Abstract
Apoptosis plays a pivotal role in the cytotoxic activity of most chemotherapeutic drugs, and defects in this pathway provide a basis for drug resistance in many cancers. Thus the ability to restore apoptosis by using small molecules could have important therapeutic implications. Using a cell-free assay to simultaneously target multiple components of the apoptosis pathway, we identified a class of compounds that activate caspases in a cytochrome c-dependent manner and induce apoptosis in whole cells. By reconstituting the apoptosis pathway with purified proteins, we determined that these compounds promote the protein–protein association of Apaf-1 into the functional apoptosome. These compounds exert cytostatic and cytotoxic effects on a variety of cancer cell lines while having little or no activity against the normal cell lines tested. These findings suggest that direct activation of the basic apoptosis machinery may be a viable mechanism to selectively target cancer.
Apoptosis is an evolutionarily conserved process that regulates development and homeostasis, and defects in this pathway are implicated in both tumorigenesis and multidrug resistance. Two distinct pathways for apoptosis have been characterized (1), both of which ultimately result in a proteolytic cascade involving cysteine aspartyl proteases, or caspases (2, 3). In the drug-induced (intrinsic) pathway, signals transmitted to the mitochondria cause the release of cytochrome c (cyto c) into the cytosol, where it binds to the scaffolding protein, Apaf-1 (4, 5). Apaf-1 is a large, multidomain protein composed of an N-terminal CARD domain, a CED4 homology domain, and a C-terminal WD40 repeat (6). Several reports have shown that in the presence of cyto c and dATP, Apaf-1 will assemble into a large oligomeric complex of ≈700 kDa termed the apoptosome, which recruits and activates procaspase-9 (Fig. 1A; refs. 6–11). Biochemical data suggest that apoptosome formation is a multistep process: Apaf-1 must bind both cyto c and dATP before it can assemble into the functional apoptosome composed of six to seven Apaf-1 monomers (6, 12–14). Active caspase-9 then activates downstream caspases, including caspase-3, ultimately leading to full implementation of the apoptotic program (15).
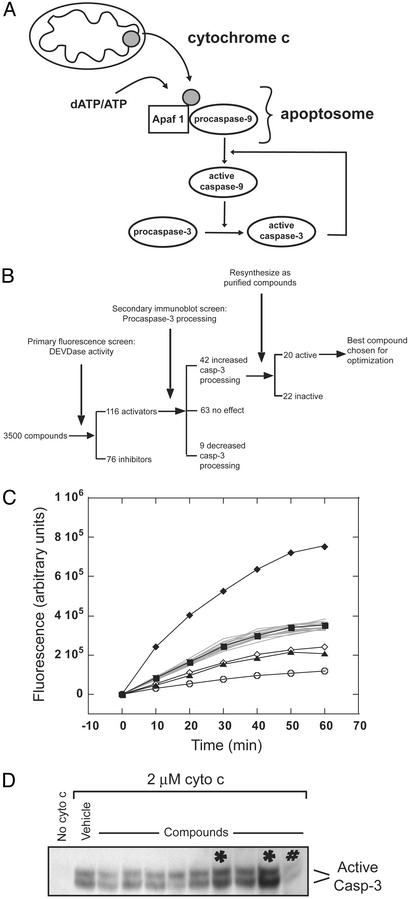
Fig. 1.
Identification of small molecules that modulate apoptosis. (A) Scheme of cyto c-induced caspase activation. (B) Scheme of compound screening process. (C) Sample plate from the primary fluorescence screen for DEVDase activity in cell lysates. Each curve represents an individual compound tested at 1 mM. The control (filled square) is the DMSO vehicle alone. The negative control (open circle) has no added cyto c. Most compounds have no effect on DEVDase activity (gray curves). An activator (filled diamond) and two inhibitors (open diamond and filled triangle) are shown. (D) A sample immunoblot from the secondary screening detecting the large subunit of active caspase-3.*, identified as activators; #, inhibitor.
Many chemotherapeutic drugs activate apoptosis as a function of their anticancer activity, and in many cases the p53 pathway has been identified as the effector of such signals. The presence of wild-type p53 correlates well with a favorable chemotherapeutic response, and DNA-damaging agents have been shown to induce apoptosis via p53 signaling in experimental systems (16, 17). Unfortunately, the p53 gene is the most frequently mutated gene in cancer, being inactivated in 30–70% of clinical tumor samples (16). Thus, in an effort to bypass the p53 pathway, recent drug discovery efforts have targeted factors further downstream in the apoptotic pathway. The majority of these efforts have focused on identifying inhibitors of antiapoptotic members of the Bcl-2 family, Bcl-2 and Bcl-XL, both of which are overexpressed in a number of cancers (18–20). To date, however, no small-molecule apoptosis activators have been identified that function subsequent to cyto c release or directly activate the basic apoptosis machinery.
The fact that chemotherapeutic agents exert their cytotoxic activity by indirectly engaging apoptosis provides a rationale for the identification of agents that can directly activate this process (21). To this end, we adapted a cell-free assay for caspase activation and screened a library of small molecules for compounds that activate apoptosis. We report the identification and characterization of a class of compounds that promote the oligomerization of Apaf-1 into the mature apoptosome. These compounds activate caspases in a cyto c-dependent manner and induce apoptosis in whole cells. When tested against a panel of normal and tumor cell lines, these compounds show strong selectivity for cancer cells, suggesting that direct activation of the basic apoptosis machinery may be a viable mechanism for targeting cancer.
Materials and Methods
Materials. Capture antibody to caspase-3 was purchased from Transduction Laboratories (Lexington, KY); detection antibody to caspase-3 was purchased from Cell Signaling Technology (Beverly, MA); antibody to caspase-9 was purchased from Cell Signaling Technology; capture antibody to Apaf-1 was purchased from Transduction Laboratories; detection antibody was purchased from Alexis Biochemicals (San Diego); and antibody to poly(ADP-ribose) polymerase (PARP) was purchased from Alexis Biochemicals. Bovine heart cyto c was purchased from Sigma, and procaspase-3 was purchased from Biomol (Plymouth Meeting, PA).
Cell-Free Apoptosis Assay. HeLa cell cytoplasmic extracts were prepared according to previously published reports (22). Compounds in DMSO were distributed into 96-well microtiter plates at a final concentration of 1 mM (final DMSO concentration was 1% vol/vol). To each well was added 250 μg of total protein from cytoplasmic extracts in HEB buffer (50 mM Hepes, pH 7.4/50 mM KCl/5 mM EGTA/2 mM MgCl), with 2 mM DTT, 2 μM cyto c, and 0.5 μM DEVD-AFC (Asp-Glu-Val-Asp–7-amino-4-trifluoromethylcoumarin) substrate in a total of 150 μl. Plates were incubated at 37°C, and fluorescence was read in a LJL Biosystems (Sunnyvale, CA) plate reader at 10-min intervals.
Caspase-3 Capture ELISA. cyto c was titrated into lysates with or without compound as detailed above. Briefly, a mouse monoclonal antibody recognizing both procaspase-3 and active caspase-3 (capture antibody) was immobilized onto a MaxiSorp plate (Nunc) at a concentration of 1 μg/ml in 50 mM Na2CO3, pH 9.0, and blocked with 5% milk. To each well was added 25 μl of the caspase activation reaction diluted with 75 μl of SuperBlock (Pierce) and incubated for 30 min. Processing of procaspase-3 was detected by a second rabbit polyclonal antibody that recognizes the large subunit of active caspase-3 (detection antibody, 1:1,000 in SuperBlock). Horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Zymed) and 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Pierce) were used for detection.
Cloning and Purification of Apaf-1 and Procaspase-9. The XL form of Apaf-1 was cloned into pFastbacH, a modified version of pFastbac with a C-terminal six-histidine tag, and expressed in insect cells. Procaspase-9 was cloned into pDest17 and expressed in Escherichia coli. Purification of Apaf-1 and procaspase-9 was performed according to Saleh et al. (10) and Srinivasula et al. (7), respectively.
Detection of Apoptosome Formation by Gel Filtration. Apaf-1 (100 μl of 2 μM) was incubated with 300 μM dATP, with or without cyto c or compounds as indicated, for 30 min at 37°C in HEB and 2 mM DTT. Samples were injected into a Superose 6 gel filtration column (Amersham Biosciences) and separated at 0.4 ml/min in HEB, and 0.8-ml fractions were collected 10 min after injection. To determine the concentration of Apaf-1 in each fraction, 100 μl from each column fraction was taken, to which was added 2-mercaptoethanol to 5 mM and Tween 20 to 0.05% final concentration. Samples were heated to 95°C for 5 min and used for the Apaf-1 capture ELISA. The capture antibody for Apaf-1 is a mouse monoclonal antibody (1 μg/ml in Na2CO3; Transduction Laboratories), and the detection antibody is rabbit polyclonal antibody (diluted 1:1,000; Alexis Biochemicals) recognizing different epitopes. The signal for each sample was normalized to the signal for 1 μg of Apaf-1. For caspase-3 activation assays, 25 μl from each fraction was added to S-100 extracts (125 μg of total protein) in a total of 100 μl, and DTT was added to give 2 mM final concentration. Reactions were incubated at 37°C for 1 h and then used for caspase-3 capture ELISA. Activation was normalized to the signal for activation at 5 μM cyto c.
Transfection Apaf-1 and Apaf-1 siRNA. Apaf-1 was cloned into pcDNA-3 and transfected into SK-OV-3 cells by using FuGENE (Roche) according to the manufacturer's instructions. For interfering RNA experiments, sense and antisense oligonucleotides corresponding to nucleotides 978–998 of Apaf-1, as reported by Lassus et al. (23), were purchased from Dharmacon. Transfection of siRNA into Jurkat cells was performed by using GeneSilencer (Gene Therapy Systems, San Diego) according to the manufacturer's instructions.
Results
Identification of Apoptosis Activators by Cell-Free Assay. To identify compounds that modulate apoptosis, we used a chemical genetics (24) approach to screen a small-molecule library against multiple components of the apoptosis pathway. We adapted an in vitro screen for apoptosis (15, 22, 25) by using cytoplasmic extracts to a 96-well format and screened a library of ≈3,500 compounds (Fig. 1B). Briefly, individual compounds were added to HeLa cell S-100 cytoplasmic extracts at a final concentration of 1 mM, and apoptosis was induced by adding bovine heart cyto c. Activation of caspase-3 was monitored by cleavage of a fluorogenic substrate, DEVD-AFC (DEVDase activity; Fig. 1C).
From this screen, we identified 76 compounds that inhibited and 116 compounds that activated DEVDase activity as compared with the DMSO vehicle (Fig. 1B). Because of their potential utility as anticancer agents, we chose to focus on the characterization and advancement of the small-molecule apoptosis activators. Because many compounds that activated DEVDase activity have intrinsic fluorescence, we used a secondary screen to directly visualize procaspase-3 processing (Fig. 1D). Of the 116 activators, 42 caused increased procaspase-3 processing in the secondary screen and were resynthesized as purified compounds. Rescreening of the 42 purified compounds in both assays identified 20 that activated procaspase-3 processing (data not shown).
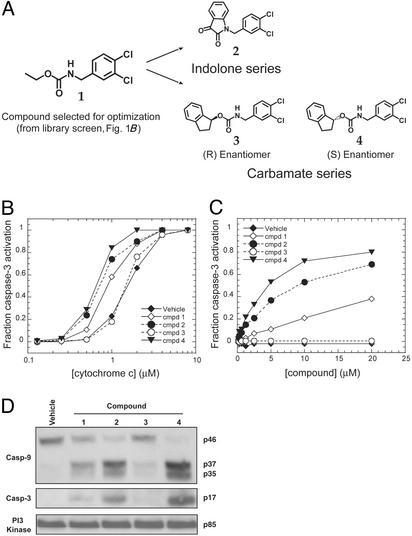
Of the 20 validated activators, compound 1 (Fig. 2A) was the most active and was chosen as a starting point for the synthesis of directed chemical libraries. Preliminary exploration into the structure–activity relationship around compound 1 showed that the positions of the two chlorines in the dichlorobenzyl moiety were critical for activity. Equally important were the nitrogen and carbonyl groups of the carbamate moiety. Libraries were synthesized that maintained these functional groups, and two classes of compounds were identified that strongly activated DEVDase activity, the carbamates and the indolones (Fig. 2 A). Of the carbamate class, compound 3, the enantiomer of compound 4, had no DEVDase activity in the in vitro fluorescence assay (data not shown), strongly suggesting that compound 4 was acting through a specific mechanism.
Fig. 2.
Compounds activate caspases in cell lysates. (A) Chemical structures of the apoptosis activators. (B) Compound activation is cyto c-dependent. cyto c was titrated into S-100 cytoplasmic extracts with vehicle alone or 20 μM compound, and procaspase-3 processing was assayed by capture ELISA. (C) Compounds or vehicle were titrated into S-100 cytoplasmic extracts at a cyto c concentration of 1.25 μM, and procaspase-3 processing was assayed by capture ELISA. (D) Immnunoblot of compound-induced caspase activation. Samples at the 20 μM compound concentration in B were probed with antibodies to caspase-9, the large subunit of active caspase-3, and phosphatidylinositol 3-kinase as a loading control.
Compounds Activate Caspases in a cyto c-Dependent Manner. To determine whether these compounds required cyto c for caspase-3 activation, we titrated cyto c into HeLa cell extract in the presence and absence of compounds, and monitored procaspase-3 processing by capture ELISA. As shown in Fig. 2B, titration of cyto c gave a dose-dependent curve for procaspase-3 processing. In most cases, the addition of compounds shifted the activation curves to the left, such that caspase-3 activation occurred at a lower concentration of cyto c. The enantiomer of compound 4, compound 3, had no effect. Furthermore, all compounds with the exception of compound 3 gave a dosedependent increase in activation when titrated into lysates at a suboptimal concentration of cyto c (1.25 μM, a concentration that produced ≈25% activation in Fig. 2B; also see Fig. 2C). The fact that the compounds require cyto c for activity suggests that they act synergistically with cyto c but cannot replace it. Significantly, the compounds do not affect the processing of procaspase-3 by active caspase-8 added directly to lysates (data not shown), indicating that they act through the cyto c/caspase-9 pathway.
To directly visualize the processing of procaspases induced by the compounds, samples at the highest compound concentration (20 μM) in Fig. 2C were examined by immunoblot for the cleavage of procaspases 9 and 3. As shown in Fig. 2D, the compounds induced the processing of procaspase-9 from the 46-kDa inactive form to give a large subunit with bands corresponding to 37 and 35 kDa. Similarly, the compounds induced the processing of procaspase-3 to give the 17-kDa large subunit of the active form. The vehicle alone showed no detectable processing of either procaspases, and only limited processing was seen with compound 3.
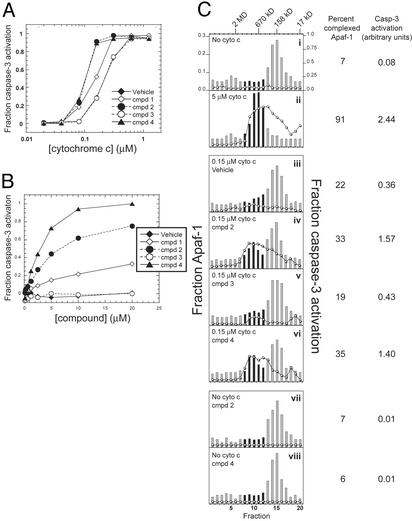
Compounds Are Active in Reconstituted Assay. Previous studies demonstrate that caspase-3 activation can be reconstituted by using four purified proteins: cyto c, Apaf-1, procaspase-9, and procaspase-3 (8, 11). The addition of dATP to all four proteins results in efficient processing of procaspase-3 (10). To determine whether one of the proteins of the reconstituted system was the target of the compounds, we cloned and expressed human Apaf-1 [Apaf-1 XL (12)] and procaspase-9; cyto c and procaspase-3 were purchased from vendors (see Materials and Methods). Addition of all four components in the presence of dATP resulted in the cleavage of procaspase-3, and the titration of cyto c gave a dose-dependent curve for the activation of caspase-3 (Fig. 3A). The addition of 20 μM compounds to the reaction caused the processing of procaspase-3 to occur at a reduced cyto c concentration as seen with the cell-free assay (Fig. 3A). In addition, we obtained dose–response curves for the compound-induced activation by titrating compounds at a cyto c concentration that produced 25% activation (0.15 μM in the purified system; Fig. 3B). These data clearly show that the compounds act on one or more of the components of the reconstituted system. The addition of compounds to either procaspase-9 or procaspase-3 alone did not induce the activation of either protein, and the addition of compounds to the active form of either caspase had no effect on proteolytic activity (data not shown). Together, these data suggest that the compounds require cyto c, Apaf-1, or both for their activity.
Fig. 3.
Compounds promote the oligomerization of Apaf-1. (A) cyto c was titrated into a reconstituted system containing 31 nM procaspase-3, 4 μM procaspase-9, 160 nM Apaf-1, and 300 μM dATP, with or without 20 μM compounds. Procaspase-3 processing was assayed by capture ELISA. (B) Compounds were titrated into reactions containing procaspase-3, procaspase-9, Apaf-1, dATP, and 0.15 μM cyto c, and procaspase-3 processing was assayed by capture ELISA. (C) Apaf-1 was incubated with dATP, with or without cyto c or 20 μM compound as indicated, and then separated by gel filtration. Individual fractions were assayed for relative Apaf-1 concentration by capture ELISA (bar graph) or the ability to activate procaspase-3 processing in lysates (line graph). The percent of Apaf-1 in the apoptosome was determined by dividing the amount of Apaf-1 in fractions 8–12 by the total amount of Apaf-1. The extent of caspase-3 activation (given in arbitrary units) corresponds to the area under the curve for fractions 8–12 in each panel. Black bars represent fractions used for calculations.
Because the binding of dATP or ATP by Apaf-1 is a requirement for the formation of the functional apoptosome (8, 22), we investigated the possibility that the compounds may be functioning as dATP mimetics. Caspase-3 activation was assayed by using the purified system at different concentrations of dATP, keeping the cyto c concentration constant at 5 μM. The processing of procaspase-3 increased as a function of the dATP concentration, but the addition of 20 μM compounds had no effect (data not shown). Thus, the compounds do not relieve or in any way affect the nucleotide requirement by Apaf-1.
Compounds Induce Apaf-1 Oligomerization. Because the compounds had no effect on caspase processing or activity in the absence of cyto c or Apaf-1, we wanted to determine whether they affected apoptosome formation. By gel filtration, Apaf-1 normally runs as a monomer of ≈140 kDa (Fig. 3Ci, bar graph). Incubation with 5 μM cyto c induced the majority of the Apaf-1 (≈90%) to form a large complex of ≈700 kDa (Fig. 3Cii). When the cyto c concentration was reduced to 0.15 μM, only 22% of Apaf-1 remained in the complex (Fig. 3Ciii). However, incubation with 20 μM compound 2 or 4 at the reduced cyto c concentration increased the fraction of Apaf-1 in the apoptosome by ≈1.5-fold to 33% and 35%, respectively (Fig. 3C iv and vi). Significantly, the addition of compound 3 (Fig. 3Cv) had no effect on Apaf-1 oligomerization (Fig. 3Ciii), nor did the addition of either compound 2 or 4 to Apaf-1 in the absence of cyto c (Fig. 3C vii and viii).
To determine whether the Apaf-1 complexes induced by the compounds are competent for caspase activation, we added 25 μl of each gel filtration fraction to cell extracts and assayed for caspase-3 activation. Fractions from samples that had been incubated with 5 μM cyto c strongly activated caspase-3 processing (Fig. 3Cii, line graph), with the maximal caspase activation corresponding to the fractions that contained the apoptosome complex. On the other hand, no fractions from the sample without cyto c activated caspase-3 processing (Fig. 3Ci). There was a small but measurable amount of caspase-3 activation when the cyto c concentration was reduced to 0.15 μM (Fig. 3Ciii). However, the extent of caspase-3 activation at the reduced level of cyto c increased ≈4-fold when either compound 2 or 4 was added to the reaction (from 0.36 arbitrary units for the vehicle alone to 1.57 and 1.4 for compounds 2 and 4, respectively; Fig. 3C iv and vi). Thus, although compounds 2 and 4 increased the oligomerization of Apaf-1 ≈1.5-fold over the vehicle alone, this effect resulted in an ≈4-fold increase in caspase-3 activation (Fig. 3C). These data are consistent with an amplification cascade where a modest effect upstream (in terms of Apaf-1 oligomerization) is magnified at later stages (in terms of caspase-3 activation).
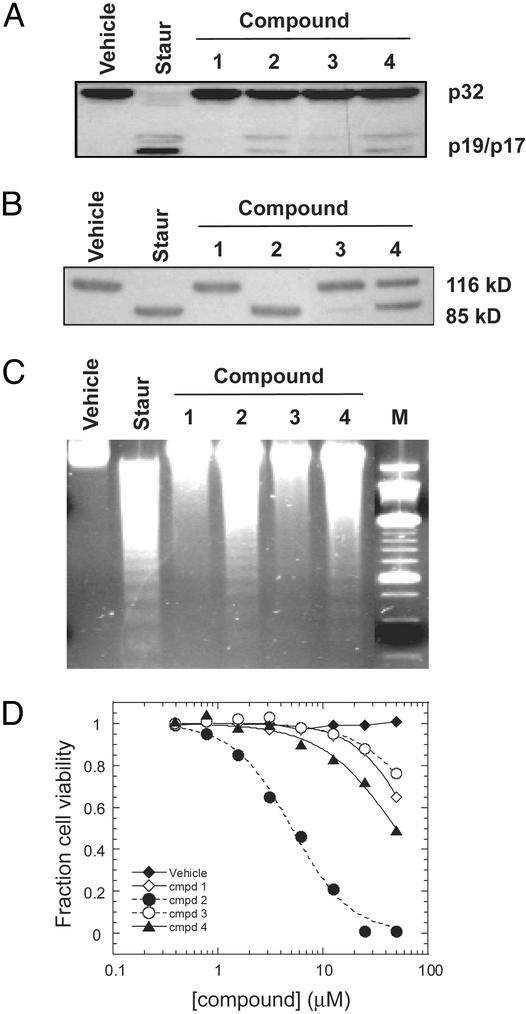
Cytotoxic Activity Results from Induction of Apoptosis. Compounds were then tested for the ability to induce apoptosis in whole cells. Jurkat cells were incubated with either compound, staurosporin (a potent, nonspecific apoptosis inducer), or vehicle and assayed for the hallmarks of apoptosis: these include the processing of procaspase-3; cleavage of poly(ADP-ribose) polymerase (PARP), a substrate for caspases 3 and 7; the fragmentation of chromosomal DNA; and cell death. As shown in Fig. 4, staurosporin potently induced apoptosis, whereas the vehicle alone had no effect. Of the compounds tested, compound 2 was the most potent in cells, strongly inducing caspase-3 activation, PARP cleavage, and DNA fragmentation, and finally killing cells with an IC50 of ≈4 μM. Although compound 4 was one of the most active in biochemical assays, it had relatively poor potency in cell assays compared with compound 2, perhaps because of poor membrane permeability.
Fig. 4.
Compounds induce apoptosis in whole cells. Jurkat cells were incubated with vehicle, 1 μM staurosporin, or 50 μM compound for 6 h and then lysed. Samples were then probed by immunoblot for cleavage of procaspase-3 (A) or PARP (B). (C) Jurkat cells were incubated with vehicle, 1 μM staurosporin, or 50 μM compound for 8 h and then lysed. The DNA was isolated and visualized by ethidium bromide staining. The last lane (M) is a 250-bp DNA molecular weight marker. (D) Jurkat cells were incubated with different concentrations of compound or vehicle for 22 h and assayed for viability by 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide (MTT) test (Sigma) according to the manufacturer's protocol.
Compound 2 Is Selective for Tumor Cells. The cytotoxic activity of compound 2 was tested against a panel of normal and transformed cell lines. Normal cell lines include peripheral blood lymphocytes (PBL) isolated from donated human blood, human mammary epithelial cells (HMEC), human prostate epithelial cells (PREC), human umbilical vein endothelial cells (HUVEC), and nontransformed mammary fibroblasts (MCF-10A). Transformed cell lines include cell lines from leukemia, breast, lung, ovarian, and epidermal cancers (Table 1). In general, we find that normal cell lines were resistant to compound 2-induced apoptosis. Of the five normal cell lines tested, compound 2 had no effect except at the highest concentration (50 μM) when tested against HMEC, PREC, or MCF-10A cells. HUVEC and PBL were slightly more sensitive to compound 2-induced apoptosis with measurable IC50 values of 43 and 50 μM, respectively (Table 1).
Table 1. Cytotoxicity of compound 2 and staurosporin against a panel of human normal and tumor cell lines.
| Cell type | Cell line | p53 status | Staurosporin IC50, μM | Compound 2 IC50, μM |
|---|---|---|---|---|
| Normal | PBL | + | 0.069 | 50 |
| MCF-10A | + | 0.126 | >50 | |
| HMEC | + | 0.109 | >50 | |
| HUVEC | + | 0.063 | 43 | |
| PREC | + | 0.364 | >50 | |
| Leukemia | Jurkat | + | 0.059 | 4 |
| Molt-4 | + | 0.054 | 6 | |
| CCRF-CEM | - | 0.069 | 9 | |
| Breast | BT-549 | - | 0.220 | 20 |
| MDA-MB-453 | + | >1 | >50 | |
| MDA-MB-468 | - | 0.357 | 44 | |
| MCF-7* | + | >1 | >50 | |
| Lung | NCI-H23 | - | 0.085 | 35 |
| Ovarian | SK-OV-3† | - | 0.198 | >50 |
| Epidermal | A431 | - | 0.059 | 40 |
Casp-3-deficient.
Apaf-1-deficient.
Of the cancer cell lines tested in the panel, cell lines of lymphoid origin (CCRF-CEM, MOLT-4, and Jurkat) were quite sensitive to compound 2-induced killing, with IC50 values ranging from 4 to 9 μM (Table 1). Breast, lung, colon, and epidermal cancer cell lines had variable sensitivity to compound 2. Significantly, the sensitivity of a particular cell line to compound 2-induced killing did not correlate with p53 status, consistent with a mechanism of action that centers at the formation of the apoptosome, an event downstream of p53 signaling. In addition, compound 2 had no effect against cell lines that are defective in the caspase-9 pathway [the ovarian cancer cell line SK-OV-3, which is deficient in Apaf-1 activity (26, 27), and the breast cancer cell line MCF-7, which is deficient in caspase-3 activity (28)].
Compound 2 Exerts Broad Cytostatic and Cytotoxic Activity on Tumor Cell Lines. Compounds 2–4 were submitted to the National Cancer Institute for screening in the cancer panel. Consistent with data generated in-house, compound 2 was the most active overall. Compound 2 exerted a cytostatic effect on the majority of cell lines tested, inhibiting cell growth by 50–100% at 10 μM in 40 of 48 cell lines tested (Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). In addition, compound 2 exerted a cytotoxic effect, reducing the cell numbers by 10–50% and 50–100% in four and eight cell lines, respectively, from the initial levels when tested at 10 μM. At 100 μM compound 2 exhibited 100% cytotoxicity in virtually all cell lines, an effect that may be due to nonspecific toxicity.
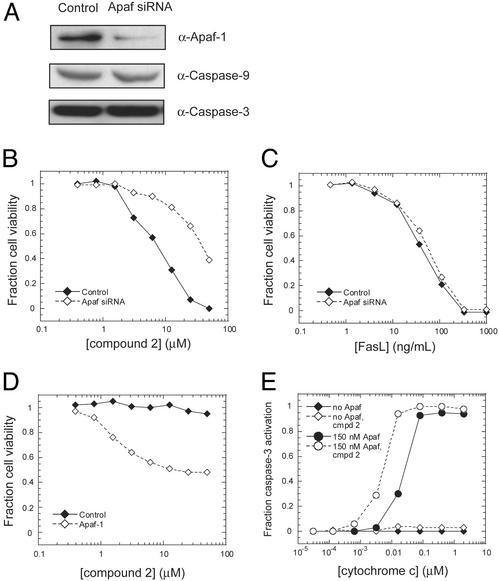
Apaf-1 Is Required for Compound 2 Cytotoxicity. To verify that compound 2 targets the apoptosome as a function of its cytotoxic activity, we used small interfering RNA (siRNA) to silence the expression of Apaf-1 in Jurkat cells. There was a significant decrease in the level of expression of Apaf-1 48 h after transfection of an Apaf-1-specific siRNA, without affecting the expression of caspases in the pathway (Fig. 5A). Reducing the expression of Apaf-1 resulted in resistance of Jurkat cells to compound 2-induced cell killing (Fig. 5B), providing strong evidence that Apaf-1 is required for the activity of compound 2 in cells. This resistance does not result from a nonspecific protective mechanism of the Apaf-1 siRNA because these cells are still sensitive to apoptosis induced by Fas ligand, which proceeds through the caspase-8 pathway (Fig. 5C).
Fig. 5.
Apaf-1 is required for compound 2 activity. (A) Jurkat cells were transfected with 20 nM Apaf-1 siRNA for 48 h, and half the cells were lysed and probed with antibodies to Apaf-1, caspase-9, and caspase-3. The other half of the cells were incubated with varying concentrations of compound 2 (B)or Fas ligand (C) for 24 h and assayed for viability by 3-[4,5-dimethylthiazol-2-yl]2,5-diphenyl tetrazolium bromide (MTT) test. (D) SK-OV-3 cells were transiently transfected with Apaf-1 or vector control for 24 h and then incubated with varying concentrations of compound 2 and assayed for viability. (E) cyto c was titrated into SK-OV-3 cell lysate in the presence of 300 μM dATP, with or without 150 nM purified Apaf-1 and 20 μM compound 2 as indicated.
If silencing Apaf-1 in Jurkat cells makes them resistant to compound 2-induced apoptosis, then we reasoned that ectopic expression of wild-type Apaf-1 in an Apaf-1-deficient cell line should render cells sensitive to compound 2. As mentioned above, the ovarian cancer cell line SK-OV-3 is deficient in Apaf-1 activity and is resistant to compound 2. Transient transfection of Apaf-1 into SK-OV-3 cells rendered them sensitive to compound 2 cytotoxicity as compared with the vector control (Fig. 5D). In several independent experiments, the viability of Apaf-1transfected SK-OV-3 cells exposed to compound 2 plateaued between 40–50%, perhaps reflecting the transfection efficiency. The requirement of Apaf-1 for compound 2 activity was also shown in experiments involving cell lysates from untransfected SK-OV-3 cells. As shown in Fig. 5E, lysates from SK-OV-3 cells do not show normal processing of procaspase-3 in response to dATP and cyto c, either in the presence or absence of compound 2. However, the addition of purified Apaf-1 to the lysates resulted in very strong processing of procaspase-3 in response to cyto c in a dose-dependent manner. The addition of 20 μM compound 2 shifted the activation curve to the left, such that activation occurred at a lower concentration of cyto c, as seen with the HeLa cell lysates. These data clearly show that Apaf-1 is required for the activity of compound 2, both in biochemical assays and whole cells.
Discussion
We report the discovery and characterization of cytotoxic agents that function by directly activating the basic apoptosis machinery. This study highlights the utility of screening against an entire signal transduction pathway, because these compounds could not have been identified by traditional screening against any single target. Biochemical dissection of the apoptosis pathway revealed that these compounds promote the oligomerization of Apaf-1 into the mature apoptosome, resulting in the activation of caspases 9 and 3. The most cell-active compound, compound 2, was shown to exert cytostatic and cytotoxic effects on a number of cancer cell lines while having little activity on the normal cell lines tested. It is intriguing that compounds that act on the basic apoptosis machinery do not induce apoptosis in all cells; this may be due to the differential expression of various proand antiapoptosis factors in normal versus cancer cells and may provide a strategy for selectively targeting cancer cells.
The majority of chemotherapeutic agents in use induce apoptosis indirectly as a by-product of either DNA damage or tubulin blockade, often transmitting signals through the p53 pathway. Thus, compounds that act downstream in the apoptosis pathway and do not require transcription for activity provide an alternative strategy for targeting cancer cells. Such compounds may provide a basis for overcoming chemoresistance by restoring the apoptosis pathway at a point downstream of the molecular defect.
The fact that these compounds do not bypass the requirement for cyto c in in vitro experiments and yet are able to induce apoptosis in whole cells in the absence of additional apoptotic stimuli is perplexing. It is possible that low but unproductive levels of cyto c may be present in the cytosol; if so, the level of cytosolic cyto c may correlate with the sensitivity of a cell line to compound 2-induced apoptosis. Alternatively, although the data from the cell-free assays and assays using purified proteins all suggest a mechanism of action centering on the apoptosome, it is formally possible that an off-target mechanism such as DNA damage may also apply in whole cells. Further studies into the effects of compound 2 on cell cycle, correlation between the expression level of various apoptosis regulators and sensitivity to compound 2, and possible synergies between compound 2 and traditional cytotoxics will provide further evidence of the mechanism of action.
As this report was being submitted, a paper appeared (29) that identified novel regulators of apoptosis by using a smallmolecule probe in cell extracts. The mechanism of action of the small molecule, α-(trichloromethyl)-4-pyridineethanol (PETCM), was not defined. We tested PETCM in our own reconstituted assay by using purified recombinant proteins and showed that it also promoted Apaf-1 oligomerization by a mechanism similar to the compounds reported here (Fig. 7, which is published as supporting information on the PNAS web site). The fact that these structurally unrelated compounds, identified in independent screens, converge on the apoptosome suggests that this complex may be a productive target for small-molecule modulation of apoptosis.
Supplementary Material
Acknowledgments
We thank the Fragment Synthesis Group at Sunesis Pharmaceuticals for making available compounds for screening; A. Braisted, R. Raimundo, A. Virgilio, and N. Ngo for assistance with chemical synthesis; T. O'Brien and M. Flanagan for experimental advice; and G. Salvesen, K. Shokat, B. Druker, D. Walker, and members of the Biology Department at Sunesis Pharmaceuticals for helpful discussions and/or critical evaluation of the manuscript. This work was supported in part by the Cancer Research Fund of Damon Runyon–Walter Winchell Foundation Fellowship DRG-1621 (to J.T.N.) and National Cancer Institute/Small Business Innovation Research Grant CA85141 (to J.A.W.).
Abbreviations: cyto c, cytochrome c; DEVD, Asp-Glu-Val-Asp; MCF, nontransformed mammary fibroblast.
References
- 1.Green, D. R. (2000) Cell 102, 1–4. [DOI] [PubMed] [Google Scholar]
- 2.Salvesen, G. S. & Dixit, V. M. (1997) Cell 91, 443–446. [DOI] [PubMed] [Google Scholar]
- 3.Song, Z. & Steller, H. (1999) Trends Cell Biol. 9, M49–M52. [PubMed] [Google Scholar]
- 4.Wang, X. (2001) Genes Dev. 15, 2922–2933. [PubMed] [Google Scholar]
- 5.Green, D. R. & Reed, J. C. (1998) Science 281, 1309–1312. [DOI] [PubMed] [Google Scholar]
- 6.Zou, H., Henzel, W. J., Liu, X., Lutschg, A. & Wang, X. (1997) Cell 90, 405–413. [DOI] [PubMed] [Google Scholar]
- 7.Srinivasula, S. M., Ahmad, M., Fernandes-Alnemri, T. & Alnemri, E. S. (1998) Mol. Cell 1, 949–957. [DOI] [PubMed] [Google Scholar]
- 8.Li, P., Nijhawan, D., Budihardjo, I., Srinivasula, S. M., Ahmad, M., Alnemri, E. S. & Wang, X. (1997) Cell 91, 479–489. [DOI] [PubMed] [Google Scholar]
- 9.Cain, K., Bratton, S. B., Langlais, C., Walker, G., Brown, D. G., Sun, X. M. & Cohen, G. M. (2000) J. Biol. Chem. 275, 6067–6070. [DOI] [PubMed] [Google Scholar]
- 10.Saleh, A., Srinivasula, S. M., Acharya, S., Fishel, R. & Alnemri, E. S. (1999) J. Biol. Chem. 274, 17941–17945. [DOI] [PubMed] [Google Scholar]
- 11.Zou, H., Li, Y., Liu, X. & Wang, X. (1999) J. Biol. Chem. 274, 11549–11556. [DOI] [PubMed] [Google Scholar]
- 12.Hu, Y., Benedict, M. A., Ding, L. & Nunez, G. (1999) EMBO J. 18, 3586–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang, X. & Wang, X. (2000) J. Biol. Chem. 275, 31199–31203. [DOI] [PubMed] [Google Scholar]
- 14.Acehan, D., Jiang, X., Morgan, D. G., Heuser, J. E., Wang, X. & Akey, C. W. (2002) Mol. Cell 9, 423–432. [DOI] [PubMed] [Google Scholar]
- 15.Slee, E. A., Harte, M. T., Kluck, R. M., Wolf, B. B., Casiano, C. A., Newmeyer, D. D., Wang, H. G., Reed, J. C., Nicholson, D. W., Alnemri, E. S., et al. (1999) J. Cell Biol. 144, 281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, P. & Oliff, A. (2001) Trends Cell Biol. 11, 343–348. [DOI] [PubMed] [Google Scholar]
- 17.Johnstone, R. W., Ruefli, A. A. & Lowe, S. W. (2002) Cell 108, 153–164. [DOI] [PubMed] [Google Scholar]
- 18.Enyedy, I. J., Ling, Y., Nacro, K., Tomita, Y., Wu, X., Cao, Y., Guo, R., Li, B., Zhu, X., Huang, Y., et al. (2001) J. Med. Chem. 44, 4313–4324. [DOI] [PubMed] [Google Scholar]
- 19.Wang, J. L., Liu, D., Zhang, Z. J., Shan, S., Han, X., Srinivasula, S. M., Croce, C. M., Alnemri, E. S. & Huang, Z. (2000) Proc. Natl. Acad. Sci. USA 97, 7124–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Degterev, A., Lugovskoy, A., Cardone, M., Mulley, B., Wagner, G., Mitchison, T. & Yuan, J. (2001) Nat. Cell Biol. 3, 173–182. [DOI] [PubMed] [Google Scholar]
- 21.Lowe, S. W. & Lin, A. W. (2000) Carcinogenesis 21, 485–495. [DOI] [PubMed] [Google Scholar]
- 22.Liu, X., Kim, C. N., Yang, J., Jemmerson, R. & Wang, X. (1996) Cell 86, 147–157. [DOI] [PubMed] [Google Scholar]
- 23.Lassus, P., Opitz-Araya, X. & Lazebnik, Y. (2002) Science 297, 1352–1354. [DOI] [PubMed] [Google Scholar]
- 24.Alaimo, P. J., Shogren-Knaak, M. A. & Shokat, K. M. (2001) Curr. Opin. Chem. Biol. 5, 360–367. [DOI] [PubMed] [Google Scholar]
- 25.Leoni, L. M., Chao, Q., Cottam, H. B., Genini, D., Rosenbach, M., Carrera, C. J., Budihardjo, I., Wang, X. & Carson, D. A. (1998) Proc. Natl. Acad. Sci. USA 95, 9567–9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, J. R., Opipari, A. W., Tan, L., Jiang, Y., Zhang, Y., Tang, H. & Nunez, G. (2002) Cancer Res. 62, 924–931. [PubMed] [Google Scholar]
- 27.Wolf, B. B., Schuler, M., Li, W., Eggers-Sedlet, B., Lee, W., Tailor, P., Fitzgerald, P., Mills, G. B. & Green, D. R. (2001) J. Biol. Chem. 276, 34244–34251. [DOI] [PubMed] [Google Scholar]
- 28.Janicke, R. U., Sprengart, M. L., Wati, M. R. & Porter, A. G. (1998) J. Biol. Chem. 273, 9357–9360. [DOI] [PubMed] [Google Scholar]
- 29.Jiang, X., Kim, H.-E., Shu, H., Zhoa, Y., Zhang, H., Kofron, J., Donnelly, J., Burns, D., Ng, S.-C., Rosenberg, S. & Wang, X. (2003) Science 299, 223–226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.