Abstract
In trypanosomes the C/D- and H/ACA-like small nucleolar RNAs (snoRNAs) are clustered and repeated in the genome. The snoRNAs studied to date are transcribed as polycistronic transcripts by RNA polymerase II and then processed, resulting in mature snoRNAs. In this study we demonstrated that snoRNA genes can be silenced in three trypanosomatid species: Leptomonas collosoma, Leishmania major, and Trypanosoma brucei. Silencing was achieved in L. collosoma and L. major by the expressing of an antisense transcript complementary to the snoRNA gene and was accompanied by the accumulation of small interfering RNA. Silencing eliminated the mature snoRNA but not its precursor and abolished the specific 2′-O-methylation guided by the snoRNA. In T. brucei, silencing was achieved by using the inducible synthesis of double-stranded RNA from T7 opposing promoters. Silencing varied between the different snoRNA genes, which may reflect the accessibility of small interfering RNA to the target RNAs. This study suggests that RNA interference can degrade snoRNAs. This study has further implications in elucidating the function of nucleolar RNAs and specific modifications guided by these RNAs in trypanosomatids and perhaps in other eukaryotes as well.
The nucleolus of eukaryotes contains small nucleolar RNAs (snoRNAs) that function in ribosomal RNA (rRNA) processing and RNA modification. The snoRNA can be divided into two main groups: the box C/D snoRNAs and H/ACA snoRNAs. Some of the snoRNAs are involved in pre-rRNA cleavage, and most function in 2′-O-ribose methylation and pseudouridine formation (1). Ribose methylation is guided by C/D snoRNAs. The C/D snoRNAs contain long stretches (>10 nt) that are complementary to their targets (2). The other group of guide RNAs are those that direct pseudouridylation; they consist of 5′ and 3′ hairpin domains connected by a single-stranded hinge and tail regions that carry the conserved H (AnAnnA) and ACA boxes, respectively. Moreover, they are characterized by two short motifs of the snoRNA that base-pair together with rRNA sequences flanking the uridine to be modified (3, 4).
Trypanosomatids constitute a diverse family of parasitic protozoans that are descendants of one of the deepest branches of the eukaryotic lineage (5). Many of the trypanosome C/D snoRNAs are organized in repeated clusters. The cluster encodes for multiple C/D snoRNAs. Studies suggest that trypanosomatids obey the +5 rule for guiding 2′-O-methylation (6–8). We have recently identified the H/ACA-like RNA in one of these C/D gene clusters, which we termed h1 (9). The h1/rRNA duplex obeys the rules for guiding pseudouridylation. h1 consists of a single hairpin structure and is the shortest H/ACA RNA described so far. Recently, we have demonstrated that SLA1, previously thought to be the trypanosome U5 homologue, belongs to the same group of H/ACA-like RNAs and directs pseudouridylation at position –12 on the spliced leader RNA (SL RNA). SLA1 is located in a cluster also encoding for C/D snoRNAs (10).
We have previously demonstrated that the expression of Leptomonas collosoma C/D snoRNA-2 depends on its orientation with respect to the resistance gene on the expression vector and that no promoter exists upstream from the individual genes within the gene cluster. All of the snoRNA clusters studied so far are transcribed by RNA polymerase II as polycistronic transcripts (7).
RNA interference (RNAi) is currently one of the most studied processes in the RNA field, not only because of its great biotechnological and medical implications but also because of interest in the basic mechanism of this exciting phenomenon (11). RNAi is a conserved posttranscriptional gene silencing mechanism that recognizes double-stranded RNA (dsRNA) as a signal to trigger the sequence-specific degradation of the homologous mRNA (11). Initiation of silencing occurs upon recognition of the dsRNA by an enzyme that cleaves the RNA to 21- to 25-nt RNAs. The cleavage to produce these small interfering RNAs (siRNAs) is mediated by RNase III (Dicer) (12, 13). These siRNA duplexes are then incorporated into a protein complex that undergoes ATP-dependent unwinding of the siRNA duplex, which remodels the complex to generate an active RNA-induced silencing complex (RISC) (14). Finally, in the last step the RISC can recognize and cleave a target RNA complementary to the guide strand of the siRNA.
The function of RNAi has been implicated in resistance to viral infection in plants (15) and in controlling transposons in trypanosomes (16) and nematodes (17). Most studies suggest that RNAi takes place in the cytoplasm, possibly on polyribosomes (11). However, accumulating evidence has indicated the existence of RNAi in the nucleus (18). More recently, in the yeast Schizosaccaromyces pombe, RNAi was shown to regulate the heterochromatin in centromeric repeats (19).
Trypanosomes were one of the first systems where RNAi was discovered (20). Several approaches were developed in trypanosomes to induce the production of dsRNA in vivo from inducible promoters, which are based either on the production of dsRNA using T7 opposing promoters or the synthesis of stem-loop RNA (21).
Here, we provide direct evidence that snoRNAs can be silenced in three trypanosomatid species. Importantly, silencing of the snoRNA reduces the specific 2′-O-methylation guided by the RNA. The silencing was achieved by either antisense RNA in L. collosoma and Leptomonas major or by the T7 opposing system in Trypanosoma brucei. However, the level of silencing varies and may depend on the accessibility of the small RNAs to the siRNAs. This study opens up the possibility of exploring the function of nucleolar RNAs in this group of parasites and perhaps in metazoa as well.
Experimental Procedures
Transfection and Preparation of Cell Lines. L. collosoma cells were grown and transfected with the plasmid carrying the tagged snoRNA-2 gene as described (7). The selection was performed with an elevated G418 concentration ranging from 25 to 600 μg/ml. Transfection of L. major was essentially the same as described (7). Transformants were selected on 1–1.2 mg/ml of G418. The T7 opposing construct was transfected into T. brucei 29–13 strain (21). Transformants were selected on 5 μg/ml of phelomycin. RNA was prepared 48 h after induction with 10 μg/ml of tetracycline.
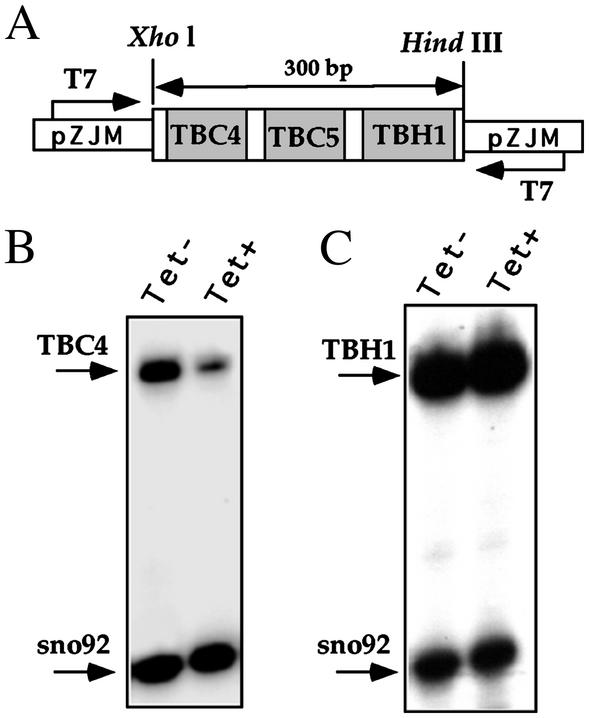
Constructs for Silencing in L. major and T. brucei. The L. major locus encoding for snoRNA 92, 270, and SLA1 was amplified by PCR using oligonucleotides LmSLA-5B and LmSLA-3B and cloned into the BamHI site of the expression vector pX-neo in an opposite orientation with respect to the neo-resistant gene. The T. brucei snoRNA cluster carrying TBC4, TBC5, and TBH1 genes was amplified by PCR with oligonucleotides Sno-1–5XH and Sno-1–3H. The fragment was cloned into the XhoI and HindIII sites of the pZJM expression vector (21) in between the two T7 opposing promoters (see Fig. 7). The construct was linearized with NotI before transfection (21).
Fig. 7.
Silencing of snoRNA in T. brucei by dsRNA. (A) Schematic representation of the opposing T7 construct. The orientation of the T7 promoters are indicated and marked with arrows. The genes in the cluster are given. (B) Primer extension analysis on TBC4. RNA from induced cells 2 days after tetracycline induction (Tet+) and uninduced cells (Tet–) were subjected to primer extension using TBC-4 and 92a oligonucleotides complementary to TBC4 and snoRNA 92 (present in another locus), respectively. (C) Primer extension analysis as in B but for TBH1 and snoRNA 92 (as a control), respectively. The extension products are marked with arrows.
Northern and Primer Extension Analysis. Total RNA was prepared from cells with TRIzol reagent (Sigma). For detecting siRNAs, 40–80 μg total RNA was separated on a 10% polyacrylamide-7 M urea denaturing gel and transferred to nylon membrane (Hybond, Amersham Pharmacia) for 5–7 h in 25 mA. The membrane was hybridized with a RNA probe complementary to snoRNA-2 at 42°C (22). After hybridization, the blot was washed twice at 45°C, for 20 min with 2× SSC and 0.1% SDS. Primer extension was performed as described (9). Primer extension for mapping the 2′-O-methylated nucleotide was performed with end-labeled antisense oligonucleotide in the presence of an elevated dNTP concentration ranging from 0.05 to 5 mM, as described (7).
RNase Protection. Total RNA (30 μg) was mixed with 100,000 cpm of gel-purified RNA probe and concentrated by ethanol precipitation. The pellet was washed and dissolved in hybridization buffer (40 mM Pipes, pH 6.4/80% formamide/0.4 M sodium acetate/1 mM EDTA). After boiling for 1 min, the samples were incubated at 45°C for 14–16 h. After the hybridization, the samples were diluted 1:10 with a solution consisting of 10 mM Tris·HCl (pH 7.5), 5 mM EDTA, 200 mM sodium acetate containing 2.5 units/ml of RNase ONE (Promega). The digestion was performed at 30°C for 1 h and terminated by proteinase K digestion. After phenol-chloroform extraction, the protected products were precipitated with ethanol in the presence of 20 μg of glycogen and analyzed on a 8% polyacrylamide-7 M urea denaturing gel (7).
Oligonucleotides. For the oligonucleotides used in these experiments, see Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org.
Results
Overexpression of the snoRNA-2 Gene in an Opposite Orientation to the neo-Resistant Gene in the Episome Silenced both Tagged and WT snoRNAs. We previously reported that the expression of the tagged L. collosoma snoRNA-2 gene from the episomal vector depends on its orientation with respect to the resistant gene on the episome (7). When the tagged snoRNA gene was expressed in a transcriptional direction of the neo gene, expression of the tagged gene was efficient. We interpreted our inability to detect the expression of the tagged gene, when cloned in the opposite orientation with respect to the neo gene, to be caused by the differential efficiency of transcription from the two strands of the episomal vector (7). Indeed, it was previously reported that episomal vectors in Leishmania are transcribed from both strands although at different rates (23).
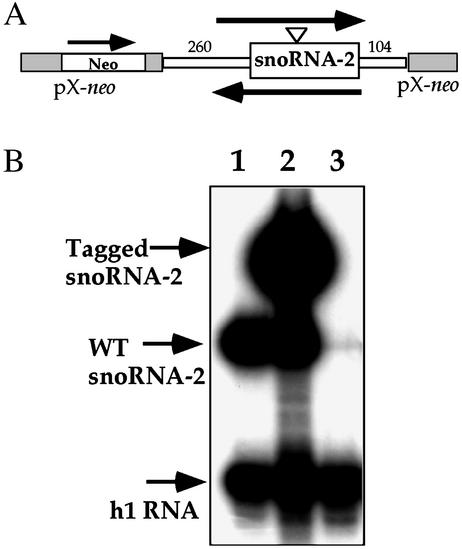
To further explore this phenomenon, we generated cell lines carrying snoRNA-2 in two orientations, as depicted in Fig. 1A.To this end, cell lines were selected with 600 μg/ml of G418 and RNA was prepared and used in primer extension experiment with an antisense probe that recognizes both the tagged and WT transcripts. The results, presented in Fig. 1B, indicate that when the snoRNA gene was in the neo direction, the tagged gene was efficiently expressed (Fig. 1B, lane 2). However, no expression was detected for the tagged gene when it was placed in the opposite orientation (Fig. 1B, lane 3). Surprisingly, we could not detect the WT transcript (Fig. 1B, lane 3).
Fig. 1.
Expression of tagged snoRNA-2 gene depends on its orientation in the pX vector. (A) Schematic representation of the constructs. A fragment containing the snoRNA-2 gene including 260 bp of 5′ and 104 bp of 3′ flanking sequences, respectively, was cloned into the BamHI site of pX-neo (7). The orientation of the snoRNA-2 inserts and neo genes are marked with arrows. (B) Primer extension analysis on RNA from WT and the cell lines carrying the snoRNA-2 gene in the two orientations. Lane 1, WT; lane 2, the cell line carrying snoRNA-2 in the orientation of the neo gene; lane 3, the cell line carrying the gene in the opposite orientation. The extension products are marked with arrows. Oligonucleotides used in extension reactions were 43362 and 16865, specific for h1 and snoRNA-2, respectively.
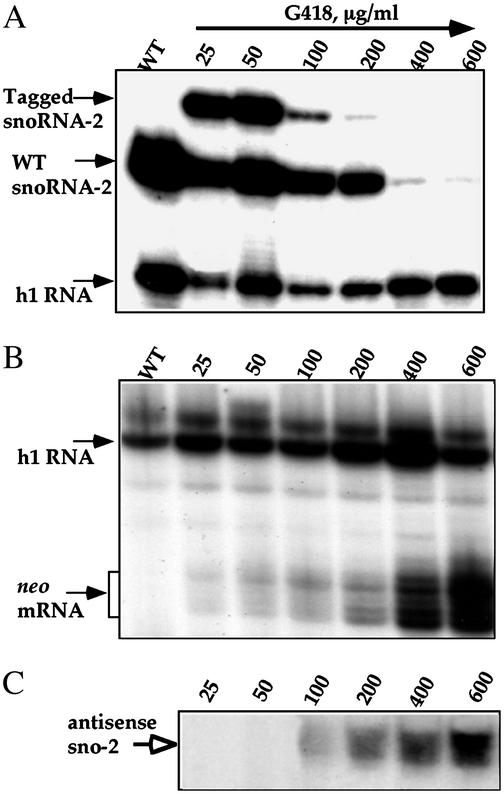
We further extended our analysis to cell lines selected at different G418 concentrations. RNA from these cell lines were subjected to primer extension, and the results are presented in Fig. 2A. At a low level of G418, the tagged gene was efficiently expressed even when present in the opposite orientation. Only when the G418 was elevated to 100–200 μg/ml did the tagged RNA begin to disappear, and at very high concentrations (400 μg/ml and above) the WT transcript was also completely eliminated, as compared with the level of h1 RNA as a control. We examined the level of neo transcripts in these different cell lines, and the results, presented in Fig. 2B, indicate that indeed the level of neo mRNA was increased. The heterogeneity in the primer extension stops results from the presence of the 5′ cap nucleotides, a hallmark of trans-spliced mRNA (24). More important for understanding the silencing phenomenon was the analysis of the level of antisense transcript, which was estimated by using a sense oligonucleotide to snoRNA-2 (Fig. 2C). Only at 100 μg/ml and above of G418 was the level of this antisense transcript detected, and it increased in accordance with the drug concentration. The results therefore indicate that the production of antisense RNA to snoRNA-2 elicits the degradation of tagged snoRNA-2. Only at a very high antisense level was the WT snoRNA-2 also eliminated.
Fig. 2.
Silencing of snoRNA-2 is achieved by increasing antisense production. (A) Effects of G418 concentration on the level of tagged and WT snoRNA-2. The level of the RNA was determined by primer extension analysis. The oligonucleotides used for primer extension were as described in Fig. 1. The concentrations at which the cell lines were selected are indicated. (B) Primer extension analysis to determine the level of neo mRNA. The extension products are marked with arrows. Primers used in the reaction were 43362 and 36815, specific for h1 snoRNA and neo mRNA, respectively. (C) Primer extension analysis to determine the level of antisense transcript to snoRNA-2. Primer extension was performed on the same RNA as in B with a sense primer 22182, specific to the snoRNA-2. The extension product is marked with an arrow.
Silencing of snoRNA-2 Is Correlated with the Production of siRNAs. It has been recently suggested that antisense effects may stem from induction of the RNAi mechanism. Indeed, antisense RNA induces the production of siRNA in a Drosophila in vitro RNAi silencing system (25).
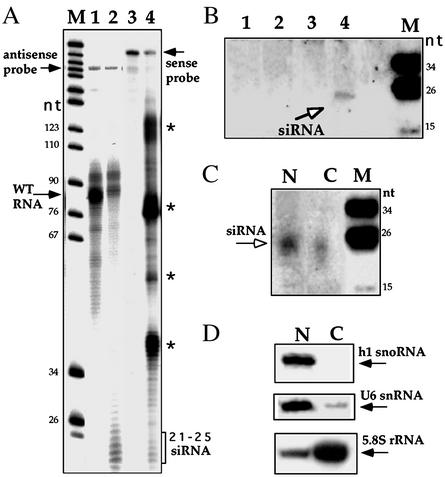
To examine whether the silencing observed in this study was carried out by a mechanism related to RNAi, we used an RNase protection assay and Northern analysis to search for siRNAs. In the RNase protection assay, two probes (sense and antisense) were used, as indicated in Fig. 3A. The results indicate that small protected fragments in the size of 21–25 nt were observed if sense or antisense probes were used. The source of the protected fragments with the sense probe is currently unknown. The pattern of siRNA-protected fragments (ranging in size from 21 to 25 nt) resembles previous related studies that used this method (26, 27).
Fig. 3.
Detection of the siRNAs. (A) RNase protection analysis to detect the existence of siRNAs. Lanes 1 and 3 are protected fragments obtained with WT RNA and labeled antisense and sense RNA probes, respectively. Lanes 2 and 4 present protected fragments with RNA from a cell line carrying the antisense snoRNA-2 construct selected at 600 μg/ml of G418 with antisense and sense probes, respectively. The protected fragments with the sense probe are marked with asterisks. The marker was a pBR322 MspI digest and the sizes are indicated in nt. (B) Northern analysis. Lane 1, total RNA from WT; lanes 2, 3, and 4 contained total RNA from cell lines expressing the antisense snoRNA-2 construct selected at 50, 200, and 600 μg/ml of G418, respectively. The membrane was hybridized with an antisense RNA probe. The siRNAs are marked with an open arrow. The size of the marker is as in A. (C) Cellular fractionation of siRNA. RNA from nuclear and cytoplasmic fractions was prepared from cell lines expressing the antisense snoRNA-2 gene grown at 600μg/ml of G418, as described (10). The membrane was hybridized with the same RNA probe as in B. The siRNAs are marked with an arrow. (D) Cellular fractionation of small RNAs. The content of U6 snRNA, h1 snoRNA, and cytoplasmic 5.8S rRNA are in the same fractions as in C. The primers used were 43362, 12407, and 19796, specific for h1 snoRNA, U6, and 5.8S rRNA, respectively. N and C designate nuclear and cytoplasmic fractions, respectively. M, the marker, as used in A.
To verify the presence of siRNA in the cell lines by a direct assay, we subjected RNA to Northern analysis and probed the blot with antisense RNA to snoRNA-2. The results, presented in Fig. 3B, suggest the existence of a clear hybridization band in a size of ≈24 nt only in RNA from the cells grown on 600 μg/ml of G418, suggesting that siRNAs are produced in detectable amounts only in cell lines where the snoRNA transcripts were almost completely eliminated.
snoRNAs are transcribed, processed in the nucleoplasm, and then migrate to the nucleolus where they function (1, 28). To investigate the distribution of siRNAs to snoRNA-2, we prepared and subjected nuclei and cytoplasmic RNA to Northern analysis with an antisense snoRNA-2 probe. The results (Fig. 3C) indicate that siRNA was present in both compartments. Using various controls to determine the quality of the fractionation (U6 for nucleoplasmic RNA, h1 for nucleolar RNA, and 5.8S for cytoplasmic RNA, Fig. 3D), we found that the amount of siRNA in the cytoplasm is higher than what would be expected solely from ``leakage'' from the nucleoplasm, suggesting that siRNAs may be produced both in the nucleus and the cytoplasm.
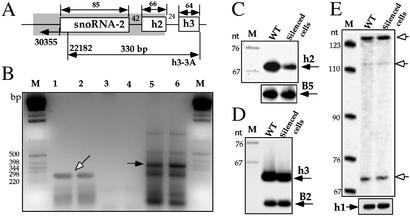
Pre-snoRNA-2 Is Not Affected in snoRNA-2-Silenced Cells. Our initial analysis of the snoRNA-2 cluster revealed only a single C/D snoRNA (6). However, inspecting the cluster for the existence of additional snoRNAs revealed the presence of two additional H/ACA RNAs, termed h2 and h3, respectively (data not shown). The gene organization within the cluster is depicted in Fig. 4A. Because the trypanosomatid guide RNAs studied so far were shown to be transcribed as polycistronic RNAs (7, 8), a quantitative RT-PCR assay was performed to determine whether the additional snoRNAs in this cluster exist as a polycistronic transcript and, most importantly, if silencing takes place at the pre-snoRNA level. The results presented in Fig. 4B suggest that indeed these two RNAs exist in a polycistronic transcript (lanes 5 and 6) and that the level of the precursor did not change, as compared with the level of another snoRNA precursor (g2), which served as a control for the amount of RNA (lanes 1 and 2) (7). Next, the levels of h2 and h3 snoRNA in WT and silenced cells were examined by primer extension, using B5 and B2 snoRNA as controls, respectively. The results (Fig. 4 C and D) indicate that the level of h2 was partially reduced in the silenced cells as expected, because h2 was also present in the antisense construct. However, no change in the level of h3 was observed, which might suggest that silencing does not take place at the precursor level. To further examine the level of the snoRNA-2 precursor, we performed a more sensitive primer extension assay with an oligonucleotide located in the intergenic region upstream from snoRNA-2, but no difference was observed between the WT and the snoRNA-2-silenced cells. These extension products may reflect stops on endonucleolytically cleaved precursor molecules. These data indicate that the pre-snoRNA-2 is not silenced by antisense RNA.
Fig. 4.
Silencing of snoRNA-2 takes place on mature molecules but not on the pre-snoRNA transcript. (A) Schematic representation of the snoRNA-2 locus. The shadow box indicates the silenced region. The newly identified H/ACA RNAs are indicated. The oligonucleotides used in the RT-PCR are indicated. The predicted size of PCR products is given. (B) The level of snoRNA precursors in WT and snoRNA-2-silenced cells. RT-PCR was performed as described (9). Lanes 1 and 2 represent RT-PCR analysis performed on RNA from WT and cell lines carrying the snoRNA-2-silenced cell lines, respectively, with oligonucleotides 26556 and 22078 to detect the level of g2 snoRNA precursor (7). Lanes 3 and 4 show PCR analysis performed on the samples described in lanes 1 and 2 to detect the DNA contamination. Lane 5 and 6 show RT-PCR analysis on the same samples with oligonucleotides 22182 and h3–3A, specific for snoRNA-2 precursor as depicted in A. The PCR products were analyzed on 2% agarose gel. The g2 precursor is marked with an open arrow, and the snoRNA-2 precursor is marked with a closed arrow. M is the 1-kb DNA ladder. (C) Silencing on h2 RNA. Primer extension was performed on the same RNA as in B with end-labeled oligonucleotides 22076 and 43388, complementary to h2 and B5 snoRNAs, respectively, encoded by a different locus. The extension products are indicated. (D) The level of h3 RNA. Primer extension was performed on the same RNA as in B with antisense primers 20406 and h3–3A, specific for B2 snoRNA and h3, respectively. The extension products are indicated. (E) The level of snoRNA-2 precursors. To detect the level of snoRNA-2 precursors, primer extension was carried out with antisense primers 30355 and 44362, specific for the snoRNA-2 sequence located upstream from the coding region (as in A) and h1 snoRNA (a control), respectively. The extension products are marked with open arrows. M is the marker as described in Fig. 3A.
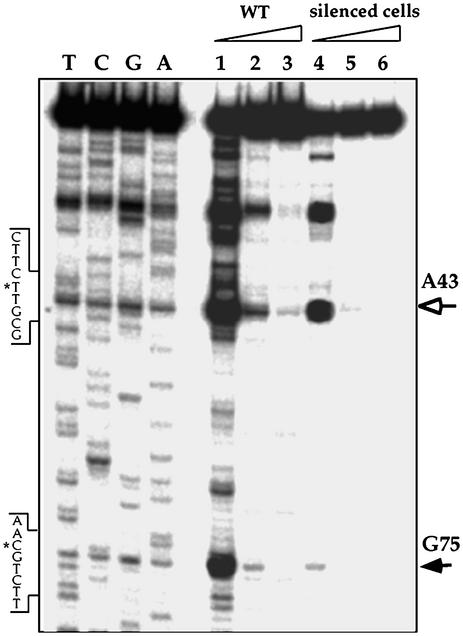
The Silencing of snoRNA-2 Eliminated Almost Completely the 2′-O-Methylation Guided by snoRNA-2. snoRNA-2 was found to guide the methylation on G75 of 5.8S rRNA (6). To determine the status of the methylation guided by snoRNA-2, we examined the modification on position G75 by reverse transcriptase primer extension assay in the presence of dNTPs from low to high concentrations. In the presence of low dNTPs, the reverse transcriptase stops 1 nt before the methylation site. The data presented in Fig. 5 indicate that the level of modification at position G75 was reduced by 87% (compare lanes 1 to 4). An additional methylation site (A43) did not change, suggesting a specific undermethylation at position G75.
Fig. 5.
Mapping of the ribose-methylated sites on 5.8S rRNA guided by snoRNA-2. Primer extension was performed in the presence of elevating dNTP concentrations (0.05, 0.5, and 5 mM) with antisense oligonucleotide 19796, specific for 5.8S rRNA. The extension products were separated on a 8% polyacrylamide/7 M urea denaturing gel next to primer extension sequencing performed with the same primer. The closed arrow indicates the reverse transcriptase stop (1 nt before the methylated site) at G75 guided by snoRNA-2. Another stop is marked by an open arrow and indicate modification on A43. Partial sequence of cDNA is presented and the methylated sites on the cDNA sequence are marked with asterisks. The triangles indicate the elevating level of dNTP. Lanes 1–3, primer extension was performed with total RNA from WT cells at elevated dNTP concentrations, and lanes 4–6, with total RNA from the cell line silenced for snoRNA-2.
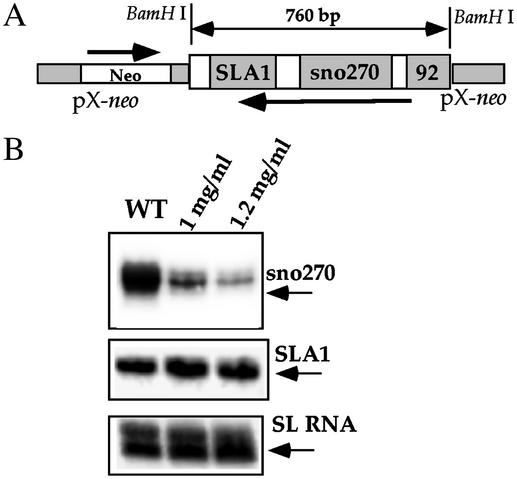
C/D snoRNA Silencing Can Be Induced in L. major. To examine whether snoRNA silencing also exists in other trypanosomatid species, we examined the silencing of snoRNAs in the SLA1 locus of L. major by expressing antisense RNA from the pX episome. The silencing construct can potentially silence both SLA1 and the 270-nt bona fide C/D snoRNA (29). Briefly, parasites expressing the antisense transcript were selected at high G418 concentrations and RNA from WT and the cell lines were subjected to primer extension with oligonucleotides complementary to either snoRNA 270 or SLA1, using the level of SL RNA as a control. The results, presented in Fig. 6, indicate silencing of snoRNA 270, suggesting that silencing can be achieved also in L. major. However, silencing was not observed for SLA1 and the level of snoRNA 92 was only partially reduced (data not shown). Note that in the snoRNA-2-silenced cell line the h2 RNA was only partially silenced as compared with the complete elimination of snoRNA-2 (Fig. 4C), suggesting that silencing is more efficient for C/D snoRNAs than for H/ACA RNAs.
Fig. 6.
Silencing of snoRNA in L. major by antisense RNA. (A) Schematic representation of the construct. The orientations of the neo-resistant gene and the snoRNA cluster are marked with arrows. The length of the insert is indicated. (B) Primer extension analysis on RNA from WT and cell lines carrying the construct expressing antisense transcript of the snoRNA cluster. The G418 concentration used for selection is indicated above the lanes. The extension products are indicated and marked with arrows. The primers used in the extension reactions are Lm270A, 31253, and 4139, complementary to the 270-nt snoRNA, SLA1, and SL RNA, respectively.
snoRNA Can Be Silenced in T. brucei by dsRNA. The silencing of snoRNA by antisense RNA is most likely mediated by a mechanism related to RNAi, because siRNAs were detected in the silenced cells (Fig. 3). To examine whether silencing of snoRNA can also be achieved in T. brucei by the expression of dsRNA, we used a well-established system for RNAi used routinely to silence protein-coding genes, based on the production of dsRNA from two T7 opposing promoters. The cluster encoding for three snoRNAs (two C/D RNAs, TBC4 and TBC5; and an H/ACA RNA TBH1) was cloned into the pZJM expression vector (21), and cloned cell lines expressing the construct were selected. In this construct, the expression of dsRNA is under the control of the tetracycline repressor. The expression of TBC4 and TBH1 in the induced and uninduced cells was examined by primer extension, using snoRNA 92 as a control, and is presented in Fig. 7. The results indicate that, as in L. major and L. collosoma, the C/D snoRNA TBC4 was silenced (85% reduction as compared with the level in uninduced cells). However, no change was observed in the level of TBH1 and only a minor reduction was observed for TBC5 (data not shown), suggesting that dsRNAs produced by the T7 opposing system are amenable to inducing the degradation of C/D snoRNA in T. brucei although at different levels.
Discussion
In this study we have provided evidence that in trypanosomatids snoRNAs can be silenced in L. collosoma and L. major by overproduction of antisense RNA, and in T. brucei by conventional RNAi using the T7 opposing promoter system. The silencing is mediated by the RNAi mechanism in T. brucei and by a related mechanism in the other trypanosomatids. The site of silencing is currently unknown but silencing takes place on a mature snoRNA and not on its precursor. The silencing reduced the level of the modification guided by the silenced snoRNA.
The silencing mechanism of snoRNA in L. collosoma and L. major can either be the same as in T. brucei (by RISC-mediated degradation) or may only involve digestion by Dicer. In the first scenario, dsRNA might be formed between the sense and antisense RNA, and siRNA generated by Dicer may induce the degradation of the mature snoRNA, possibly by RISC. However, snoRNA cleavage may also take place by Dicer acting on the dsRNA formed between the mature snoRNA and the antisense transcript without the need for RISC. However, in T. brucei snoRNA silencing may involve the RISC complex as in the silencing of mRNA induced by the same method. Interestingly, RNAi in Leishmania using dsRNA was not reported but antisense RNA was shown to silence protein-coding genes (30). The distinction between these different silencing mechanisms awaits the identification of the Dicer and RISC components in these parasites. It will be most interesting if such differences can be found. Note, however, that so far all of the molecularly unique characteristics (trans-splicing, editing, and others) of this family are shared by all species.
The cleavage to produce siRNAs can take place either in the nucleus, the cytoplasm, or both (Fig. 3C). However, cleavage can take place either in the nucleus immediately after the processing that is performed in the nucleoplasm (28) or in the nucleolus, because mature snoRNAs accumulate in this compartment and silencing was observed only on mature snoRNAs. Pre-snoRNA may be immune to degradation because it is protected by the processing machinery. In Caenorhabditis elegans early studies suggest that silencing of pre-mRNA does not take place (31). However, it was reported that silencing can occur at the pre-mRNA level, most likely because of slow processing (32).
Most studies suggest that siRNA production by Dicer takes place in the cytoplasm, possibly on polyribosomes (33). However, recent studies also suggest the existence of a variant nuclear RISC that carries a chromatin remodeling complex (18). In addition, Dicer and RISC were implicated in silencing centromeric repeats, suggesting that siRNAs can be produced and actually operate in the nucleus (19). Because to date none of the proteins involved in the RNAi silencing of trypanosomes have been characterized, we could not yet determine whether the silencing machinery also exists in the nucleoplasm or nucleolus.
The reduction of snoRNA-2 was more efficient than that of h2 in L. collosoma. Degradation of snoRNA 270 but not SLA1 was observed in L. major and TBC4 was degraded by RNAi but not TBH1 in T. brucei, suggesting that snoRNAs are differently silenced. The amount of siRNAs or antisense RNAs derived from the same construct with the potential to silence these small RNAs should be almost the same, because in each case the RNAs were present in the same transcript. The differential silencing may therefore stem from the accessibility of siRNAs or antisense RNAs to the mature snoRNAs. Note that although RNAi can potentially be used to silence this group of guide RNAs, the extent of the silencing varies considerably. Most C/D snoRNAs have an open unpaired secondary structure, whereas the H/ACA-like RNAs are highly base-paired. This structural difference may explain why the silencing of C/D snoRNA is more efficient than that of the H/ACA RNAs.
A most intriguing question is why trypanosomes possess RNAi machinery amenable to digest snoRNAs. In recent years, a variety of macromolecules were shown to exist in the nucleolus, with no apparent function in ribosome assembly. A small portion of human telomerase RNA is also localized to the nucleolus (34); methylation of U6 RNA is guided by two snoRNAs, and snRNPs accumulate in Cajal bodies near the nucleolus (35). Perhaps the RNAi in the nucleolus or other distinct nuclear compartments functions in the quality control of small RNPs. If not properly assembled, the RNA will be exposed and susceptible to degradation by RNAi.
This study provides a potentially very powerful tool for studying the role of guide RNAs in processing and modification. In trypanosomes almost all genes encoding for snoRNAs are present in reiterated clusters; it is almost impossible to knock out the gene clusters. RNAi can be used to silence specific modifications on rRNAs and snRNAs. However, as stated throughout the article, it is hard to predict whether any particular snoRNA will be amenable to degradation by RNAi. Based on our experience this methodology is efficient only for small RNAs that have accessible domains that can interact with other RNAs such as C/D snoRNA and to a lesser extent the H/ACA RNA. Note that RNAi seems to be specific for these guide RNAs, because we were unable to silence other small RNAs such as 7SL RNA and SL RNA by using the T7 opposing system (Y. Lustig, X-h.L., and S.M., unpublished data).
To summarize, this study provides direct evidence that RNAi can degrade snoRNAs.
Supplementary Material
Acknowledgments
We thank Paul Englund for providing the construct to perform RNAi in T. brucei and Elisabetta Ullu for many helpful and stimulating discussions. This research was initially supported in part by a grant from the Israeli Ministry of Health, an International Research Scholars grant (to S.M.) from the Howard Hughes Medical Institute, and a grant from the German–Israel Foundation (to S.M.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: snoRNA, small nucleolar RNA; rRNA, ribosomal RNA; RNAi, RNA interference; dsRNA, double-stranded RNA; siRNA, small interfering RNA; RISC, RNA-induced silencing complex; SL RNA, spliced leader RNA.
References
- 1.Kiss, T. (2002) Cell 109, 145–148. [DOI] [PubMed] [Google Scholar]
- 2.Kiss-Laszlo, Z., Henry, Y., Bachellerie, J. P., Caizergues-Ferrer, M. & Kiss, T. (1996) Cell 85, 1077–1088. [DOI] [PubMed] [Google Scholar]
- 3.Ganot, P., Bortolin, M. L. & Kiss, T. (1997) Cell 89, 799–809. [DOI] [PubMed] [Google Scholar]
- 4.Ni, J., Tien, A. L. & Fournier, M. J. (1997) Cell 89, 565–573. [DOI] [PubMed] [Google Scholar]
- 5.Sogin, M., Elwood, H. & Gunderson, J. (1986) Proc. Natl. Acad. Sci. USA 83, 1383–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levitan, A., Xu, Y., Ben-Dov, C., Ben-Shlomo, H., Zhang, Y. & Michaeli, S. (1998) Nucleic Acids Res. 26, 1775–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu, Y., Liu, L., Lopez-Estrano, C. & Michaeli, S. (2001) J. Biol. Chem. 276, 14289–14298. [DOI] [PubMed] [Google Scholar]
- 8.Dunbar, D. A., Chen, A. A., Wormsley, S. & Baserga, S. J. (2000) Nucleic Acids Res. 28, 2855–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang, X. H., Liu, L. & Michaeli, S. (2001) J. Biol. Chem. 276, 40313–40318. [DOI] [PubMed] [Google Scholar]
- 10.Liang, X. H., Xu, Y. & Michaeli, S. (2002) RNA 8, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hannon, G. J. (2002) Nature 418, 244–251. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. (2001) Nature 409, 363–366. [DOI] [PubMed] [Google Scholar]
- 13.Ketting, R. F. (2001) Gene Dev. 15, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nykanen, A., Haley, B. & Zamore, P. D. (2001) Cell 107, 309–321. [DOI] [PubMed] [Google Scholar]
- 15.Baulcombe, D. (1999) Arch. Virol. 15, Suppl., 189–201. [DOI] [PubMed] [Google Scholar]
- 16.Djikeng, A., Shi, H., Tschudi, C. & Ullu, E. (2001) RNA 7, 1522–1530. [PMC free article] [PubMed] [Google Scholar]
- 17.Tabara, H., Sakissian, M., Kelly, W. G., Fleenor, J., Grishok, A., Timmons, L., Fire, A. & Mello, C. C. (1999) Cell 99, 123–132. [DOI] [PubMed] [Google Scholar]
- 18.Mette, M. F., Aufsatz, W., van der Winden, J., Matzke, M. A. & Matzke, A. J. (2000) EMBO J. 19, 5194–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volpe, T., Kidner, C., Hall, I., Teng, G., Grewal, S. & Martienssen, R. (2002) Science 297, 1833–1837. [DOI] [PubMed] [Google Scholar]
- 20.Ngo, H., Tschudi, C., Gull, K. & Ullu, E. (1998) Proc. Natl. Acad. Sci. USA 95, 14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, Z., Morris, J. C., Drew, M. & Englund, P. T. (2000) J. Biol. Chem. 275, 40174–40179. [DOI] [PubMed] [Google Scholar]
- 22.Michaeli, S., Roberts, T. G., Watkins, K. P. & Agabian, N. (1990) J. Biol. Chem. 265, 10582–10588. [PubMed] [Google Scholar]
- 23.Laban, A., Tobin, J. F., Curotto de Lafaille, M. A. & Wirth, D. F. (1990) Nature 343, 572–574. [DOI] [PubMed] [Google Scholar]
- 24.Perry, K. L., Watkins, K. P. & Agabian, N. (1987) Proc. Natl. Acad. Sci. USA 84, 8190–8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zamore, P. D., Tuschl, T., Sharp, P. A. & Bartel, D. P. (2000) Cell 101, 25–33. [DOI] [PubMed] [Google Scholar]
- 26.Sijen, T., Vijn, I., Rebocho, A., van Blokland, R., Roelofs, D., Mol, J. & Kooter, J. (2001) Curr. Biol. 11, 436–440. [DOI] [PubMed] [Google Scholar]
- 27.Sijen, T., Fleenor, J., Simmer, F., Thijssen, K. L., Parrish, S., Timmons, L., Plasterk, R. H. & Fire, A. (2001) Cell 107, 465–476. [DOI] [PubMed] [Google Scholar]
- 28.Samarsky, D. A., Fournier, M. J., Singer, R. H. & Bertrand, E. (1998) EMBO J. 17, 3747–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunbar, D. A., Wormsley, S., Lowe, T. M. & Baserga, S. J. (2000) J. Biol. Chem. 275, 14767–14776. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, W. W. & Matlashewski, G. (2000) Mol. Biochem. Parasitol. 107, 315–319. [DOI] [PubMed] [Google Scholar]
- 31.Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 32.Bosher, J. M., Dufourcq, P., Sookhareea, S. & Labouesse, M. (1999) Genetics 153, 1245–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammond, S. M., Boettcher, S., Caudy, A. A., Kobayashi, R. & Hannon, G. J. (2001) Science 293, 1146–1150. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell, J. R., Cheng, J. & Collins, K. (1999) Mol. Cell. Biol. 19, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sleeman, J. E. & Lamond, A. I. (1999) Curr. Biol. 9, 1065–1074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.