Abstract
Acetyl-CoA carboxylase 1 (ACC1) catalyzes the formation of malonyl-CoA, the C2 donor for de novo synthesis of long-chain fatty acids. We have identified 64 exons, including 7 alternatively spliced minor exons (1A, 1B, 1C, 3, 5A′, 5A, and 5B) in human ACC1 gene (≈330 kb). The gene is regulated by three promoters (PI, PII, and PIII), which are located upstream of exons 1, 2, and 5A, respectively. PI is a constitutive promoter and has no homology with the PI sequences of other mammalian ACC1. PII is regulated by various hormones. PIII is expressed in a tissue-specific manner. The presence of several alternatively spliced exons does not alter the translation of the 265-kDa ACC1 protein starting from an ATG present in exon 5. Translation of PIII transcripts from exon 5A generates a 259-kDa isoform in which the N-terminal 75 aa of 265-kDa ACC1 are replaced with a new sequence of 17 aa. Interestingly, the inclusion of exon 5B between 5A and 6 in PIII transcripts would yield a third 257-kDa isoform, which is translated from an ATG in exon 6. However, the presence of exon 5B in PI and PII transcripts leads to an in-frame stop codon that results in an ACC1-related 77-aa peptide. The presence of alternatively spliced exons and three isoforms of ACC1 could contribute to overall ACC1 activity either by influencing the mRNA stability and translational efficiency or by increasing the stability and specific activity of the ACC1 protein, respectively.
Acetyl-CoA carboxylase 1 (ACC1) catalyzes the carboxylation of acetyl-CoA to form malonyl-CoA, the donor of the two-carbon units in the synthesis of long-chain fatty acids (1). ACC1 is highly expressed in lipogenic tissues such as liver, adipose, and lactating mammary gland (2), and its activities are regulated at various levels. Short-term regulation of enzymatic activities occurs by allosteric interactions with cellular metabolites such as citrate, CoA, and palmitoyl-CoA (1, 3) and by reversible phosphorylations, which are influenced by diet and hormones (4–7). Long-term regulation of ACC1 by diet and hormones also occurs at the level of transcription (8, 9).
The regulation of ACC1 gene in rat is under control by two promoters (PI and PII), which are located upstream of exons 1 and 2, respectively (3, 10, 11). Alternative usage of these promoters leads to the heterogeneity of the 5′-untranslated region (10). Because all of these transcripts contain the translational start codon located in exon 5, the mRNAs generated by both promoters produce the same 265-kDa ACC1 protein. PI transcripts are principally expressed in adipose tissues, but are expressed in the liver only under lipogenic conditions (10). Rat PI contains 28 repeats of CA dinucleotide, which act as a repressor element (12). PII, on the other hand, contains several regulatory elements and is activated in lactating rat mammary gland (11–15).
A third promoter (PIII) has been identified in bovine and ovine ACC1 genes (14, 16). PIII transcribes the mRNA from exon 5A, which is located in intron 5. Lactation dramatically stimulates expression of PIII in ovine and bovine mammary glands (16, 17). Exon 5A encodes 17 residues that constitute the N terminus of a 259-kDa isoform of ACC1 (17). However, neither the existence of the isoform of ACC1 protein nor the physiological role of the N-terminal alteration of the enzyme has been demonstrated (16, 17).
The regulation of ACC1 gene of rat (1, 3, 18), ovine (16, 19, 20), and bovine (14, 15, 17) have been extensively investigated. Except for a description that there are two kinds of 5′-untranslated regions in human ACC1 mRNA population (21), not much is known about the regulation of human ACC1 gene. We describe here the characterization of the human ACC1 gene, identification of seven alternative exons located in introns 1 and 5, the presence of three promoters, and the complex heterogeneity of the 5′-region of human ACC1 mRNA. The potential existence of three isoforms of ACC1 protein is also described.
Materials and Methods
RT-PCR. Total RNAs were extracted from different cell lines and tissues by using TRIzol Reagent (Invitrogen). First-strand cDNA was synthesized by using 5 μg of total RNA, 250 ng of hexamer random primer, and SuperScript II RNase H– Reverse Transcriptase (Invitrogen). Usually, 2–10% of the yield from first-strand cDNA synthesis was used as template for subsequent PCR amplifications using Taq DNA Polymerase (Invitrogen) and ``touch-down'' amplification protocols (14). The primer sequences used in RT-PCR and other amplification described below are listed in Table 1.
Table 1. Oligonucleotides used for various PCR amplification.
| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| RT-PCR | |
| ex1f | CTGCTCGTGGATGAACCAGAC |
| ex2f | TGTCAGCCATCGCCCGAGC |
| ex5f | CTGTAAGAGCTCATTTTGGAGGA |
| ex5Af1 | GGCAGCCAGGAGAGGCAG |
| ex5Af2 | TAAAGGAGAGCTGCTAAGATCTAC |
| ex6r2 | CCACAGTGAAATCTCGTTGAGA |
| ex6f | gagctcaCATGTCTGGCTTGCACCTAGTAA |
| ex12r | agaTCTGAGGCCTTGATCATTACTG |
| 5′-RACE | |
| ex4r | TTCTTACTGTCTGAGTAGATATCC |
| ex1r | CTGATTGAAACGCACCCTCTTC |
| ex2r | CTCAGGGTGGCAACGTGTG |
| ex5A-6r | CAGTCTGGACCTTTGAAGCATG |
| ex5Ar2 | TTACACACACAATTAGTTGATTTC |
| Promoter specific | |
| P1f1 | GAGGCTGAGGCATGAGAGTTG |
| ex1r2 | gaaggcctGCACGGGGACAGCAGCAGG |
| P2f1 | ggggtaccGGGCGACAGAGCGAGACTCC |
| ex2r2 | gaaggcctGATGGCTGACAGGCGTGCAG |
| P3f1 | ggggtaccAGCTGGTTGTCATGTTGGGTG |
| P3f2 | ggggtaccGAGTTTATCAAGATCACAGGGTG |
| ex5Ar3 | gagatatcCTGCCTCTCCTGGCTGCCAG |
The primer names indicate which exon or promoter they are specific for, whether they are forward or reverse, and whether more than one primer was designed from the same region. The sequences shown in uppercase are based on human ACC1 gene sequence, and the sequences indicated in lowercase contain restriction enzyme sites used for cloning the amplified products.
5′-RACE. The 5′-RACE amplifications were carried out with Marathon human mammary gland cDNA Amplification Kit with adaptor primers AP1 and AP2 (CLONTECH). To amplify the 5′-end of human ACC1 mRNA, AP1 and exon 4-specific reverse primer ex4r (Table 1) were used to amplify all of the cDNAs encoding PI/PII-generated transcripts. The resulting primary PCR products were diluted 50-fold and used as templates for amplification of cDNAs specific for PI-generated transcripts by using primers AP2 and ex1r. The cDNAs encoding PII-generated transcripts were amplified by using AP2 and ex2r. To determine the 5′-end of exon 5A, the primary PCR was carried out by using AP1 and ex5A-6r, the secondary PCR by using AP2 and ex5Ar2. The RACE products were cloned into pGEM-T Easy vector (Promega) and sequenced.
Cloning of Human ACC1 Promoters and Construction of Reporter Gene Plasmids. The bacterial artificial chromosome (BAC) genomic clone 24B7 was isolated by screening a human BAC library with ACC1 exon 1 primers. By using this BAC DNA as template, high-fidelity PCR was used to amplify ACC1 promoters (PI, PII, and PIII) with Platinum Pfx DNA Polymerase (Invitrogen) as described by the manufacturer's instructions. The primers (Table 1) were designed based on the ACC1 genomic sequence (GenBank accession no. AC068400). The PI PCR product amplified by using P1f1/ex1r2 primer pair was digested with SstI/StuI and cloned into SstI/blunted HindIII sites of pGL3-Basic, a promoter-less firefly luciferase reporter gene vector (Promega), generating the plasmid PI(–2019/+53)Luc, which contains 2,019 bp of PI and 53 bp of exon 1. The PII PCR product obtained by using primers P2f1 and ex2r2 was digested with KpnI/StuI, cloned into KpnI/blunted HindIII sites of pGL3-Basic, resulting in the plasmid PII(–1965/+30)Luc. The PIII PCR-amplified products obtained by using primer pair P3f1/ex5Ar3 or P3f2/ex5Ar3 were digested with KpnI/EcoRV, cloned into KpnI/blunted HindIII sites of pGL3-Basic. The resulting PIII constructs were designated as PIII(–3041/+18)Luc and PIII(–998/+18)Luc, respectively. Another two PIII reporter gene constructs PIII(–3041/+182)Luc and PIII(–998/+182)Luc were generated by using the primer pairs P3f1/ex5Ar2 and P3f2/ex5Ar2, respectively. A series of deletion constructs of the three promoters were generated from the longest promoter containing reporter plasmids by exploiting available restriction sites.
Cell Culture, Transient Transfection, and Luciferase Assay. Human cell lines HepG2 and T-47D were maintained as described (22–24). The human lung cell line H441 was maintained in the same medium as HepG2. Human breast cancer ZR-75-1 cells were maintained in RPMI medium 1640, supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin. For transient transfections, cells were split into 24-well plates (5 × 104 cells per well) 1 day before transfection. A mixture of 0.5 μg of the respective ACC1 promoter firefly luciferase reporter construct, and 40 ng of the control plasmid pRL-TK (Promega), which expresses Renilla luciferase under the control of TK promoter, were cotransfected by using LipofectAMINE 2000 (Invitrogen) as described by the manufacturer. Two days after transfection, firefly luciferase and Renilla luciferase activities were measured in relative light units by using Dual-Luciferase Assay Kit (Promega) and Monolight 2010 Luminometer (Analytical Luminescence Laboratory, San Diego). The firefly luciferase activities were normalized against the Renilla luciferase activities. In each experiment, each construct was transfected in duplicate, and the transfection experiments were repeated at least three times. The conditions used for studying the regulation of ACC1 promoters by T3 and sterol were as described (23, 24).
In Vitro Transcription and Translation. The plasmids containing human ACC1 cDNA sequences were used as templates to synthesize the protein by using TNT reticulocyte transcription and translation kit (Promega). Translation reactions were performed by using [35S]methionine (Amersham Pharmacia), and the radioactive proteins were resolved by using 4–12% NuPAGE gel (Invitrogen) using Mes buffer and detected by autoradiography.
Results and Discussion
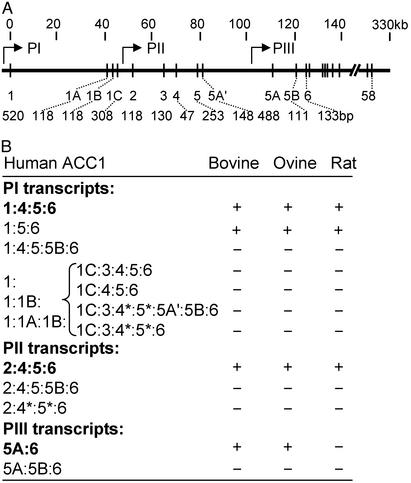
Genomic Structure of Human ACC1 Gene. Human ACC1 gene (Fig. 1A) is ≈330 kb in length (GenBank accession no. NT_010795). There are a total 64 exons, including 7 additional exons (1A, 1B, 1C, 3, 5A′, 5A, and 5B), that we have identified in this study. ACC1 protein contains three functional domains. ATP- and biotin-binding sites are encoded by exons 9 and 22, respectively. Acyl-CoA binding regions are encoded by both exons 50 and 51. Exon 5 contains the ATG codon as in other animal ACC1.
Fig. 1.
Genomic structure of the 5′-region of human ACC1 gene and heterogeneity of 5′-region of the mRNA. (A) The depiction of the exon-intron organization of human ACC1 gene is shown. Only the first six major exons, and additional exons, sizes of the exons, and the promoter locations are indicated. Each vertical line represents an exon. Promoters are designated as PI, PII, and PIII. (B) The exon compositions of 5′-region of ACC1 mRNA transcribed from three promoters are shown. The major exon compositions are indicated in bold. The asterisks (*) indicate the splice-variants found within the exons 4 and 5. For comparison, the existence of the corresponding transcripts in bovine, ovine, and rat is also listed on the right.
The sequence of exon 1 has no homology with the exon 1 sequences of rat, bovine, or ovine ACC1 genes (14, 21). Exon 3 (130 bp) is unique and has no homology with the rat and bovine exon 3 sequences. Rat ACC1 exon 3 is 61 bp (10), whereas bovine ACC1 exon 3 is 47 or 54 bp, depending on the alternative splicing (14). Exon 5A, found in human ACC1 gene, is highly homologous to that described in bovine and ovine ACC1 genes (16, 17). However, this exon is not present in rat or mouse ACC1 genes. Exon 32 is only 24 bp and encodes 8 aa and is alternatively spliced. The presence of exon 32-encoded 8 aa in ACC1 protein inhibits the in vitro phosphorylation of Ser-1200 by cAMP-dependent protein kinase in rat ACC1 (25). However, in lactating ovine, the ratio of mRNA lacking exon 32 to those containing exon 32 increases significantly, and hence this exon is alternatively spliced during lactation to increase the overall activity of ACC1 (26). This discrepancy needs further clarification.
Heterogeneity of 5′-Region of Human ACC1 mRNA. RT-PCR amplification of human ACC1 mRNA isolated from ZR-75-1 cells revealed unique varieties of the 5′-unstranslated region. Many of the transcripts did not exist in other species (Fig. 1B). In most transcripts, exon 4 (nucleotides 1–47) was spliced to exon 5 (nucleotides 1–253) whereas, in some transcripts, only nucleotides 1–31 of exon 4 were spliced to nucleotides 91–253 of exon 5 (represented as exons 4* and 5* in Fig. 1B). The exon composition before exon 1C is either 1, [1:1B], or [1:1A:1B]. Fig. 1B summarizes the transcripts that we have characterized so far; however, it seems that other exon compositions of the 5′-untranslated region might exist.
Determination of the Number of Human ACC1 Promoters. We used RACE and RT-PCR to determine the transcription initiation site by using exons 1, 2, or 5A specific reverse primer (Table 1, Fig. 1B). Based on the sequence analysis, exon 1 is 520 bp, which is 278 bp longer than that estimated by Ha et al. (21). Exon 2 is 118 bp, which is 18 bp longer than that previously reported (21). Exon 5A is 488 bp, similar to bovine ACC1 exon 5A, which is 493 bp (17). We did not detect any other exon sequence preceding exons 1, 2, or 5A. Based on these results, regulation of human ACC1 is under control of three promoters (PI, PII, and PIII). PI transcribes the mRNA from exon 1, PII from exon 2, and PIII from exon 5A. The sequence of exon 1 to exon 5A, which spans 110 kb, is one-third of the whole length of ACC1 gene and can be considered as the regulatory region of ACC1 gene (Fig. 1 A). We do not know yet the upstream limits of PI. The interesting finding is that human exon 1 and its preceding PI sequences have no homology with exon 1 of ACC1 in other mammalian species. However, we found that exon 1C of human ACC1 is highly homologous to exon 1 of rat, bovine, and ovine ACC1 genes (70% identity). We have not detected by the 5′-RACE any transcripts starting with exon 1C; hence, the sequences upstream of exon 1C do not seem to be functional as a promoter in human ACC1.
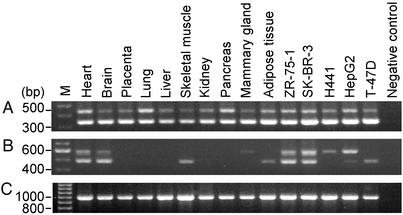
Evaluation of Translational Products of mRNAs Containing Exon 5B. All of the animal ACC1 cDNAs contain a highly conserved exon 5, which harbors the start codon of ATG (2, 14, 27, 28). We have identified an alternatively spliced exon 5B, in intron 5, in the human ACC1 transcripts present between exons 5A and 6 (Fig. 1), which was not found in the bovine, ovine, or rat. When exon 5B is included in transcripts, it disrupts the translation initiated in exon 5 of PI- and PII-generated mRNAs, and the translation initiated in exon 5A of PIII-generated mRNA (Fig. 1). In these mRNA populations, only the ATG codon located in exon 6 can initiate the translation of an ACC1 protein of 257 kDa instead of 265 kDa. Hence, we analyzed for the presence of exon 5B containing ACC1 mRNAs in various tissues. The PI- and PII-generated mRNAs containing exons [5:5B:6] were found to exist in all human tissues and various cell lines (Fig. 2A) whereas PIII transcripts containing [5A:5B:6] mRNA are not ubiquitous and show tissue-specific expression (Fig. 2B).
Fig. 2.
RT-PCR analysis of human ACC1 mRNA in various tissues and cell lines. First-strand cDNA was reverse transcribed from RNA isolated from different cell lines (ZR-75-1, SK-BR-3, HepG2, H441, or T-47D) by using hexamer random primers and used as template for PCR. In addition, multiple human cDNA panel (heart, brain, placenta, lung, skeletal muscle, kidney, pancreas) and Marathon-Ready cDNAs of mammary gland and adipose tissues (CLONTECH) were also used as templates. (A) Shown are PCR products obtained by using primers ex5f and ex6r2, which will amplify both PI and PII transcripts (Table 1). Both the PCR products were sequenced to confirm their exonic compositions. The two bands of PCR products contain exons [5:5B:6] and [5:6], respectively. (B) Same as in A, except that the primers used were ex5Af1 and ex6r2, which will amplify the PIII-specific transcripts. The two bands of PCR products contain exons [5A:5B:6] and [5A:6], respectively. (C) Glyceraldehyde 3-phosphate dehydrogenase (G3PDH)-specific primers (CLONTECH) were used to amplify 1 kb of cDNA, for semiquantitative comparison. M represents 100-bp DNA ladder.
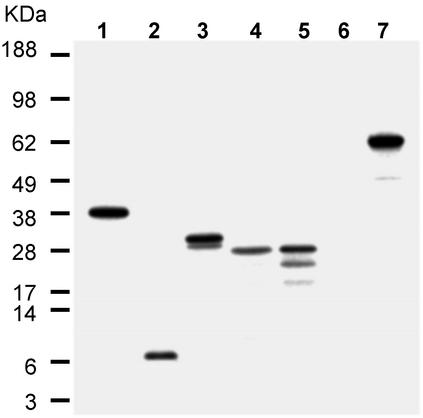
To determine how the presence of exon 5B affects translation of downstream exons, we performed in vitro transcription and translation with various plasmid constructs that contain or lack this exon. The human ACC1 cDNAs containing sequences of exons [5:6–12], [5:5B:6–12], [5A:6–12], [5A:5B:6–12], or [6–12] were amplified by RT-PCR using a common exon 12 reverse primer ex12r in combination with exon-specific forward primers ex5f, ex5Af1, or ex6f, respectively (Table 1), and cloned into pET32a (Novagen) at XbaI/XhoI restriction sites. The resulting plasmids were then in vitro transcribed by using T7 RNA Polymerase and translated as described under Materials and Methods. The results showed that the RNA containing exons [5:6–12] gave rise only to one band of 35.4-kDa protein, which is the expected size for a translation product initiating in exon 5 (Fig. 3, lane 1). The RNA containing exons [5:5B:6–12] produced an 8.3-kDa short peptide (77 aa), by using the same ATG located at the exon 5 and the stop codon in exon 5B (Fig. 3, lane 2). This peptide is actually a part of 265-kDa ACC1, in that 75 aa are translated from exon 5. However, further verification of its existence in vivo is required. The RNA containing exons [5A:6–12], as expected, produced one major protein and one minor protein, due to translation initiations with two different inframe ATGs located in exon 5A (Fig. 3, lane 3). The RNA containing exons [5A:5B:6–12] was translated into one major protein (27 kDa) by using the ATG in exon 6 (Fig. 3, lane 4), and that the translation initiated in exon 6 was confirmed by the 27-kDa translation product (Fig. 3, lane 5) obtained by using the control RNA containing exons [6–12].
Fig. 3.
In vitro transcription and translation of human ACC1 cDNA constructs containing or lacking exon 5B coding sequences. The details of in vitro transcription are described in Materials and Methods. Autoradiogram of [35S]methionine-labeled translation products were fractionated on 4–12% polyacrylamide-gradient gels. Lanes 1–5 correspond to the five constructs containing different exon compositions of human ACC1 gene: lane 1, [5:6–12]; lane 2, [5:5B:6–12]; lane 3, [5A:6–12]; lane 4, [5A:5B:6–12]; lane 5, [6–12]; lane 6, no plasmid DNA (negative control); lane 7, luciferase T7 DNA control plasmid (positive control, 61 kDa).
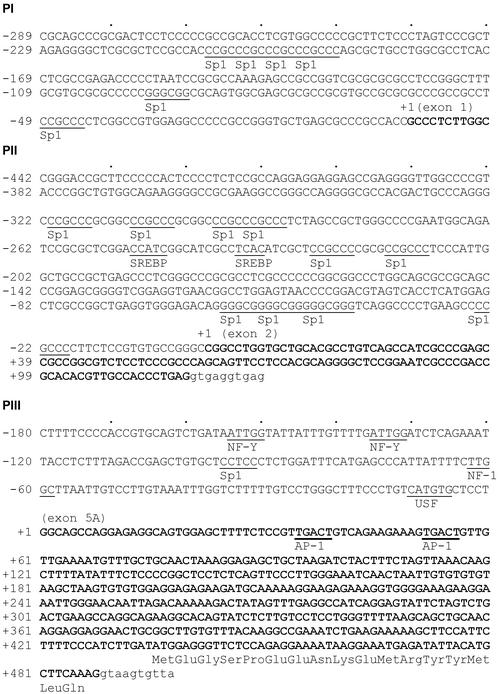
Characterization of the Three Human ACC1 Promoters. We have cloned the three human ACC1 promoters: 2.0 kb of PI, 1.9 kb of PII, and 3.0 kb of PIII. The GC content from –400 to –1 of PI is very high (83%), and it gradually decreases to ≈67% near –1000. In this region, there are four repeat sequence (CCCG) instead of three in the sequence available in GenBank Database with accession no. AC068400 or NT_010795 (Fig. 4). The sequence of PI is unique to human and bears no homology with the PI sequences of other mammalian species. Moreover, the rat, bovine, and ovine PIs are not GC-rich promoters. In addition, the human ACC1 sequence of exon 1 does not have any similarities to those of rat, bovine, and ovine exon 1. But, we have identified in the mouse ACC1 gene (GenBank accession no. NM_000039) a highly homologous sequence (181 bp) to human exon 1 (84% identity); however, we could not find any PI sequence homology upstream of this putative exon. The GC content of PII is also very high (78.2%), between –400 to –1, and decreases to 65.2% near –1000. The GC content of PIII from –400 to –1 is only 41.5%; therefore, it is not a GC-rich promoter similar to ovine and bovine PIII (16, 17). Sequence comparisons showed that both PII and PIII are highly conserved with the corresponding bovine and ovine ACC1 promoter sequences.
Fig. 4.
Promoter sequences of human ACC1 gene. The core promoter sequences, the transcription initiation site, and the partial promoter-specific first exon sequences (boldfaced) are indicated. The lowercase letters represent the intron sequences. The potential binding sites for some of the transcription factors such as Sp1, SREBP, NF-Y (nuclear factor-Y), USF (upstream stimulatory factor), and AP-1 (activator protein-1) are underlined. The repeat sequence (CCCG)4 at positions –208 to –193 in PI, and the 17 amino acid residues encoded by exon 5A, in the PIII transcript are also shown.
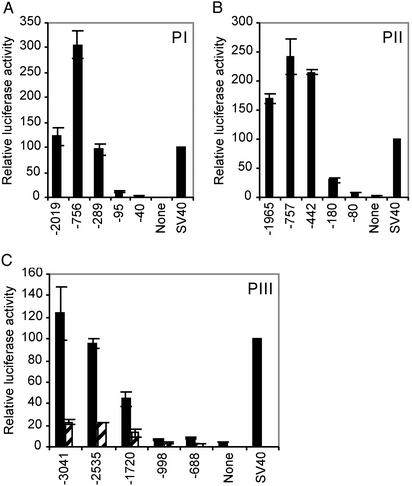
Determination of the Core Promoter Regions of the Three Promoters of Human ACC1 Gene. To test the activities of the three ACC1 promoters, we generated luciferase reporter gene constructs PI(–2019/+53)Luc, PII(–1965/+30)Luc, and PIII(–3041/+182)Luc. Transient transfections with these constructs in HepG2, H441, and T-47D showed that PI and PII are very active in all three cell lines. However, PIII is barely active in HepG2 and H441, but it is as active as SV40 reporter gene in T-47D cell line (data not shown). Several 5′ deletions of these three promoters were used to determine the core promoter region. PI reporter gene deletion constructs were tested in HepG2 cells. Deletion of the region –2019 to –756 increases luciferase activities by 150% (Fig. 5A), whereas deletion to –289 results in a decrease of the luciferase to original activities of (–2019/+53)Luc. Further deletion of the 194 bp between –289 and –95 leads to a dramatic decrease (≈87%) in luciferase activity. Luciferase activities remain very low until deletion of bases up to –40. Hence, the core promoter is probably located between –289 to –1 of PI. The sequence of this region contains several putative Sp1 sequences (Fig. 4).
Fig. 5.
Deletion analysis of human ACC1 promoters. Several 5′-deletion constructs were generated from the reporter plasmids PI(–2019/+53)Luc, PII(–1965/+30)Luc, PIII(–3041/+18)Luc, or PIII(–3041/+182)Luc as described in Materials and Methods. The reporter gene plasmids of PI and PII were transiently transfected into HepG2 cells, and the relative luciferase activities determined were shown in A and B, respectively. PIII plasmids were transfected in T-47D cells, and the promoter activities were shown in C. For PIII, we have tested two kinds of reporter plasmids; one contains exon 5A sequence from 1–182 (black bars) and the other contains exon 5A sequence from 1–18 (stippled bars). The plasmid pRL-TK was cotransfected in all experiments and used as internal control to normalize the transfection efficiencies. The relative luciferase activities of various promoter constructs were determined as described in Materials and Methods, and were further normalized with SV40 promoter activities, which were set at 100. The values are the mean ± SD calculated from at least three independent experiments. None and SV40 represent the pGL3-Basic and pGL3-Promoter vectors (Promega), respectively.
PII reporter gene is highly active in HepG2 (>1.6-fold of SV40 promoter; Fig. 5B). Based on the deletion analysis of the region between –442 to –80, the core promoter region of PII resides within –442 to –1 region (Fig. 5B). As shown in Fig. 4, this promoter region contains several putative Sp1-binding sites, two sterol regulatory element-binding protein (SREBP) binding sites, and an NF-Y binding site.
Because PIII activities were very low in HepG2 and H441, as described above, we used T-47D, a human mammary cancer cell line for transient transfection experiments. The reporter gene construct containing –3041 to +182 showed highest promoter activity. The reporter gene constructs containing serial 5′-deletions of the region between –3041 and –688, showed progressive decrease of luciferase activities (Fig. 5C), suggesting that the major regulatory elements are located in the distal region of PIII. This result is different from PI and PII, where the regulatory elements are located in the promoter proximal regions. Interestingly, as shown in Fig. 5C, inclusion of the 5′ sequence of exon 5A in the PIII reporter gene constructs enhances this promoter function. If the sequence between +19 and +182 was deleted, the promoter activities were dramatically decreased in T-47D (81%), as well as in H441(51%), and in HepG2 (85%) (data not shown). The relatively high expression of PIII in mammary gland cell lines (Figs. 2B and 5C) suggests that this promoter plays an important role in the production of milk lipids that contain both short- and long-chain fatty acids.
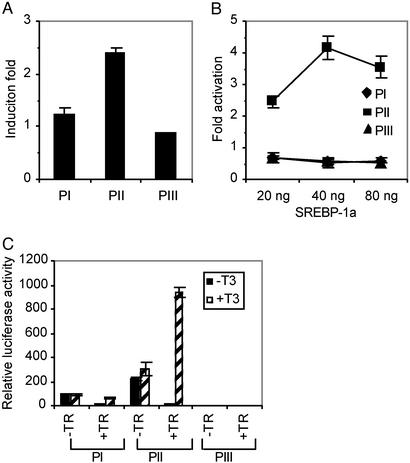
Cholesterol Response of Human ACC1 Promoters. Cholesterol represses lipogenic enzymes such as fatty acid synthase and ACC1 (24, 29). To test which promoter of the human ACC1 responds to cholesterol, HepG2 cells were transiently transfected with PI(–2019/+53)Luc, PII(–1965/+30)Luc or PIII(–3041/+182)Luc, and switched to DMEM with 10% lipoprotein-deficient serum either lacking or containing cholesterol and 25-hydroxyl cholesterol (24). Based on the reporter gene assays performed in the presence and absence of these sterols, we found that only human ACC1 PII, which showed ≈2.5-fold stimulation, responds to cholesterol mediated regulation (Fig. 6A). When these promoter constructs were cotransfected with pCMX-SREBP1a, only promoter PII was activated by SREBP1a, based on a 2.5- to 4.0-fold increase in the reporter gene activity (Fig. 6B). This result is consistent with those observed with PII of rat ACC1 (29).
Fig. 6.
Cholesterol- and thyroid hormone-mediated regulation of human ACC1 promoters. PI(–2019/+53)Luc, PII(–1965/+30)Luc, PIII(–3041/+18)Luc, or PIII(–3041/+182)Luc were transiently transfected into HepG2 cells, and their response to cholesterol (A and B) and T3 (C) was determined and expressed as relative luciferase activities. (A) Cholesterol response was measured as the fold-activation promoter expression, which was derived by dividing the relative light unit (RLU) values obtained when the transfected cells grown in the inducing medium (DMEM with 10% lipoprotein-deficient serum) with the RLU values obtained when the transfected cells were grown in the repressing medium (DMEM with 10% lipoprotein-deficient serum, plus 10 μg/ml cholesterol and 1 μg/ml 25-hydroxyl cholesterol). (B) Activation of human ACC1 promoters by SREBP-1. HepG2 cells were transfected with PI, PII, and PIII reporter gene plasmid along with 20, 40, or 80 ng of pCMX-SREBP1a plasmid DNA. The transfected cells were grown in culture medium containing 10% FBS for 20 h, and luciferase activities were determined. Fold activation is the ratio of RLUs obtained with and without pCMX-SREBP1a-transfected cells. (C) HepG2 cells were transfected with PI, PII, or PIII constructs, together with or without pRSV-TRβ. Cells were then treated with or without 1 μM T3 for 24 h. One representative transfection experiment from three to five independent experiments was shown. The plasmid pRL-TK was cotransfected in all experiments and used as internal control to normalize the transfection efficiencies.
Triiodothyronine (T3) Response of Human ACC1 Promoters. To determine which of the three promoters respond to T3, we transiently transfected the three longest promoter reporter gene constructs described above into HepG2 cells. As shown in Fig. 6C, the luciferase reporter gene activities of all these three promoters are not affected by T3, suggesting lack of thyroid hormone receptor (TR) in HepG2 cells (23). When reporter gene constructs were cotransfected with a human TRβ expression plasmid pRSV-hTRβ (23), luciferase activity of only the PII construct dramatically increased 78-fold in the presence of T3 (Fig. 6C), whereas PI and PIII did not respond to T3 significantly. Further, only the PII promoter activities were dramatically repressed when cotransfected with TRβ plasmid in the absence of T3, suggesting that this promoter contains thyroid hormone response elements (23).
The results described above indicate that the presence of multiple promoters and alternatively spliced exons is a common feature of all mammalian ACC1 genes thus far analyzed. In human ACC1 gene, we have identified three promoters. Interestingly, human PI has no homology with rat, bovine, and ovine ACC1 PI sequences. However, PI of human ACC1 is constitutively and highly expressed, unlike that in other mammalian species. The PII sequence of human ACC1 has similarities with those of rat, bovine, and ovine ACC1, and also functions as a major regulatable promoter of ACC1. It has been shown that bovine and ovine PIIIs are highly activated in mammary glands during lactation (16, 17). It is well established that ACC1 is highly expressed in human breast cancer tissues. In addition, breast cancer tissues also express several lactating mammary gland specific proteins. The observation that PIII-reporter gene activity is highly stimulated in human mammary cancer cell line T-47D, compared with its activity in HepG2 cells, suggests similar tissue-specific expression of human ACC1 PIII. Hence, it seems that in humans, ACC1 PIII plays a very critical role during lactation to generate milk fat needed for neonatal development.
Due to the absence of 3′ splice site recognition sequences in the introns preceding the first exons of PII (exon 2) and PIII (exon 5A), PI transcripts do not contain exon 2 or 5A. Similarly, PII transcripts do not contain exon 5A (Fig. 1B). In addition, the highly conserved exons 4 and 5 are alternatively spliced, and there are internal splice sites within both exons (Fig. 1B). Presence of these and other splice sites in introns 1 and 5 generates a complex set of human ACC1 mRNAs with 5′ heterogeneity (Fig. 1B). Preliminary analysis of ACC1 genomic DNA sequence suggests that some of these alternative exons have week 3′ or 5′ splice junction sequences, and in some cases the flanking introns lack the putative splicing enhancers. These observations might explain why these alternative exons are only occasionally included in the ACC1 mRNA. However, all of the transcripts produced by PI and PII contain exon 5 (Fig. 1B), which is highly conserved in all of the reported cDNA sequences of mammalian (14, 27, 28) and the human ACC1 (2). The putative amino acid sequence of 265-kDa ACC1 starts with the ATG located in the sequence coded by exon 5. Although a majority of the PI and PII transcripts contain exons [1:4:5:6–58] and [2:4:5:6–58], respectively (Fig. 1B), the contribution of the minority of the transcripts that contain various alternative 5′ exons to ACC1 protein expression is difficult to assess. However, all mammalian ACC1 genes do produce transcripts that contain a variety of alternative exons upstream of exon 5 (14, 21, 27, 28). In these transcripts, the presence of alternative exons might influence either the stability or translational efficiency of these mRNAs. However, a majority of the PIII transcripts [5A:6–58] contain an ORF coding for a 259-kDa ACC1 isoform, which initiates with an ATG located in exon 5A (Fig. 1B). However, the presence of an alternative exon 5B that contains translation termination codons in all three reading frames, located downstream of exon 5A, and is present occasionally in all of the three promoter transcripts results in premature termination of the ACC1 ORF. Based on the in vitro transcription and translation experiments, the presence of exon 5B in PI and PII transcripts, the translation from exon 5 terminates at exon 5B and generates a short peptide of 8.3 kDa (Fig. 3), a result that can be explained by the lack of reinitiation of translation with ATG codon in exon 6. However, when exon 5B is present in PIII transcripts, the Met codon located in exon 6 is used for translation initiation, resulting in a truncated 257-kDa isoform of ACC1 (Fig. 3). This result can be explained by assuming that the secondary structure of mRNA containing exons 5A and 5B allows only the ATG codon in exon 6 to be accessible for translation initiation. These observations strongly suggest that, even though the transcripts that contain alternative exons present in either 5′ of exon 5 or downstream exon 5A do not constitute a respectable portion of all of the ACC1 mRNAs, their effects on translational efficiencies and the isoforms they produce could have an impact on the activity of ACC1. These observations suggest that ACC1 activity is regulated by posttranscriptional and translational mechanisms, in addition to transcriptional and posttranslational regulations.
Acknowledgments
We thank A. Craig Chinault and Weimei Di for screening the BAC library. We also thank Lutfi Abu-Elheiga and Andrew McCullough for helpful discussion. This work was supported in part by grants from the Clayton Foundation and the National Institutes of Health (GM-63115).
Abbreviations: ACC, acetyl-CoA carboxylase; T3, triiodothyronine; TR, thyroid hormone receptor; SREBP, sterol regulatory element-binding protein; BAC, bacterial artificial chromosome.
References
- 1.Wakil, S. J., Stoops, J. K. & Joshi, V. C. (1983) Annu. Rev. Biochem. 52, 537–579. [DOI] [PubMed] [Google Scholar]
- 2.Abu-Elheiga, L., Jayakumar, A., Baldini, A., Chirala, S. S. & Wakil, S. J. (1995) Proc. Natl. Acad. Sci. USA 92, 4011–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim, K.-H. (1997) Annu. Rev. Nutr. 17, 77–99. [DOI] [PubMed] [Google Scholar]
- 4.Thampy, K. G. & Wakil, S. J. (1988) J. Biol. Chem. 263, 6447–6453. [PubMed] [Google Scholar]
- 5.Thampy, K. G. & Wakil, S. J. (1988) J. Biol. Chem. 263, 6454–6458. [PubMed] [Google Scholar]
- 6.Mabrouk, G. M., Helmy, I. M., Thampy, K. G. & Wakil, S. J. (1990) J. Biol. Chem. 265, 6330–6338. [PubMed] [Google Scholar]
- 7.Hardie, D. G. (1989) Prog. Lipid Res. 28, 117–146. [DOI] [PubMed] [Google Scholar]
- 8.Iritani, N. (1992) Eur. J. Biochem. 205, 433–442. [DOI] [PubMed] [Google Scholar]
- 9.Girard, J., Perdereau, D., Foufelle, F., Prip-Buus, C. & Ferre, P. (1994) FASEB J. 8, 36–42. [DOI] [PubMed] [Google Scholar]
- 10.Lopez-Casillas, F. & Kim, K.-H. (1989) J. Biol. Chem. 264, 7176–7184. [PubMed] [Google Scholar]
- 11.Lopez-Casillas, F., Ponce-Castaneda, M. V. & Kim, K.-H. (1991) Endocrinology 129, 1049–1058. [DOI] [PubMed] [Google Scholar]
- 12.Tae, H.-J., Luo, X. & Kim, K.-H. (1994) J. Biol. Chem. 269, 10475–10484. [PubMed] [Google Scholar]
- 13.Luo, X. & Kim, K.-H. (1990) Nucleic Acids Res. 18, 3249–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mao, J., Marcos, S., Davis, S. K., Burzlaff, J. & Seyfert, H.-M. (2001) Biochem. J. 358, 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao, J. & Seyfert, H.-M. (2002) Biochim. Biophys. Acta 1576, 324–329. [DOI] [PubMed] [Google Scholar]
- 16.Barber, M. C. & Travers, M. T. (1998) Biochem. J. 333, 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao, J., Molenaar, A. J., Wheeler, T. T. & Seyfert, H.-M. (2002) J. Mol. Endocrinol. 29, 73–88. [DOI] [PubMed] [Google Scholar]
- 18.O'Callaghan, B. L., Koo, S.-H., Wu, Y., Freake, H. C. & Towle, H. C. (2001) J. Biol. Chem. 276, 16033–16039. [DOI] [PubMed] [Google Scholar]
- 19.Travers, M. T. & Barber, M. C. (1999) J. Mol. Endocrinol. 22, 71–79. [DOI] [PubMed] [Google Scholar]
- 20.Travers, M. T., Vallance, A. J., Gourlay, H. T., Gill, C. A., Klein, I., Bottema, C. B. & Barber, M. C. (2001) Biochem. J. 359, 273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ha, J., Daniel, S., Kong, I.-S., Park, C. K., Tae, H.-J. & Kim, K.-H. (1994) Eur. J. Biochem. 219, 297–306. [DOI] [PubMed] [Google Scholar]
- 22.Hsu, M. H., Chirala, S. S. & Wakil, S. J. (1996) J. Biol. Chem. 271, 13584–13592. [DOI] [PubMed] [Google Scholar]
- 23.Xiong, S., Chirala, S. S., Hsu, M. H. & Wakil, S. J. (1998) Proc. Natl. Acad. Sci. USA 95, 12260–12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong, S., Chirala, S. S. & Wakil, S. J. (2000) Proc. Natl. Acad. Sci. USA 97, 3948–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong, I.-S., Lopez-Casillas, F. & Kim, K.-H. (1990) J. Biol. Chem. 265, 13695–13701. [PubMed] [Google Scholar]
- 26.Barber, M. C., Pooley, L. & Travers, M. T. (2001) J. Mol. Endocrinol. 27, 349–356. [DOI] [PubMed] [Google Scholar]
- 27.Luo, X., Park, K., Lopez-Casillas, F. & Kim, K.-H. (1989) Proc. Natl. Acad. Sci. USA 86, 4042–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barber, M. C. & Travers, M. T. (1995) Gene 154, 271–275. [DOI] [PubMed] [Google Scholar]
- 29.Lopez, J. M., Bennett, M. K., Sanchez, H. B., Rosenfeld, J. M. & Osborne, T. F. (1996) Proc. Natl. Acad. Sci. USA 93, 1049–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]