Abstract
The blood coagulation scheme for the puffer fish, Fugu rubripes, has been reconstructed on the basis of orthologs of genes for mammalian blood clotting factors being present in its genome. As expected, clotting follows the same fundamental pattern as has been observed in other vertebrates, even though genes for some clotting factors found in mammals are absent and some others are present in more than one gene copy. All told, 26 different proteins involved in clotting or fibrinolysis were searched against the puffer fish genome. Of these, orthologs were found for 21. Genes for the ``contact system'' factors (factor XI, factor XII, and prekallikrein) could not be identified. On the other hand, two genes were found for factor IX and four for factor VII. It was evident that not all four factor VII genes are functional, essential active-site residues having been replaced in two of them. A search of the genome of a urochordate, the sea squirt, Ciona intestinalis, did not turn up any genuine orthologs for these 26 factors, although paralogs and/or constituent domains were evident for virtually all of them.
Keywords: blood clotting, domain shuffling
Blood clotting follows the same fundamental pattern in all vertebrates, from the early diverging jawless fishes to mammals (1). In all cases the principal event is the thrombin-catalyzed conversion of a soluble plasma protein, fibrinogen, into an insoluble polymeric fibrin clot. Thrombin is a serine protease, itself the product of a series of proteolytic events. It is well established that all groups of fish (cyclostomes, elasmobranches, and teleosts) generate thrombin by pathways involving vitamin K-dependent factors, exhibit factor XIII-dependent fibrin cross-linking, and manifest a fibrinolysis that is inhibited by the same agents as inhibit fibrinolysis in mammals (1–3). In contrast, thrombin-generated fibrin clotting has not been reported in nonvertebrate chordates or other invertebrate animals.
Because such a convoluted pathway could not have evolved in one fell swoop, it was long ago realized that a series of gene duplications must lie at the heart of the complex set of interactions observed in mammalian clotting. In this regard, past sequence comparisons of serine proteases have led to the suggestion that certain of the clotting factors (particularly those constituting the ``contact system'' involving factors XI and XII, and prekallikrein) must have made their appearance more recently in evolution than some of the other clotting factors and would likely be absent in lower vertebrates (4).
The recent publication (5) of the genome sequence for the puffer fish, Fugu rubripes, makes it possible to test that prediction. Additionally, the availability (6) of the complete genome sequence for a urochordate, the sea squirt, Ciona intestinalis, allows a direct comparison of two early diverging chordates. The results confirm that the main lines of the vertebrate clotting pathway were evolved in the interval between the last common ancestor of these two creatures, a period thought to be significantly less than a hundred million years.
In the present study, 26 proteins involved in mammalian blood clotting (Table 1) were examined to see whether they have counterparts in the puffer fish and/or the sea squirt. The processes of fibrin formation and destruction are inextricably linked, and the proteins selected include both lytic factors and inhibitors. Because many paralogs could be involved (the result of recent gene duplications), stringent criteria were set for deciding whether or not a gene for a given coagulation factor was present. In the end, 21 orthologs of the coagulation factor genes were found in the puffer fish genome, but not one authentic ortholog was identified in the sea squirt genome.
Table 1. Blood clotting factors searched against puffer fish genome.
| Clotting component* | Pufferfish scaffold† | Hit regions | Back search‡ | Percent identity§ | No. aa (Puf/Mam)¶ | Introns (Puf/Mam)∥ | Accessory domains** |
|---|---|---|---|---|---|---|---|
| Factor VII | 2859 | 13975<17516 | T | 42 | 461/467 | 6/6 | GLA, EGFs |
| Factor VII | 2859 | 13975<17516 | T | 44 | 419/467 | 6/6 | |
| Factor VII | 2859 | 13975<17516 | T | 42 | 418/467 | 6/6 | |
| Factor IX | 917 | 50760<53103 | T | 43 | 481/461 | 7/6 | GLA, EGFs |
| Factor IX | 1343 | 44331>46796 | T | 46 | 473/461 | 7/6 | |
| Factor X | 2859 | 5850<13900 | T | 41 | 472/488 | 6/5 | GLA, EGFs |
| Protein C | 8062 | 3394<5430 | T | 45 | 448/461 | 6/6 | GLA, EGFs |
| Prothrombin | 403 | 10447<14481 | T | 52 | 614/622 | 13/12 | GLA, KRs |
| Protein S | 2356 | 2811>6972 | T | 51 | 647/650 | 12/14 | GLA, EGFs |
| Factor XI | 512 | 41260>48333 | F | — | — | — | (apples) |
| Factor XII | 2128 | 34416<35292 | F | — | — | — | (FN2, FN1, EGFs, KR) |
| Prekallikrein | 512 | 41260>48333 | F | — | — | — | (apples) |
| Factor XIIIA | 3692 | 14397>17526 | T | 48 | 742/732 | 13/13 | |
| Factor XIIIB | 310 | 69650<78059 | F | — | — | — | (10 sushis) |
| Factor V | 3405 | 13245>4228 | T | 41 | 1846/2224 | 22/24 | F5/8-As, F5/8-Cs |
| Factor VIII | 2929 | 2009<10768 | T | 42 | 1583/2351 | 24/24 | F5/8-As, F5/8-Cs |
| ϕ Alpha†† | 3291 | 1007>4785 | T | 37 | 697/741 | 8/na | |
| ϕ Beta†† | 6262 | 1478<3684 | T | 57 | 467/461 | 11/7 | |
| ϕ Gamma†† | 39 | 57384>59627 | T | 51 | 416/411 | 7/7 | |
| Plasminogen | 1979 | 45704>48368 | T | 56 | 747/810 | 18/18 | PAN, KRs |
| 9368 | 1>4148 | T | |||||
| 1457 | 60610<60452 | T | |||||
| t-PA | 191 | 53968>57860 | T | 56 | 524/524 | 11/11 | FN2, EGF, KRs |
| u-PA | 4367 | 6096<8169 | T | 42 | 392/431 | 7/8 | EGF, KR |
| 3932 | 5755<8546 | T | 41 | 454/431 | 8/8 | EGF, KR | |
| Tissue factor | 8956 | 1219<2489 | T | 33 | 237/295 | 5/5 | FN3 |
| Thrombomodulin | 195 | 8338>9357 | T | 29 | 472/575 | 0/0 | LEC, EGFs |
| Thrombomodulin | 195 | 11078>12352 | T | 37 | 542/575 | 0/0 | LEC, EGFs |
| TFI | 1267 | 33102>34123 | T | 44 | 241/235 | 4/4 | Kunitz |
| TFI | 611 | 69717>76256 | T | 38 | 242/304 | 4/4 | Kunitz |
| TAFI | 123 | 149764<152808 | T | 45 | 416/425 | 9/10 | |
| α2-Antiplasmin | 1092 | 62744<64057 | T | 34 | 460/488 | 6/8 | Serpin |
| Antithrombin III | 1063 | 73580<71488 | T | 55 | 437/464 | 7/5 | Serpin |
| PAI-1 | 1754 | 50591>52865 | F | 44 | 406/415 | 6/6 | Serpin |
TAFI, thrombin-activated fibrinolysis inhibitor; PAI-1, plasminogen activator inhibitor 1. KR, kringle; LEC, lectin.
Sequences for the human clotting factors were used as probes.
Puffer fish scaffolds range in size from 670 kb for scaffold 1 to 2 kb for scaffold 12,381.
The back-searching of candidates was against GenBank. T=True, means a reciprocal match. F=False, means that the candidate sequence retrieved a protein other than the probe.
Percent identity between the putative puffer fish (Puf) and human (Mam) proteins.
The number of amino acids in the puffer fish and human proteins is compared.
The number of introns in the puffer fish and human genes is compared.
Domain characteristics of the clotting factor are listed.
The symbol ϕ denotes fibrinogen. In the case of the fibrinogen α chain, the comparison is with the α chain of chicken fibrinogen.
On the other hand, the wherewithal for the evolutionary assembly of all of the coagulation factor genes seems to be present in the sea squirt genome in the form of various and sundry domains that in ancestral lines were duplicated and rearranged into the genes for the present-day vertebrate clotting factors. Many of these accessory domains occur at the amino termini of zymogens for serine proteases. Paramount among them is the ``GLA domain,'' an entity that contains multiple γ-carboxy-glutamic acid residues whose synthesis depends on vitamin K (7). The blood coagulation proteins that contain GLA domains include prothrombin, factors VII, IX, and X, protein C, and the nonprotease protein S. Other peripheral domains found in association with clotting proteases include ``kringles'' (8), fibronectin ``finger'' domains (FN-1 and FN-2; refs. 9 and 10), fibronectin domain III (FN-3; ref. 11), epidermal growth factor (EGF) domains (9, 12), and ``apple domains,'' four copies of which are found at the amino termini of mammalian factor XI and prekallikrein (13). Apple domains are actually part of a larger group now referred to as PAN domains (14). Additionally, there are sundry other domains associated with clotting proteins, including the F5/8-A and -C domains found in factors V and VIII, two proteins that themselves are evolved from ceruloplasmin (15), although the latter protein is composed exclusively of three A domains. The F5/8-C domain is also known as ``discoidin.''
Many of these accessory domains are active in localizing the clotting process. GLA domains, for example, serve to bind the vitamin K-dependent factors to platelets or thrombocytes, and other domains serve to bind the factors to each other. In passing, it should be noted that platelets are not found in nonmammals, their role being enacted by a class of white cells designated ``thrombocytes.''
Methods
Publicly accessible sequence data were downloaded from the Fugu and Ciona web sites: fugu.hgmp.mrc.ac.uk and www.jgi.doe.gov/ciona.
Puffer Fish. The puffer fish DNA data are available in the form of 12,381 scaffolds ranging in size from 657 to 2 Kb. These sequences amount to 332.5 Mb containing ≈30,000 potential genes and make up >95% of the puffer fish genome (6). Candidate genes were initially identified by searching the sequence of each human coagulation protein against the puffer fish scaffolds with the tblastn program (16).
In each case, a prioritized list of candidate scaffolds was examined. The approximate matching regions were extracted from scaffolds, and the program genescan (17) was used in an effort to link up exons. In general, genescan was ≈75% effective, missing about a quarter of all exons. The roughly translated sequence was aligned with the appropriate human protein, after which missing regions were identified with the aid of the program inspect (18). Standard blast (16) was then used to back-search the full amino sequence against GenBank. If the candidate was indeed the ortholog of the factor under study, the same factor should be returned (T = True). If that was not the case (F = False), the process was repeated on the next candidate on the list, and so on, until a reasonable conclusion about presence (T) or absence (F) could be reached. The method is not foolproof. Occasionally the best match found in the puffer fish genome gave rise to a much better match upon back-searching, indicating that the candidate was not an ortholog. In some other cases, the matches were reciprocal, but the resemblance was not particularly strong. In this regard, most of the genuine orthologs were >40% identical with their human counterparts (Table 1). Valid orthologs were also expected to have the same arrangement of modular components. Intron locations were also compared. Additionally, for those factors that belong to well characterized families, phylogenetic trees were constructed (19). Pairwise alignments of 21 putative puffer fish proteins with human counterparts and with intron positions marked are published as supporting information on the PNAS web site, www.pnas.org.
Sea Squirt. A similar strategy for the same 26 factors was used for searching the sea squirt genome. The 160-Mb genome encompasses 16,000 genes assembled into 2,501 scaffolds (7). The downloaded sequences were subjected to the regimen described above with tblastn (16), genescan (17), inspect (18), and finally blast to back-search candidates against GenBank. The same criteria for orthologs were used as was applied to puffer fish: namely, (i) the highest resemblance to any protein in GenBank with the back-search had to be the coagulation factor, and (ii) the modular arrangement of domains had to be the same as occurs in the mammalian factor.
Results
Puffer Fish. Vitamin K-dependent factors. Orthologous genes were identified for each of the five vitamin K-dependent serine proteases (prothrombin, factors VII, IX, and X, and protein C). Remarkably, two genes were found for factor IX, both of which may be functional. The two proteins are 51% identical with each other, a stronger resemblance than is observed with either to mammalian factor IX, indicating that the gene duplication leading to the two homologs occurred after fish diverged from the lineage leading to tetrapods.
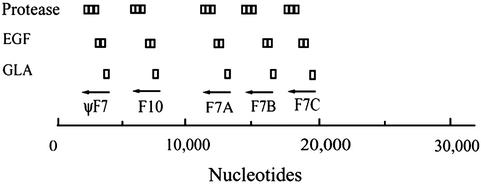
Four genes (not all intact) were found for factor VII, all on a single scaffold together with a gene for factor X (Fig. 1). Of the four, the homolog labeled f7B most likely represents the genuine factor, its putative protein product being 46% identical with mammalian factor VII. Homologs f7A and f7C are 42% and 43% identical with the mammalian factor VII, respectively; in homolog f7A, the expected active-site serine codon has been changed to that of aspartic acid. The gene denoted ψF7 in Fig. 1 is in an obvious state of evolutionary decay, the sequence in the region of the expected active site being greatly fragmented.
Fig. 1.
Diagrammatic depiction of genes for factor VII and factor X on FUGU scaffold 2859 (32,129 nt) from the puffer fish (F. rubripes) genome. The section denoted by ψF7 lacks the catalytic site characteristic of serine proteases and is likely not a functional entity. In F7A, the serine at the active site has been replaced by an aspartic acid.
The single factor X gene is 41% identical to human factor X. That the puffer fish factor X and factor VII genes are adjacent to each other (Fig. 1) may be significant in that, among all of the major human coagulation factors, only these two are found at the same locus on the same chromosome (20).
An additional GLA-containing homolog with two EGF domains was found on scaffold 6546, the sequence of which doesn't fall within any of the four subfamilies with this arrangement of domains (factors VII, IX, or X, or protein C). The gene contains a canonical signal sequence, and its activation site and active site are both intact. All indications are that this could be a fully active gene for the precursor of a serine protease. In contrast, a GLA-containing gene on scaffold 468 most closely resembles factor X, but it lacks the critical active-site constellation of residues needed for catalysis, as well as the key basic-residue cleavage site needed for activation.
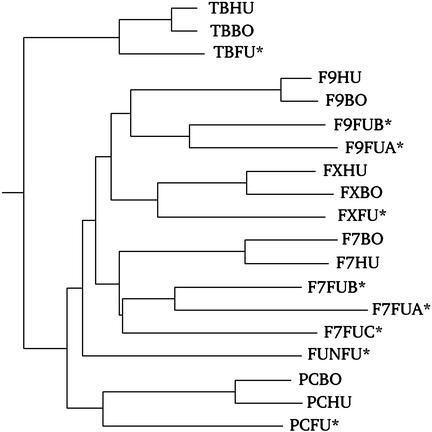
A phylogenetic tree based on the serine protease portions of the vitamin K-dependent proteases from human, bovine, and puffer fish clustered all except one of the puffer fish factors with their mammalian counterparts, the one exception being the unassigned factor on scaffold 6546 (Fig. 2). The tree indicates that most of the extra genes for the various factors are the results of duplications that have occurred after fish and tetrapods diverged.
Fig. 2.
Phylogenetic tree constructed from serine protease portions of the vitamin K-dependent proteases (prothrombin, protein C, factor VII, factor IX, and factor X) of human (HU), bovine (BO), and puffer fish (FU). Puffer fish entries are also marked with asterisks. The puffer fish entry denoted FUNFU (from scaffold 6546) does not fall into any of the usual groups.
Protein S. The GLA-containing steroid-binding protein known as protein S (21) was present, and like its human counterpart, it is situated near a similar homolog, which in humans is denoted as a growth arrest-specific gene (22).
Tissue factor. A single gene was found for tissue factor. The putative protein is only 34% identical with human tissue factor. Tissue factor is a quite rapidly changing entity, however, and the orthologous mammalian proteins are only 60–70% identical one to another. The five introns are situated at the same positions in the puffer fish and human genes, and the identification seems certain.
The contact system. Genes for factor XI and prekallikrein were not found in puffer fish, the best match found on back-searching in both cases being a mosquito protease. In line with these observations, no apple domains were found to be associated with serine proteases, although several such domains were found on a relatively short scaffold (scaffold 11138) that could conceivably be associated with some protease on another scaffold.
A gene for factor XII could not be found, the closest candidate on back-searching in this case being salmon trypsin. A search for scaffolds containing both kringles and EGF domains did not reveal any proteases with domainal arrangements similar to what occurs in mammalian factor XII.
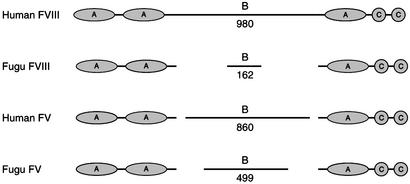
Factors V and VIII. The genes for factors V and VIII, themselves paralogs descended from ceruloplasmin (15), were readily identified. These very large proteins are composed of three A domains and two carboxyl-terminal discoidin domains (denoted C in Fig. 3). The B region between the second and third A domains varies greatly between the puffer fish and human proteins, as it does between factors V and VIII from any species. This region has been found to be expendable in human factor VIII expression systems (23).
Fig. 3.
Schematic comparison of human and puffer fish factors V and VIII. The regions denoted B are variable from species to species; numbers indicate residues in the B segment.
Fibrinogen genes. The β- and γ-chains of puffer fish fibrinogen are 58% and 52% identical with their human counterparts. In the case of the β-chain gene, the puffer fish has four additional introns, all quite short, besides the ones in common with the human gene. In the γ-chain gene, all introns were found at approximately the same positions as occur in the human gene except for one missing from the puffer fish that marks the splice site near the carboxyl terminus of mammalian γ-chains. This intron, which was previously found to be absent from the lamprey γ-chain gene (24), is what in mammals leads to a minor form of fibrinogen with γ′ chains that serve as binding sites for factor XIII (25).
The fibrinogen α-chain in puffer fish has a similar arrangement to what is found in chickens, none of the central repeats found in mammalian or lamprey α chains being present. Additionally, several large deletion events are apparent in the αC domain. The alternatively spliced αCE domain characteristic of the minor extended form of fibrinogen known as fibrinogen-420 is encoded in the downstream region of the gene, just as occurs in birds and mammals (26). In lamprey, the minor form of the α-chain is encoded in an entirely separate gene denoted α2 (27).
Factor XIII. The A chain of factor XIII was obvious (48% identical to human factor XIII); a second somewhat truncated version of the protein was found on another scaffold and was only 43% identical. A gene for the factor XIII accessory B chain was not found. In mammals, the B chain is composed of 10 ``sushi'' domains (also known as complement-control modules or β2-glycoprotein domains). These domains are quite common in mammalian proteins where they occur in numerous components of the complement system. The domains are also common in the puffer fish genome, but no appropriate string of ten was found that corresponds to factor XIII B. The best match occurred between a cluster of a dozen or more sushi domains on scaffold 310 that on back-searching identified a mouse polydomain protein containing 18 such domains.
Fibrinolytic proteases. Orthologous genes for plasminogen, t-PA (tissue plasminogen activator) and u-PA (urokinase-type plasminogen activator), were readily identified. The plasminogen gene actually spans three different scaffolds. The main body of the protein, including kringles 4 and 5, is encoded on scaffold 9368 (4148 nt), but the carboxyl-terminal region is encoded at the 5′-end of scaffold 1457 (160,920 nt), and the amino terminus, including a PAN domain and kringles 1–3, at the 3′-end of scaffold 1979 (48,368 nt).
The t-PA gene has the expected FN-1 domain (Fig. 4A), an EGF domain, and two kringles. In the case of u-PA, the terminal EGF domain seems to be fragmented, but the single kringle is intact. In all cases, introns separate the peripheral domains from each other and the serine protease portions of the genes.
Fig. 4.
(A) Alignment of FN1 sequences from human t-PA and factor XII with two homologous sequences from puffer fish. No homologs were found in the sea squirt genome. (B) Alignment of FN2 sequences from human factor XII, puffer fish scaffold 1677, and sea squirt scaffold 883.
Serpins. A gene for antithrombin III was found whose putative protein is 55% identical with the human protein. The protein has two introns not found in the human gene, as well as five that they have in common.
The search for plasminogen activator inhibitor 1 (PAI-1) resulted in a failed back-search. The initial search of the puffer fish genome with human PAI-1 as a probe identified a gene on scaffold 1754 whose gene product was 44% identical. When that sequence was back-searched against GenBank, however, the best hit, at 59% identity, was leucocyte elastase inhibitor. Contrarily, when the human α2 antiplasmin sequence was searched against puffer fish, a best match was found with a gene whose protein product was only 34% identical. Nonetheless, the back-search against GenBank turned up α2 antiplasmin as the best hit. For the moment, we have cautiously labeled this as an ortholog, even though the resemblance is lower than expected.
Other inhibitors. Two genes for tissue factor inhibitor (TFI) were identified, just as is observed in humans. The best match among these was 44% identical. As for other inhibitors, a gene for the carboxypeptidase known as thrombin-activated fibrinolysis inhibitor was found, the putative protein sequence of which is 45% identical with its human counterpart.
Two genes that correspond to thrombomodulin were found adjacent to each other on puffer fish scaffold 195; an analogous situation of two adjacent genes occurs in humans, in which case one of the genes is thrombomodulin and the other is thought to be an accessory protein for complement factor C1Q (28). As is also the case in humans, neither of the two puffer fish genes has introns.
Peripheral domain inventory. All told, 19 GLA domains were found in the puffer fish genome (Table 2), 11 of which were associated with the vitamin K-dependent serine protease clotting factors. Two others were parts of protein S and its homolog. None of the remaining six were associated with EGF domains or serine protease domains.
Table 2. Some domains in puffer fish and sea squirt genomes*.
| Domain type | Puffer fish | Sea squirt |
|---|---|---|
| GLA | 19 | 4 |
| Kringle | 34 | 50 |
| Apple (PAN) | 27 | 0 |
| F5/8-A | 12 | 3 |
| F5/8-C (discoidin) | 27 | 23 |
| FN1 | 2† | 0 |
| FN2 | 19 | 16 |
| FN3 | >100 | >100 |
| EGF | >100 | >75 |
Domains were identified with TBLASTN using cutoff E = 10-3.
Cutoff E-1; see also Fig. 4A.
The total number of kringles found was 34, of which 5 were in plasminogen, 2 were in t-PA, 1 was in u-PA, and 2 were in prothrombin, leaving about two dozen unaccounted for. Many of these are likely associated with nonclotting proteins like hepatocyte growth factor (29), but we did not pursue the matter further.
Only two FN1 domains were found in the puffer fish genome (Fig. 4A), but at least 19 FN2 domains are present (Table 2). None of the 27 apple domains found in the puffer fish genome were associated with proteases, although, as noted above in some cases, the scaffolds were small, and associated proteins may not have been recovered in the assembly process. We also found 12 F5/8-A domains and 27 F5/8-C (discoidin) domains (Table 2).
Sea Squirt Genes. None of the 26 coagulation factors turned up a convincing case for an orthologous gene. Nonetheless, almost all of the constituent domains that are generally associated with these factors were found in one context or other, including GLA, sushi, FN2, FN3, EGF, and F5/8-A and -C domains. The two exceptions were the FN1 and apple (PAN) domains, neither of which were found.
Only four GLA domains were found to be encoded in the sea squirt genome. None of these were associated with kringles, making it unlikely that there is a prothrombin-like gene. On the other hand, all of the GLAs are associated with multiple EGF domains, but none of these is near a serine protease domain. In one case, the GLA domain is near an EGF domain but in the wrong order; i.e., the EGF is on the amino-terminal side of the GLA.
All told, 50 kringles were found in sea squirt genome, including eight situations where multiple kringles are found with serine proteases. These included four sets of two, three sets of four, and one set of five. However, neither plasmin, t-PA, nor u-PA survived the back-search process.
Several multikringle serine proteases were found that might be construed as ``plasminogen-like.'' That the best of these was not an authentic ortholog was attested to by the lack of a terminal PAN domain (13), the first 120 residues being instead more like a domain found in thrombospondins (30). Moreover, the putative serine protease portion failed the back-search test, being more similar to a trypsin from a sponge than to any plasmin; indeed, plasmin was not among the top 100 hits.
A number of sequences were identified that are homologous to the carboxyl-terminal domains of fibrinogen, not unexpected because these domains are widespread in animals (31); however, no full-length genes were found with the potential for the constituent coiled coils that are hallmarks of fibrinogen.
One intriguing situation involved a gene on scaffold 87 conceivably related to factors V and VIII, which, as noted above, are descended from ceruloplasmin (15). A putative protein was identified that has both A domains and a C domain. The A domains are much more similar to ceruloplasmin than they are to either factor V or factor VIII, however, even though in vertebrates (including puffer fish) ceruloplasmin has not here-tofore been found to have a C domain.
The sea squirt genome has a transglutaminase, but there was no evidence of the signal peptide required for a circulating factor XIII, and it is likely an ordinary tissue transglutaminase. Similarly, a number of serpins are encoded in the genome, but they could not be matched with antithrombin. The sea squirt also has a carboxypeptidase paralogous to thrombin-activated fibrinolysis inhibitor, as well as numerous domains that are similar to those found in tissue factor, tissue factor inhibitor, and thrombomodulin.
Discussion
Late-Appearing Factors. The presence of many genes for mammalian clotting proteins in the puffer fish genome was anticipated, a number of them having been isolated and/or cloned from more primitive fish in the past. For example, prothrombin has been isolated from lamprey blood plasma (2) and cloned from the hagfish (32). Moreover, the principal ingredient of blood clots, fibrin(ogen), has been fully characterized (sequenced, cloned, and fragments crystallized) from lamprey (1–3, 33). As such, the events involving the thrombin-catalyzed conversion of fibrinogen to fibrin and its subsequent cross-linking and lysis were not an issue. The exact pathways of thrombin generation were still unclear, however, and it remained uncertain whether all of the vitamin K-dependent factors are present in lower vertebrates, although it should be noted that factor VII, as well as prothrombin, has been identified in zebrafish (34, 35).
It was expected that some components of the ``contact phase'' system (factors XI and XII, and prekallikrein) would be missing from lower animals because amino acid sequence comparisons showed that factor XI and prekallikrein are so similar that the gene duplication leading to them must have been very recent (4). Nonetheless, the complete absence of the contact phase system from the puffer fish genome is noteworthy. Of course it cannot be ruled out that one or all of the absent genes do not reside among the 5% of DNA sequence data that are not yet available from the puffer fish. This absence aside, it is remarkable how stable the rest of the coagulation scheme has been over the course of the last 400 million years.
Assembling the Scheme. It is thought that 50–100 million years separate the appearances of urochordates (which include the sea squirt) and vertebrates. During that time the machinery for thrombin-catalyzed fibrin formation had to be concocted by gene duplication and the shuffling about of key modular domains. The relative times of duplicative events can be estimated by various means, the most obvious being the presence or absence of a gene in earlier diverging organisms, although it must be kept in mind that lineages may lose genes. Another way to gauge events is from the relative positions of various gene products on phylogenetic trees, earlier branching implying earlier appearance. In this regard, (pro)thrombin invariably appears lower on the phylogenetic trees than do the other vitamin K-dependent factors (Fig. 2).
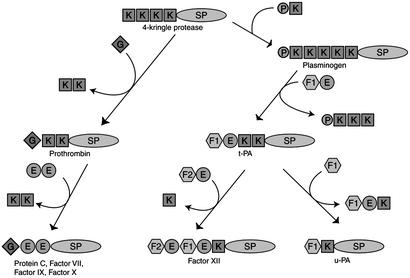
The order of events can also be inferred by considering the most parsimonious route to assembling the various clusters of peripheral domains. Nine of the proteases under discussion can be accounted for by six domain-swapping events (Fig. 5). Indeed, the presence of a multiple-kringle protease in the sea squirt genome provides a reasonable model for a step-by-step parallel evolution of the clotting and lysis systems. It should be noted that a serine protease with only one kringle has been found in the ascidian Herdmania momus (36). Although numerous scenarios have been offered in the past about how modular exchange was involved in generating these schemes (refs. 4, 12, and 37–41, inter alia), the new genomic data now provide a realistic set of starting materials.
Fig. 5.
Putative evolution of nine proteases involved in generation and destruction of fibrin clots by a small number of domainal exchanges. The proposed root protein is a four-kringle protease, prototypical genes for which are found in the sea squirt genome. A three-kringle protease root could equally well have been used. G, GLA; E, EGF; K, kringle; P, PAN domain; F1, fibronectin domain 1; F2, fibronectin domain II; SP, serine protease. All of these domains are found in the sea squirt genome, but not associated in the arrangements found in clotting factors. Factor XII does not appear in the puffer fish genome and likely appeared after the divergence of fish and tetrapods.
The timing of duplicative events can also be approximated from ortholog–paralog comparisons. As an example, human and puffer fish factor V are 41% identical, and human and puffer fish factor VIII are 42% identical (not counting the variable B regions). On the average, the two factors themselves (in this region) are 38% identical, implying that the gene duplication that led to them occurred only a relatively short while before the common ancestor of fish and mammals. The difference is so small (42% vs. 38%) that it may turn out that the earlier diverging jawless fish will have only the preduplication gene. A genome study devoted to the lamprey or hagfish would settle the point.
Supplementary Material
Acknowledgments
We are grateful to Da-Fei Feng for his help and encouragement during the course of this project. This work was supported by National Institutes of Health Grant HL-26873.
Abbreviations: t-PA, tissue plasminogen activator; u-PA, urokinase-type plasminogen activator; EGF, epidermal growth factor; FN, fibronectin; GLA, γ-carboxy-glutamate; TFI, tissue factor inhibitor.
References
- 1.Doolittle, R. F. & Surgenor, D. M. (1962) Am. J. Physiol. 203, 964–970. [DOI] [PubMed] [Google Scholar]
- 2.Doolittle, R. F., Oncley, J. L. & Surgenor, D. M. (1962) J. Biol. Chem. 237, 3123–3127. [PubMed] [Google Scholar]
- 3.Doolittle, R. F. (1990) Adv. Exp. Med. 281, 25–37. [DOI] [PubMed] [Google Scholar]
- 4.Doolittle, R. F. & Feng, D. F. (1987) Cold Spring Harbor Symp. Quant. Biol. 52, 869–874. [DOI] [PubMed] [Google Scholar]
- 5.Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J. M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. (2002) Science 297, 1301–1310. [DOI] [PubMed] [Google Scholar]
- 6.Dehal, P., Satou, Y., Campbell, R. K., Chapman, J., Degnan, B., De Tomaso, A., Davidson, B., Di Gregorio, A., Gelpke, M., Goodstein, D. M., et al. (2002) Science 298, 2157–2167. [DOI] [PubMed] [Google Scholar]
- 7.Stenflo, J. & Suttie, J. (1977) Annu. Rev. Biochem. 46, 157–172. [DOI] [PubMed] [Google Scholar]
- 8.Sotterup-Jensen, L., Claeys, H. Zajdel, M., Petersen, T. E. & Magnusson, S. (1978) Prog. Chem. Fibrinolysis Thromb. 3, 191–209. [Google Scholar]
- 9.Banyai, L., Varadi, A. & Patthy, L. (1983) FEBS Lett. 163, 37–41. [DOI] [PubMed] [Google Scholar]
- 10.Ozhogina, O. A., Trexler, M., Banyai, L. Llinas, M. & Patthy, L. (2001) Protein Sci. 10, 2114–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bazan, J. F. (1990) Immunol. Today 11, 350–354. [DOI] [PubMed] [Google Scholar]
- 12.Doolittle, R. F., Feng, D. F. & Johnson, M. S. (1984) Nature 307, 558–560. [DOI] [PubMed] [Google Scholar]
- 13.Fujikawa, K., Chung, D. W., Hendrickson, L. E. & Davie, E. W. (1986) Biochemistry 25, 2417–2424. [DOI] [PubMed] [Google Scholar]
- 14.Tordai, H., Banyai, L. & Patthy, L. (1999) FEBS Lett. 461, 63–67. [DOI] [PubMed] [Google Scholar]
- 15.Vehar, G. A., Keyt, B., Eaton, D., Rodriguez, H., O'Brien, D. P., Rotblat, F., Opperman, H., Keck, R., Wood, W. I., Harkins, R. N., et al. (1984) Nature 312, 337–342. [DOI] [PubMed] [Google Scholar]
- 16.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burge, C. & Karlin, S. (1997) J. Mol. Biol. 268, 78–94. [DOI] [PubMed] [Google Scholar]
- 18.Doolittle, R. F. (1987) Of Urfs and Orfs: A Primer on How to Analyze Derived Amino Acid Sequences (University Science Press, Mill Valley, CA).
- 19.Feng, D.-F. & Doolittle, R. F. (1996) Methods Enzymol. 266, 368–372. [DOI] [PubMed] [Google Scholar]
- 20.Koeleman, B. P., Reitsma, P. H., Bakker, E. & Bertina, R. M. (1997) Thromb. Haemostasis 77, 873–878. [PubMed] [Google Scholar]
- 21.Walker, F. J. (1980) J. Biol. Chem. 255, 5521–5524. [PubMed] [Google Scholar]
- 22.Varnum, B. C., Young, C., Elliott, G., Garcia, A., Bartley, T. D., Fridell, Y. W., Hunt, R. W., Trail, G., Clogston, C., Toso, R. J., et al. (1995) Nature 373, 623–626. [DOI] [PubMed] [Google Scholar]
- 23.Toole, J. J., Pittman, D. D., Orr, E. C., Murtha, P., Wasley, L. C. & Kaufman, R. J. (1986) Proc. Natl. Acad. Sci. USA 83, 5939–5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan, Y. (1992) Ph.D. dissertation (Univ. of California, San Diego).
- 25.Siebenlist, K. R., Meh, D. A. & Mosesson, M. W. (1996) Biochemistry 35, 10448–10453. [DOI] [PubMed] [Google Scholar]
- 26.Fu, Y., Cao, Y., Hertzberg, K. M. & Grieninger, G. (1995) Genomics 30, 71–76. [DOI] [PubMed] [Google Scholar]
- 27.Pan, Y. & Doolittle, R. F. (1992) Proc. Natl. Acad. Sci. USA 89, 2066–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nepomuceno, R. R., Henschen-Edman, A. H., Burgess, W. H. & Tenner, A. J. (1997) Immunity 6, 1119–1129. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura, T., Nishizawa, T., Hagiya, M., Seki, T., Shimonishi, M., Sugimura, A., Tashiro, K. & Shimizu, S. (1989) Nature 342, 440–443. [DOI] [PubMed] [Google Scholar]
- 30.Lawler, J & Hynes, R. O. (1986) J. Cell Biol. 103, 1035–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doolittle, R. F., Spraggon, G. & Everse, S. J. (1997) in Plasminogen Related Growth Factors, eds. Bock, G. R. & Goode, J. A. (Wiley, New York), pp. 4–23.
- 32.Banfield, D. K. & MacGillvray, R. T. A. (1992) Proc. Natl. Acad. Sci. USA 89, 2779–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, Z., Spraggon, G., Pandi, L., Everse, S. J., Riley, M. & Doolittle, R. F. (2002) Biochemistry 41, 10218–10224. [DOI] [PubMed] [Google Scholar]
- 34.Jagadeeswaran, P., Gregory, M., Zhou, Y., Zon, L. and Padmanabhan, K. (2000) Blood Cells Mol. Dis. 26, 479–489. [DOI] [PubMed] [Google Scholar]
- 35.Sheehan, J., Temple, M., Gregory, M., Hanumanthaiah, R., Troyer, D., Phan, T., Thankavel, B. and Jagadeeswaran, P. (2001) Proc. Natl. Acad. Sci. USA 98, 8768–8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnold, J. M., Kennett, C., Degnan, B. M. & Lavin, M. F. (1997) Dev. Genes Evol. 206, 455–463. [DOI] [PubMed] [Google Scholar]
- 37.Patthy, L. (1985) Cell 41, 657–663. [DOI] [PubMed] [Google Scholar]
- 38.Doolittle, R. F. (1985) Trends Biochem. Sci. 10, 233–237. [Google Scholar]
- 39.Patthy, L. (1990) Semin. Thromb. Hemostasis 16, 245–254. [DOI] [PubMed] [Google Scholar]
- 40.Krem, M. W. & Di Cera, E. (2002) Trends Biochem. Sci. 27, 67–74. [DOI] [PubMed] [Google Scholar]
- 41.Gherardi, E., Manzano, R. G., Cottage, A., Hawker, K. & Aparicio, S. (1997) in Plasminogen Related Growth Factors, eds. Bock, G. R. & Goode, J. A. (Wiley, New York), pp. 24–41. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.