the phylogenetically conserved core of the RNA. The protein is found to dimerize through an extensive, well-ordered interface. This dimerization may reflect a mechanism of thermal stability of the protein before assembly with the RNA moiety of the holoenzyme.
RNase P catalyzes the divalent metal-dependent hydrolysis of a specific phosphodiester bond in pre-tRNAs, to release the 5′ precursor sequences and produce the mature 5′ ends of the tRNAs (see refs. 1–4 for reviews). All RNase P species studied to date are complex holoenzymes composed of one RNA and at least one protein component. The bacterial RNase P typically consists of a large RNA (350–400 nt; 100–130 kDa) and one small (≈120 aa; 12–13 kDa) protein subunit. The active site of the bacterial RNase P is contained within the RNA. This is shown by the catalytic activity of the RNA in the absence of the protein moiety in vitro, at high ionic strength (5). At physiological ionic strength and in vivo, however, the protein component is required for pre-tRNA processing (5, 6).
The role of the bacterial RNase P protein in the RNase P reaction is not entirely clear. The fact that it confers high activity on the RNA at physiological ionic strength suggests that the protein stabilizes the holoenzyme against electrostatic distortion (7). In vitro, the protein component of bacterial RNase P has been implicated in increasing the turnover rate of the enzyme (7), possibly by contribution to specific binding of the substrate over the product (8). Some mutational, enzymatic, and photoaffinity crosslinking studies have been interpreted to indicate that in the bacterial holoenzyme RNase P protein may form direct contacts with the 5′ precursor sequence of the substrate pre-tRNA (9–11). The structural basis of any such interaction is not known. Moreover, there is no consistent structural model for the interaction between the protein and the RNase P RNA, despite several crosslink and footprint studies (11–14).
Phylogenetic comparative analysis has identified two major structural types of bacterial RNase P RNA (15). Both types have a homologous core structure that consists of about half the sequence lengths of the RNAs, but about half the sequence of each type of RNA has no homologous counterpart in the other RNA. The ancestral type (A type) is phylogenetically predominant; most bacterial RNase P RNAs conform to the A type of structure. The Bacillus type (B type) structure is found only in the phylogenetic group of low G+C Gram-positive bacteria. Structural differences between A- and B-type RNase P RNAs are substantial. Only ≈60% of either kind of the RNA is found in the other. The sequence similarities among the RNase P proteins are low, with ≈20–30% amino acid identity. Nonetheless, A- and B-type proteins both can activate the other type of RNA in vitro (5). Additionally, the B-type RNAs can complement the A-type protein in vivo (16). Common features in the structures of the two types of proteins are expected, therefore, to provide insight into the elements that participate in the interaction with and activation of the catalytic RNA.
Structural studies of RNase P proteins have so far been limited to B-type proteins, from the low G+C Gram-positive bacteria Bacillus subtilis (17) and Staphylococcus aureus (18). These proteins have a high degree of similarity, but are closely related and so do not provide much comparative perspective. We report here a high-resolution x-ray crystal structure of the RNase P protein from Thermotoga maritima, a hyperthermophilic bacterium with the A-type RNase P RNA.
Methods
Crystallization and Data Collection. The expression, purification, and crystallization of the RNase P protein from T. maritima have been described (19). Briefly, the protein was overexpressed in Escherichia coli as a GST fusion, the fusion polypeptide was digested with thrombin to release GST, and RNase P protein was purified by a combination of denaturing and nondenaturing ion-exchange chromatography. Plate-like protein crystals were grown by vapor diffusion from 100 mM NaOAc (pH 4.8–5.2), 12–18% PEG 1,500, 200 mM K2SO4, and 3 mg/ml protein at ambient temperature (23°C). To incorporate an anomalously scattering atom into the RNase P protein, an L41M mutation was introduced into the fusion expression construct, and the mutant protein was overexpressed in E. coli strain BL21(DE3)pLysS grown in a selenomethionine-containing medium (20). The selenomethionine-substituted mutant protein was purified and crystallized by vapor diffusion as described for the native version of the protein, except 1 mM DTT was added where appropriate to reduce oxidation of selenomethionine. Nucleation of the mutant crystal growth was promoted by introducing seed microcrystals of native protein. Crystals of the selenomethionine-substituted mutant protein were highly isomorphous to the crystals of nonmodified protein, indicating that no significant structural change was introduced by the selenomethionine substitution.
Before data collection, crystals were transferred for at least 5 min into a cryoprotectant solution [100 mM NaOAc (pH 5.2), 35% PEG 1,500, 20 mM K2SO4, and 1 mM DTT] and flash-cooled in a 1.8-ml vial of liquid propane that was immediately immersed into liquid nitrogen. The crystals then were stored inside a solid propane ``popsicle'' immersed in liquid nitrogen for up to a few weeks. Data were collected from a single crystal at cryogenic temperature (100 K) at beam line 5.0.2, Lawrence Berkeley National Laboratory. To minimize noise in measurements of the anomalous differences an inverse-beam strategy was used (21). All data were processed with mosf Lm and merged with scala programs from the ccp4 suite (22). Data redundancy and Rp.i.m. were calculated by using the computer program rmerge (23). The processing statistics are given in Table 1.
Table 1. Data collection and processing.
| Data set | λ2 (Peak) | λ1 (Inflection) | ||
|---|---|---|---|---|
| Space group | P21 | P21 | ||
| Unit cell, Å, ° | a = 56.23 | a = 56.27 | ||
| b = 64.14 | b = 64.17 | |||
| c = 68.30 | c = 68.37 | |||
| α = γ = 90 | α = γ = 90 | |||
| β = 102.01 | β = 102.01 | |||
| Wavelength, Å | 0.9792 | 0.9794 | ||
| Resolution, Å | 1.20 (1.35-1.20) | 30-1.35 | 1.20 (1.35-1.20) | 30-1.35 |
| Total reflections | 774,818 (67,203) | 707,615 | 775,753 (67,666) | 708,087 |
| Unique reflections | 118,678 (16,982) | 101,786 | 118,822 (16,897) | 101,925 |
| Redundancy | 6.53 (4.01) | 6.94 | 6.53 (4.00) | 6.95 |
| Completeness, % | 80.0 (38.7) | 97.5 | 79.8 (38.7) | 97.5 |
| deff, Å | 1.29 | 1.29 | ||
| dopt, Å | 1.22 | |||
| I/Σ(I) | 5.3 (1.1) | 5.6 | 5.2 (0.9) | 5.8 |
| Rmerge, %* | 0.059 (0.565) | 0.061 | 0.061 (0.699) | 0.065 |
| Rr.i.m.† | 0.070 (0.772) | 0.070 | 0.072 (0.954) | 0.077 |
| Rp.i.m.‡ | 0.027 (0.372) | 0.026 | 0.027 (0.456) | 0.026 |
| PCV, %§ | 8.40 (95.8) | 8.50 | 8.70 (119.5) | 9.20 |
| Overall B, Å2 | 13.688 | 13.860 |
Rmerge = ΣhklΣi|Ihkl,i - <I>hkl|/ΣhklΣiIhkl,i.
Rr.i.m. = Σhkl[N/(N - 1)]1/2Σi|Ihkl,i - <I>hkl|/ΣhklΣiIhkl,i.
Rp.i.m. = Σhkl[1/(N - 1)]1/2Σi|Ihkl,i - <I>hkl|/ΣhklΣiIhkl,i.
PCV (pooled coefficient of variation) = Σhkl{[1/(N - 1)[Σi|Ihkl,i - <I>hkl|2}1/2/ ΣhklΣiIhkl,i.
The completeness and the signal-to-noise ratio of the available data deteriorated beyond 1.35 Å. However, both high redundancy and low precision-indicating merging R factor (Rp.i.m.) indicated that these high-resolution data would be useful in the refinement. Indeed, the anisotropic refinement with shelx (see below) and the subsequent refinement steps did not notably progress unless all of the observed data (up to 1.20 Å) were used. The ``effective'' (deff, ref. 23) and optical (dopt, ref. 24) resolution values indicate that the resolution of the refined structure extends beyond 1.30 Å (Table 1).
Crystallographic Phases. The Harker section of anomalous difference Patterson's map (calculated for either λ1 or λ2 data set) exhibited three strong peaks (data not shown), suggesting that three selenium sites per asymmetric unit are well ordered. A search for the selenium sites using a combination of reciprocal and direct-space methods implemented in the cns suite (25) identified three well-ordered sites; one more site was identified upon inspection of a log-likelihood gradient map and added to the anomalous scatterers model (26, 27).
To solve the crystallographic phases, two separate phase probability distributions were generated to 2.50-Å resolution by single-wavelength anomalous diffraction phasing in cns using data sets at wavelengths λ2 (peak of Se fluorescence) and λ1 (inflection point). These phase probability distributions were then combined and a centroid phase probability distribution was calculated. The centroid phase probability distribution had a low overall figure-of-merit value (36.3%), but subsequent density modification by solvent flipping significantly improved the phases (overall figure-of-merit value 92.6%) and resulted in an interpretable experimental electron density map.
Model Building and Refinement. An initial model of the protein content of the asymmetric unit was built automatically with the arp/warp software package (28), using structural amplitudes from the λ2 data set and experimental phases extended to 1.50-Å resolution. Six starting models were averaged and refined by torsion dynamics. A total of 415 of 472 protein residues in the asymmetric unit were built in this manner to yield a reasonably good model (R = 32.9%, Rfree = 33.1%).
Subsequent refinement with cns included crystallographic refinement to 1.50-Å resolution, rounds of manual rebuilding and automated building of ordered solvent molecules, and extension of crystallographic refinement to 1.35-Å resolution. Manual rebuilding of the model was performed in o (29) and was assisted by combined 2 Fo – Fc and Fo – Fc electron density maps. The final round of refinement with cns reached R = 21.82% and Rfree = 23.68% for the data to 1.35 Å. Further refinement steps were performed with shelxl (30) against the data to 1.20 Å and included anisotropic refinement of individual ADPs, modeling of multiple conformations for 68 of 433 protein residues with xtalview (31), adjustments to the solvent model, and addition of the ``riding'' hydrogens to the protein model; the refinement reached R = 16.25%, Rfree = 20.92%. The last round of refinement was performed against all of the data (including cross-validation set) and reached R = 16.34%. Stereo-chemical quality of the model was assessed with the program procheck (32).
Results
Structure Determination. The structure was determined by using a single crystal of selenomethionine-substituted mutant (L41M) T. maritima RNase P protein. Data collection, model building, and refinement are outlined in Methods. Although the available data were incomplete beyond 1.35 Å (Table 1), refinement extended the resolution to 1.2 Å. The final model of the asymmetric unit (Table 2) consists of 433 protein residues, 533 ordered water molecules, and 16 sulfate ions.
Table 2. Refinement statistics.
| Resolution range of the reflection used | 50-1.20 Å |
| No. of reflections used | 117,875 |
| No. of least squares parameters | 41,196 |
| Observations/parameters | 2.86 |
| Solvent content fraction | 0.359 |
| Vm | 1.92 Å3/Da |
| No. of protein residues | 433 |
| Occupancy sum of non-H atoms | 4,230.30 |
| No. of water molecules | 533 |
| Average temperature factors | |
| Protein main chain | 16.65 Å2 |
| Solvent and side chains | 24.44 Å2 |
| R factor | 16.34% |
| Goodness of fit (SHELX) | 2.085 |
| Restrained goodness of fit | 1.737 |
| Protein stereochemistry | |
| rms bond lengths | 0.014 Å |
| rms angle | 2.715 |
| rms ΔBiso bonded | 2.92 |
| Overall average G factor (PROCHECK) | -0.10 |
Four molecules of the T. maritima protein (named molecules A, B, C, and D) occupy the asymmetric unit and form two asymmetric dimers. The four monomer elements of the asymmetric unit adopt similar, but not identical, conformations. A ribbon representation of the peptide backbone of one of the protein units is shown in Fig. 1A. Differences between the four monomers in the asymmetric unit are mainly in the conformations of the termini, loops, and surface side chains. This is illustrated by the superimposition of the four structures in Fig. 1B. Eight to 11 N-terminal residues in each protein molecule are sufficiently disordered that they cannot be traced reliably in the electron density maps. Thirty-eight surface side chains (mostly R and K) also are disordered. Atoms for which there was no observable electron density at the final refinement steps were not included in the model. The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB ID code 1NZ0).
Fig. 1.
Structure of the RNase P protein from T. maritima. (A) Ribbon representation of the backbone conformation of the protein. Helical regions are red; β-sheets are blue. Elements of the secondary structure are numbered in the order of their occurrence from the N terminus. N and C termini of the protein model are indicated. (B and C) Similarities among the conformations of the four T. maritima RNase P protein molecules in the asymmetric unit (molecules A, B, C, and D). The molecules were superimposed to minimize the rms deviations among their backbone atoms. (B) Wire representation of the four superimposed molecules. Molecule A, red; molecule B, blue; molecule C, cyan; molecule D, green. The orientation of the superimposed models is approximately the same as in A. (C) rms deviations (Å) between the backbone atoms of molecule A and those of the other three molecules plotted against the residue number. Regions of the secondary structure are indicated.
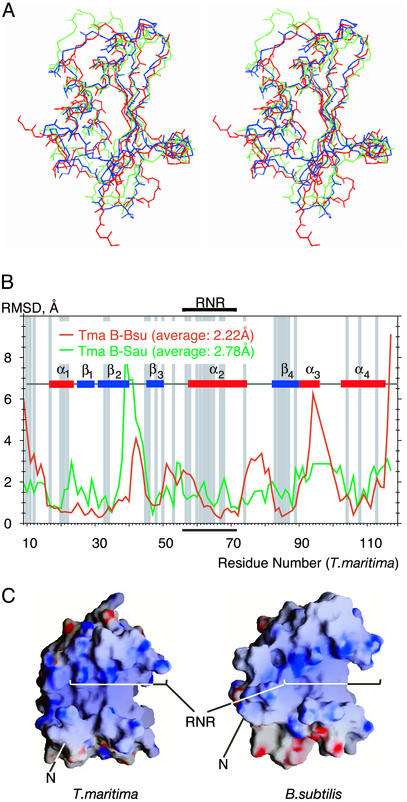
Structural Elements and Conservation. The structure of the RNase P protein from T. maritima is remarkably similar to the recently determined structures of the homologous proteins from B. subtilis (by crystallography, ref. 17) and S. aureus (by NMR, ref. 18). This is shown by the superimposition of the backbone structures of the different proteins in Fig. 2A. The rms deviations are minimal among the coordinates of the secondary structures. Indeed, differences in the T. maritima and B. subtilis structures are comparable to differences in the backbone conformations of the four different T. maritima protein molecules in the asymmetric crystal unit (compare Figs. 1C and 2B). Overall, the proteins adopt an α-β sandwich fold with a topology αβββαβα and a globular structure of approximate dimensions 40 × 35 × 30 Å. The central four-stranded β-sheet (T. maritima residues 25–29, 31–39, 45–49, and 81–89) is arranged with the topology β1-β2-β4-β3 and is surrounded by four α-helices (residues 17–23, 57–73, 90–93, and 101–113). One face of the β-sheet forms the hydrophobic core of the protein by packing together with helices α2 and α4. The other face of the β-sheet, with the helix α1 packed against it, forms a cleft that has been suggested, based on photoaffinity crosslinking and NMR studies, to interact with the 5′ leader sequence of the substrate pre-tRNA (11, 17, 18). The structural similarities of these phylogenetically diverse proteins extend to the distribution of surface charges, as shown in Fig. 2C for the electropositive faces of the proteins that potentially interact with RNA (see Discussion). Most of the conserved, positively charged amino acid residues (R7, R10, K22, A57, R60, N61, K62, K64, R65, R68, and K113) occur on the surface of the protein, mainly in an 11-aa patch that forms a highly conserved ``RNR'' sequence motif.
Fig. 2.
Similarities among the backbone conformations of RNase P proteins from T. maritima, B. subtilis (PDB ID code 1A6F), and S. aureus (PDB ID code 1D6T). The molecules were superimposed to minimize the rms deviations among their backbone atoms. (A) Cross-eyed stereo view of a wire representation of the superimposed backbones. Red, T. maritima (molecule B was chosen arbitrarily); blue, B. subtilis; green, S. aureus. The orientation of the superimposed models is related to the orientation of the models in Fig. 1 by ≈90° rotation around a vertical axis in the plane of view. (B) rms deviations (Å) between the backbone atoms of the T. maritima protein (molecule B) and those of the B. subtilis and S. aureus proteins plotted against the residue number. Regions of the secondary structure are indicated. Conserved residues (18) are shadowed in gray. The highly conserved RNR motif is bracketed. (C) Charge distribution on the surface of RNase P proteins from T. maritima and B. subtilis. Molecular surfaces of RNase P proteins were calculated from the protein x-ray structures. The electrostatic potentials were calculated from the same structures and mapped on the molecular surfaces with the grasp software package (46). Blue, positive charge; red, negative charge. The orientation of the protein molecules is the same as in A. Note that nine N-terminal residues in T. maritima protein are disordered, creating an illusion of a longer and deeper cleft on the protein surface.
The core of the T. maritima RNase P protein, as in the case of the other proteins, is well structured and composed of residues with phylogenetically conserved properties, mainly hydrophobic. Particularly conserved core residues include I20, V34, L46, I48, V50, L63, V67, F71, I84, V86, V104, and L108. Core amino acids also include R45 and D83, which form a highly conserved intramolecular salt bridge that probably is a significant stabilizing element of the folded protein conformation. Overall, the high degree of conservation of structure and residues between the A- and B-type RNase P proteins indicates a strong evolutionary pressure for conservation of the globular shape. This finding suggests that the protein is not unfolded upon interaction with the RNA subunit of the holoenzyme.
We observe 16 sulfate ions bound to the surfaces of the four protein monomers in the asymmetric unit. Four of these sulfate ions mediate crystal contacts. Although most of the sulfate ions are bound by at least one conserved protein residue, there is no specific sulfate-binding site, because each sulfate in the asymmetric unit interacts with the protein molecules in a different manner. Binding of sulfate ions to the B. subtilis RNase P protein previously was observed crystallographically (17) and suggested to contribute to conformational stability of the protein (33). Comparison of the sulfate-binding sites in the B. subtilis and T. maritima RNase P protein structures reveals no similarities, however, so we conclude that the bound sulfate anions probably have no specific biological significance.
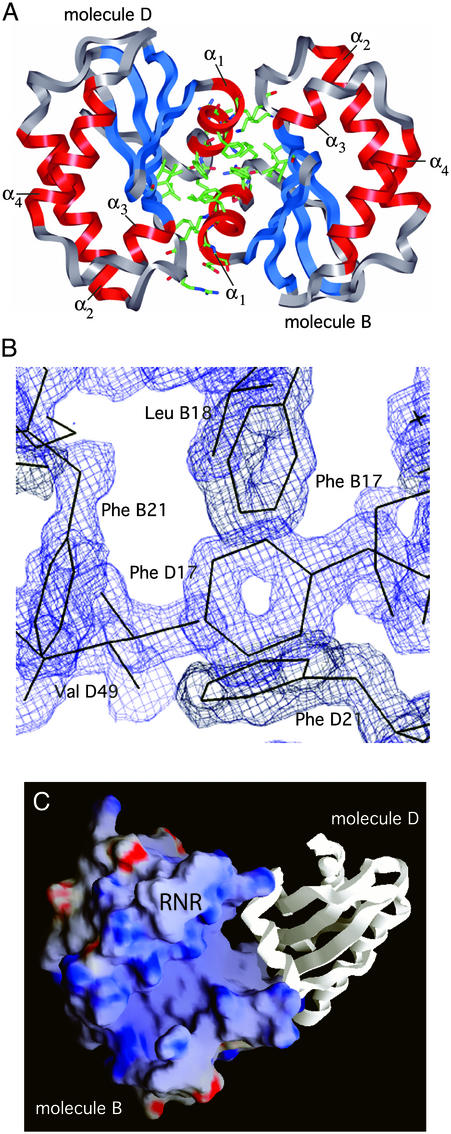
Protein Dimers. Each of the monomer units in the T. maritima RNase P protein crystal structure is involved in dimerization, shown for one of the pairs in Fig. 3A. The structure is highly resolved at the face between the monomers, as illustrated by the electron density map in Fig. 3B, part of that interface. The interaction asymmetrically packs together hydrophobic faces of the monomers and involves ≈10% of the surface area of each monomer. The two dimers in the unit cell do not have identical dimerization interactions, but they are very similar. About 80% of the atomic contacts (within 4 Å) made by the monomers with one another are the same in the two dimers. Similar dimer formation was not observed in crystals of the B. subtilis RNase P protein (17) or at high solution concentrations used for NMR analysis of the S. aureus protein (18). We do not know whether or not dimer formation in the case of the T. maritima protein has physiological relevance. The fact that the interface between the monomer units in the dimers is so extensive suggests some biological importance, perhaps for thermal stability (see Discussion).
Fig. 3.
Dimerization of the RNase P protein from T. maritima. (A) One of the two protein dimers in the asymmetric unit, formed by molecules B and D. Protein backbones are represented by ribbons. Side chains of the protein residues involved in dimerization are shown. (B)A2 F0 – FC electron density map at 1.2-Å resolution (contoured at 1.5 σ) illustrates a high degree of order at the dimerization interface. (C) Potential RNase P RNA-binding surfaces are exposed in the protein dimer. Electrostatic potentials were calculated and mapped onto protein molecular surfaces as described in the Fig. 2 legend. (C) View of the potential RNase P RNA-binding face of the protein molecule B of the B-D dimer (molecule D is shown in ribbon representation). The location of the highly conserved RNR motif is indicated. Orientation of the molecule B is related to the orientation of the molecules in Fig. 2 by counterclockwise rotation of ≈30° around the axis normal to the plane of view.
Discussion
The A- and B-type RNase P proteins are from organisms that represent a wide breadth of bacterial phylogenetic diversity. The structural similarity between RNase P proteins from T. maritima (A-type RNA) and B. subtilis and S. aureus (B-type RNA) is remarkable considering the substantial evolutionary distance between these groups of bacteria and the relatively low sequence identity among the proteins. In the light of this similarity between the A and B types of proteins, we can expect that any bacterial RNase P protein has a similar structure. This expectation does not extend beyond the phylogenetic domain of bacteria, however. Although eukaryotes and archaea possess an RNase P RNA that is a clear homolog of the bacterial version, no homolog of the bacterial RNase P protein has been identified in genome searches. Instead, both eukaryotes and archaea have substituted a suite of several proteins, all distinct from the bacterial RNase P protein (34).
It has been suggested that phylogenetically conserved, positively charged surface side chains in the protein constitute recognition determinants for the RNase P RNA (17, 18). One conspicuous feature is the RNR motif of helix α2 (indicated in Figs. 1 A and 2). This most highly conserved portion of the protein forms, together with the N-terminal part of the protein and strand β3 of the β-sheet, a positively charged surface that is highly similar between A- and B-type proteins (Fig. 2C). In the case of the E. coli RNase P holoenzyme, a photoagent attached to a residue on this surface was found to crosslink to the RNase P RNA (14), consistent with the orientation of this positively charged surface toward the RNase P RNA in the holoenzyme. The RNR motif is part of an unusual left-handed βαβ-crossover, which occurs among RNA-binding motifs in the ribosomal protein S5 superfamily (17). This superfamily includes, besides RNase P protein, the ribosomal protein S9, domain IV of ribosomal elongation factor G, and the C-terminal domain of ribosomal protein S5. In the cases of S5 and EF-G proteins, the left-handed βαβ-crossover is known to contain residues critical for RNA binding (35–38). This correlation between the different proteins provides indirect support for a role of the RNR motif of RNase P protein in RNA binding (17).
The striking conservation of shape and distribution of charges on the surface of the RNase P proteins indicates conservation of the complementary protein-binding surface among RNase P RNAs. Only about half the length of any particular RNase P RNA occurs in all bacterial RNAs. This conserved component constitutes the core of the folded RNA structures. The high degree of similarity of the different protein structures indicates that RNase P protein binds RNase P RNA primarily through direct interactions with the phylogenetically conserved core of the RNA (39). The structure of this interaction remain to be uncovered.
T. maritima is a hyperthermophilic organism with optimum growth at 85–90°C, and the RNase P protein is correspondingly expected to be more stable to denaturation than the RNase P proteins of mesophiles (40). Proteins that are part of holoenzyme complexes need to be structurally stable both within the holoenzyme and before the assembly into the holoenzyme. Structural stability before assembly is expected to be required for specific recognition of the binding site on the RNase P RNA.
Enhanced thermostability of proteins is commonly attributed to a number of factors, including a higher number of ion pairs per residue (41), shorter loops and a higher number of proline residues in the loops (42), decreased usage of uncharged polar amino acids, and an increase in the average residue volume that correlates with increased residue hydrophobicity (43). The number of observed ion pairs per residue is not significantly different between the T. maritima (0.043–0.051) and mesophilic B. subtilis (0.053) RNase P protein homologs and therefore cannot be considered a significant thermostability factor. If ionic interactions with the ligands (sulfate and zinc ions) were included, the number of ion pairs per residue increases dramatically to a value typical of the hyperthermophilic proteins, but still is not particularly different between the T. maritima (0.085–0.090) and B. subtilis (0.088) homologs. Given the strong dependence of thermodynamic stability of the RNase P protein on the concentration of anionic species (33), such a high number of ionic contacts perhaps is a primary mechanism for overcoming the unfavorable surface charge density associated with an RNA-binding protein not bound to RNA (Fig. 2C).
Because of the high evolutionary pressure for conservation of the shape of the surface, RNase P proteins, with few exceptions, exhibit little variation in the length of the loops. Thus, loop size and composition evidently do not contribute to thermostability. The one region of sequence length variation in the proteins from T. maritima and B. subtilis is between the strand β4 and the helix α4. In the B. subtilis protein structure, these elements are connected with a fairly large loop, eight residues in length. In the T. maritima protein structure, this linker extends to 12 residues, but four residues in this region adopt a conformation of a classic α-helix (helix α3). The average (per residue) B-factor values in this region of the B. subtilis protein are 0.60 SD below the average value for the whole protein, but in the T. maritima protein the corresponding values are 0.67 SD above the average. Therefore, even the acquisition of a small secondary structure element does not appear to contribute to the thermal stability of the protein in this case, because this element is more disordered (compared with the rest of the protein) than the corresponding loop of the mesophilic homolog. The number of proline residues in the loops of mesophilic RNase P proteins varies from zero (Borellia burgdorferi, SWISS PROT accession no. P50069) to 10 per protein (Chlamydia pneumoniae, SWISS PROT accession no. Q9Z6X2); two proline residues in the case of the T. maritima protein therefore are not expected to make a significant contribution to thermal stability.
The amino acid composition of the T. maritima protein is generally consistent with its thermophilic nature. The protein is low in uncharged polar amino acid content (only 12% of total amino acids, compared with 17–22% in the mesophilic homologs). The fraction of hydrophobic residues in T. maritima protein (47.9%), however, is similar to that of mesophiles (e.g., S. aureus, 50.5%). The ratio of the number of specific amino acid occurrences that correlate with thermophilicity (43) to the number that do not correlate is higher than in mesophilic RNase P proteins, but there is only a modest increase in the average residue volume (150.4 Å3 in the T. maritima protein vs. 137.0–144.7 Å3 in mesophilic proteins).
One property of the T. maritima RNase P protein that may have significant impact on thermal stability is the formation of the dimers seen in the crystal structure. The B. subtilis RNase P holoenzyme has been reported to dimerize in solution at high concentrations (44, 45), but it is not clear that this dimerization is physiologically relevant. We have not studied the protein dimerization process in solution, but the extensive fit of the interface between the monomer units suggests some biological role. Dimerization of the T. maritima RNase protein is expected to impart some, perhaps a considerable, degree of thermal stability. Side chains involved in dimerization are mostly hydrophobic, well ordered (Fig. 3B), and inaccessible to solvent. The overall surface area consumed by dimerization is large (≈1.4 × 103 Å2), ≈10% of the surface area of each of the monomers. In essence, dimerization of the T. maritima RNase P protein results in the formation of a classic hydrophobic core that is comparable in size to the hydrophobic core of a monomer. The free energy gain associated with the folding of this additional hydrophobic core could be a major factor contributing to thermal stability of the T. maritima RNase P protein before docking onto the RNA subunit. The proposed RNase P RNA-binding surfaces of the proteins in the dimer are both solvent-accessible (Fig. 3), consistent with the possibility that the dimer can interact with RNA. Because active bacterial RNase P holoenzyme is thought to consist of one RNA and one protein molecules (45), one protein in the dimer would need to dissociate during maturation of the holoenzyme. Thus, in this speculative model one of the monomers in the protein dimer would serve as a molecular chaperone to deliver the other monomer, in the folded state, to the docking site on the RNA.
Acknowledgments
We thank the staffs of the x-ray facility at the Department of Chemistry and Biochemistry (University of Colorado, Boulder) and of beam line 5.0.2 at the Lawrence Berkeley National Laboratory for their time and efforts. This work was supported by National Institutes of Health Grant GM34527 (to N.R.P.).
Abbreviations: A type, ancestral type; B type, Bacillus type.
Data deposition: Atomic coordinates and experimental intensities have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1NZ0).
References
- 1.Frank, D. N. & Pace, N. R. (1998) Annu. Rev. Biochem. 67, 153–180. [DOI] [PubMed] [Google Scholar]
- 2.Gopalan, V., Vioque, A. & Altman, S. (2002) J. Biol. Chem. 277, 6759–6762. [DOI] [PubMed] [Google Scholar]
- 3.Gopalan, V., Talbot, S. J. & Altman, S. (1994) in RNA-Protein Interactions, eds. Nagai, K. & Mattaj, I. W. (IRL, Oxford), pp. 103–126.
- 4.Harris, M. E., Frank, D. N. & Pace, N. R. (1998) in RNA Structure and Function, eds. Simons, R. W. & Grunberg-Manago, M. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 309–337.
- 5.Guerrier-Takada, C., Gardiner, K., Marsh, T., Pace, N. & Altman, S. (1983) Cell 35, 849–857. [DOI] [PubMed] [Google Scholar]
- 6.Schedl, P. & Primakoff, P. (1973) Proc. Natl. Acad. Sci. USA 70, 2091–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reich, C., Olsen, G. J., Pace, B. & Pace, N. R. (1988) Science 239, 178–181. [DOI] [PubMed] [Google Scholar]
- 8.Crary, S. M., Niranjanakumari, S. & Fierke, C. A. (1998) Biochemistry 37, 9409–9416. [DOI] [PubMed] [Google Scholar]
- 9.Loria, A. & Pan, T. (1998) Biochemistry 37, 10126–10133. [DOI] [PubMed] [Google Scholar]
- 10.Kurz, J. C., Niranjanakumari, S. & Fierke, C. A. (1998) Biochemistry 37, 2393–2400. [DOI] [PubMed] [Google Scholar]
- 11.Niranjanakumari, S., Stams, T., Crary, S. M., Christianson, D. W. & Fierke, C. A. (1998) Proc. Natl. Acad. Sci. USA 95, 15212–15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biswas, R., Ledman, D. W., Fox, R. O., Altman, S. & Gopalan, V. (2000) J. Mol. Biol. 296, 19–31. [DOI] [PubMed] [Google Scholar]
- 13.Rox, C., Feltens, R., Pfeiffer, T. & Hartmann, R. K. (2002) J. Mol. Biol. 315, 551–560. [DOI] [PubMed] [Google Scholar]
- 14.Sharkady, S. M. & Nolan, J. M. (2001) Nucleic Acids Res. 29, 3848–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haas, E. S. & Brown, J. W. (1998) Nucleic Acids Res. 26, 4093–4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waugh, D. S. & Pace, N. R. (1990) J. Bacteriol. 172, 6316–6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stams, T., Niranjanakumari, S., Fierke, C. A. & Christianson, D. W. (1998) Science 280, 752–755. [DOI] [PubMed] [Google Scholar]
- 18.Spitzfaden, C., Nicholson, N., Jones, J. J., Guth, S., Lehr, R., Prescott, C. D., Hegg, L. A. & Eggleston, D. S. (2000) J. Mol. Biol. 295, 105–115. [DOI] [PubMed] [Google Scholar]
- 19.Krivenko, A. A., Kazantsev, A. V., Adamidi, C., Harrington, D. J. & Pace, N. R. (2002) Acta Crystallogr. D 58, 1234–1236. [DOI] [PubMed] [Google Scholar]
- 20.Doublie, S. (1997) Methods Enzymol. 276, 523–530. [PubMed] [Google Scholar]
- 21.Hendrickson, W. A. & Ogata, C. M. (1997) Methods Enzymol. 276, 494–523. [DOI] [PubMed] [Google Scholar]
- 22.Collaborative Computational Project No. 4 (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 23.Weiss, M. S. (2001) J. Appl. Crystallogr. 34, 130–135. [Google Scholar]
- 24.Vaguine, A. A., Richelle, J. & Wodak, S. J. (1999) Acta Crystallogr. D 55, 191–205. [DOI] [PubMed] [Google Scholar]
- 25.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 26.Bricogne, G. (1984) Acta Crystallogr. A 40, 410–445. [Google Scholar]
- 27.Grosse-Kunstleve, R. W. & Brunger, A. T. (1999) Acta Crystallogr. D 55, 1568–1577. [DOI] [PubMed] [Google Scholar]
- 28.Perrakis, A., Morris, R. & Lamzin, V. S. (1999) Nat. Struct. Biol. 6, 458–463. [DOI] [PubMed] [Google Scholar]
- 29.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 30.Sheldrick, G. M. & Schnider, T. R. (1997) in Methods Enzymol. 277, 319–343. [PubMed] [Google Scholar]
- 31.McRee, D. E. (1999) J. Struct. Biol. 125, 156–165. [DOI] [PubMed] [Google Scholar]
- 32.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 33.Henkels, C. H., Kurz, J. C., Fierke, C. A. & Oas, T. G. (2001) Biochemistry 40, 2777–2789. [DOI] [PubMed] [Google Scholar]
- 34.Xiao, S., Scott, F., Fierke, C. A. & Engelke, D. R. (2002) Annu. Rev. Biochem. 71, 165–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Czworkowski, J. & Moore, P. B. (1997) Biochemistry 36, 10327–10334. [DOI] [PubMed] [Google Scholar]
- 36.Nissen, P., Kjeldgaard, M., Thirup, S., Polekhina, G., Reshetnikova, L., Clark, B. F. & Nyborg, J. (1995) Science 270, 1464–1472. [DOI] [PubMed] [Google Scholar]
- 37.Czworkowski, J., Wang, J., Steitz, T. A. & Moore, P. B. (1994) EMBO J. 13, 3661–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramakrishnan, V. & White, S. W. (1992) Nature 358, 768–771. [DOI] [PubMed] [Google Scholar]
- 39.Pace, N. R. & Brown, J. W. (1995) J. Bacteriol. 177, 1919–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paul, R., Lazarev, D. & Altman, S. (2001) Nucleic Acids Res. 29, 880–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan, M. K., Mukund, S., Kletzin, A., Adams, M. W. & Rees, D. C. (1995) Science 267, 1463–1469. [DOI] [PubMed] [Google Scholar]
- 42.Watanabe, K., Chishiro, K., Kitamura, K. & Suzuki, Y. (1991) J. Biol. Chem. 266, 24287–24294. [PubMed] [Google Scholar]
- 43.Haney, P. J., Badger, J. H., Buldak, G. L., Reich, C. I., Woese, C. R. & Olsen, G. J. (1999) Proc. Natl. Acad. Sci. USA 96, 3578–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fang, X. W., Yang, X. J., Littrell, K., Niranjanakumari, S., Thiyagarajan, P., Fierke, C. A., Sosnick, T. R. & Pan, T. (2001) RNA 7, 233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barrera, A., Fang, X., Jacob, J., Casey, E., Thiyagarajan, P. & Pan, T. (2002) Biochemistry 41, 12986–12994. [DOI] [PubMed] [Google Scholar]
- 46.Nicholls, A., Sharp, K. & Honig, B. (1991) Proteins Struct. Funct. Genet. 11, 281–296. [DOI] [PubMed] [Google Scholar]