Abstract
African trypanosomes are ancient eukaryotes that cause lethal disease in humans and cattle. Available drugs are inadequate and the need for new therapeutic targets is great. Trypanosoma brucei and related pathogens differ strikingly from higher eukaryotes in many aspects of nucleic acid structure and metabolism. We find yet another example of this in their unusual DNA topoisomerase IB. Type IB topoisomerases relieve the supercoils that accumulate during DNA and RNA synthesis, and are of considerable importance as the target for antitumor camptothecins. Dozens of type IB topoisomerases sequenced from eukaryotes, bacteria, and pox viruses are all encoded by a single gene that predictably contains a highly conserved DNA binding domain and C-terminal catalytic domain, linked by a nonconserved hydrophilic region. We find that topoisomerase IB in T. brucei is encoded by two genes: one for the DNA-binding domain and a second for the C-terminal catalytic domain. In keeping with this, highly purified fractions of native T. brucei topoisomerase IB catalytic activity contain two proteins, of 90 and 36 kDa. The native enzyme is conventional in its Mg2+-independence, ability to relax positive and negative supercoils, and inhibition by camptothecin. Camptothecin promotes the formation of a covalent complex between 32P-labeled substrate DNA and the small subunit. This unusual structural organization may provide a missing link in the evolution of type IB enzymes, which are thought to have arisen over time from the fusion of two independent domains. It also provides another basis for the design of selectively toxic drug candidates.
The African trypanosome, Trypanosoma brucei, is a flagellated protozoan that causes sleeping sickness in humans. This tsetse fly-transmitted meningoencephalitis is of increasing incidence and is fatal if not treated (1). Closely related organisms cause Chagas' disease and leishmaniasis. Available therapies for sleeping sickness are widely acknowledged to be inadequate: they require multiple parenteral doses for cure, are expensive, toxic, and are losing efficacy against drug-resistant parasites. Trypanosomes and leishmania are ancient eukaryotes whose distinctive features include structurally and metabolically unusual DNAs. In African trypanosomes, the nuclear genome is comprised of 11 chromosome pairs, several additional chromosomes of unknown ploidy, and ≈100 minichromosomes, each containing a single gene that encodes a variable surface protein (2). Surface protein genes can be activated by recombination-based transfer from minichromosomes to transcriptionally active sites in larger chromosomes. Large polycistronic transcripts are trans-spliced to form mRNAs with a 5′-leader sequence and 3′ poly(A) tail. Even more unusual is their mitochondrial DNA, termed kinetoplast or kDNA, which is in the form of a single gigantic network of interlocked relaxed DNA circles (3, 4). Mature mitochondrial messages are created by an editing process that involves systematic insertion or removal of U residues.
DNA topoisomerases are enzymes essential for the orderly synthesis and structural maintenance of nucleic acids (topoisomerases reviewed in refs. 5–7). Type I enzymes, which make a single-strand break in substrate DNA, are further divided into IA and IB classes that are distinct from one another in sequence, structure, catalytic mechanism, substrate specificity, and inhibitor susceptibility. The IB enzymes are monomers whose highly conserved structure contains a central DNA-binding core domain and a C-terminal catalytic domain with its active site tyrosine (Fig. 1A). Type I topoisomerases have been isolated from a number of kinetoplastids, including T. cruzi (8), Crithidia fasciculata (9), and Leishmania donovani (10). In general their activities are consistent with class IB enzymes (11) and purified native proteins are reportedly monomers of 65–79 kDa: smaller than expected for eukaryotes (90–135 kDa) but larger than for vaccinia virus (36 kDa).
Fig. 1.
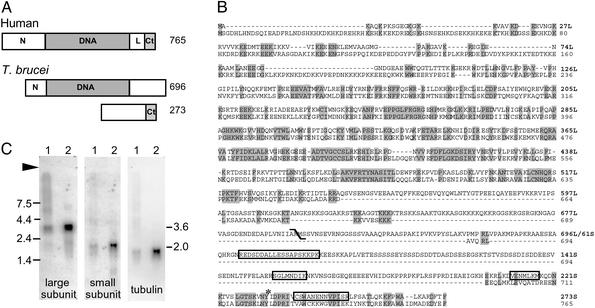
T. brucei topoisomerase IB gene and its expression. (A) Type IB topoisomerases contain four domains within a single protein (91-kDa human protein depicted): a nonconserved hydrophilic N terminus (N), a highly conserved core DNA-binding domain (DNA), a short nonconserved linker (L), and a conserved C-terminal catalytic domain that contains the active site tyrosine (Ct). T. brucei topoisomerase IB contains all of these elements, but they are divided between two proteins as shown. Drawings are to scale, the numbers indicate predicted amino acid residues. (B) Predicted protein sequences for T. brucei (first line) and human (second line) topoisomerase IB. The diagonal is at the boundary of T. brucei large and small subunits; shaded residues are identical; boxed residues were obtained from peptide microsequencing; asterisks indicate active site tyrosines; large and small subunits are numbered independently. (C) Northern blot of 12.6μg total RNA from insect forms (lane 1) and 1.6 μg mRNA from bloodstream forms (lane 2) was probed as indicated. An arrow indicates the origin, size markers are given on the left, and calculated sizes for large and small subunit mRNAs are given on the right.
Type IB topoisomerases are the sole molecular target for the camptothecins, an important class of antitumor agents (12). These compounds are uncompetitive inhibitors that trap the enzyme in an immobile ternary complex with drug and substrate DNA. When disrupted (e.g., by SDS or alkali) such complexes yield a denatured enzyme covalently linked, via the active site tyrosine, to the 3′ end of the enzyme-induced break in substrate DNA. In previous work with African trypanosomes we found that camptothecin promotes the formation of covalent protein–DNA complexes with both nuclear and mitochondrial DNA, and inhibits DNA biosynthesis (13). Furthermore, it is cytotoxic to three pathogenic kinetoplastids: T. brucei, T. cruzi, and L. donovani. Structure-activity studies revealed that trypanosomes and mammalian cells differ in their susceptibility to camptothecin analogs (14). We were intrigued by the possibility that these differences might reflect structural dissimilarities in the host and parasite enzymes. To explore this possibility we set out several years ago to obtain the gene sequence for T. brucei topoisomerase I.
In this report we describe a unique topoisomerase from trypanosomes that is IB-like in its sequence, catalytic properties, and susceptibility to camptothecin, but which differs in that it is a heterodimer whose subunits are encoded by different genes. Available genome databases and very recent findings from another lab suggest that this unusual structure is shared by other kinetoplastid pathogens, including T. cruzi and Leishmania species (15).
Materials and Methods
Growth and Isolation of Cells. Studies were conducted with cells in exponential growth. Insect form T. b. brucei (Mitat 1.2, strain 427–60) were grown at 27°C in SDM-79 medium with 15% FBS (16); and bloodstream forms (Mitat 1.2, strain 427) at 37°C in phenol red-free Iscove's modified Dulbecco's medium (Mediatech) supplemented as described (17).
Large Subunit Sequence. A 726-bp HindIII–AvrII fragment of human topoisomerase I recognized a 5-kb band in EcoRI-restricted T. brucei genomic DNA. Gel-isolated DNA of this size was ligated into pBKCMV and electroporated into DH10B E. coli. Clones were selected and both strands of a 2.7 kb region encompassing the entire ORF were sequenced (very few trypanosome genes have introns, ref. 2). The stop codon was confirmed in an RT-PCR-amplified segment sequenced with 5′ and 3′ primers (coding nucleotides 1372–1390 and 3′ untranslated nucleotides +39 to +49, respectively). Sequences were assimilated in SEQMAN 3.57 (DNASTAR, Madison, WI) and the predicted protein sequence was analyzed by NCBI Conserved Domain Search with RPS-BLAST 2.2.5 (www.ncbi.nlm.nih.gov:80/blast/), by PSORTII (http://psort.ims.u-tokyo.ac.jp/form2.html), and by PROTPARAM (http://us.expasy.org/tools/protparam.html). This gene has been assigned to chromosome 4 (GeneDB; www.genedb.org).
Large Subunit Recombinant Protein and Antibodies. A fragment encoding amino acids 108 to 696 was inserted into BamHI–SacI of PET 28. Transformed BL21 DE3 E. coli were treated with IPTG, pelleted, and lysed in 6 M guanidine·HCl. The N-terminal His-tagged recombinant protein was isolated by cobalt matrix chromatography (Talon, CLONTECH) and SDS/PAGE. The prominent band at 75 kDa was excised, emulsified, and used to generate polyclonal antibodies in mice.
Purification of Topoisomerase I Activity from Trypanosomes. Solutions and pH determinations were made at room temperature; all subsequent steps were at 4°C. DTT and protease inhibitors were added just before use. Unless indicated otherwise all buffers contained 1 mM phenylmethanesulfonyl fluoride, 10 μg/ml E–64, 100 μg/ml Pefabloc, 0.01× protease inhibitor cocktail (Roche). Solutions included lysis buffer (10 mM Tris·HCl, pH 7.5/1 mM EDTA/0.1 mM EGTA/5 mM DTT/10 mM 2-mercaptoethanol/0.2% Nonidet P-40/0.5 mM spermidine-HCl/0.1× protease inhibitor cocktail); Buffer A (50 mM Tris·HCl, pH 7.5/200 mM NaCl/20% glycerol/1 mM EDTA/0.5 mM DTT/10 mM 2-mercaptoethanol); Buffer B (50 mM Tris·HCl, pH 7.5/20% glycerol/0.1 mM EDTA/0.5 mM DTT); Buffer C (50 mM Tris·HCl, pH 7.5/15% glycerol/0.1 mM EDTA/0.5 mM DTT/0.5 mM phenylmethanesulfonyl fluoride); and Buffer D (75 mM potassium phosphate, pH 7.0/10% glycerol/0.5 mM DT T/0.5 mM phenylmethanesulfonyl fluoride).
Insect form trypanosomes (3.5 × 107 per ml in 12 liters) were harvested and stored at –70°C. Pellets were thawed in 30 ml of lysis buffer, disrupted with six strokes (Dounce A homogenizer), incubated on ice for 15 min, and centrifuged (9,500 rpm, Sorvall GS3, 30 min). The ``nuclear'' pellet was resuspended (40 ml Buffer A, six strokes), centrifuged (11,500 rpm, Sorvall GS3, 30 min) and applied to DEAE Sepharose (Pharmacia Fast Flow; 60-ml bed) equilibrated in Buffer A. The first 15 ml of eluate was discarded; remaining flow-through and 80 ml of Buffer A wash were collected, pooled, and added to a 50-ml slurry of phosphocellulose (Whatman P11) in Buffer A. The slurry was applied to phosphocellulose (75-ml bed; 3 ml/min) equilibrated in Buffer A. The column was washed with 160 ml of 250 mM NaCl in Buffer B, developed with 400 ml each 400 mM then 1 M NaCl in Buffer B. Fractions containing peak activity were pooled, made 1.5 M in NaCl, and applied to phenylsepharose (Pharmacia Fast Flow low substitution; 25-ml bed) equilibrated with 1.5 M NaCl in Buffer C. Flow-through fractions containing activity were pooled, dialyzed (50 mM Tris·HCl, pH 7.5/150 mM NaCl/10% glycerol/0.1 mM EDTA/0.5 mM DTT), made 1 M in NaCl, and applied to HA Ultragel (BioSepta; 7-ml bed) equilibrated in 50 mM Tris-HCl, pH 7.5/1 M NaCl/10% glycerol/0.5 mM DTT. The column was washed with 75 mM potassium phosphate in Buffer D (30 ml) and developed by successive addition of 15 ml 300, 600, 800 mM potassium phosphate in Buffer D. Peak fractions were concentrated 10-fold in Millipore filtration devices and loaded on Superdex 200 HR10/30 FPLC (Amersham Pharmacia; 0.25 ml/min) equilibrated and run with 50 mM Tris-HCl, pH 7.5/100 mM KCl/10% glycerol/3 mM DTT/0.1 mM phenylmethanesulfonyl fluoride. Elution volumes of sizing standards (BioRad) were determined immediately before (and sometimes also after) the unknown. Active fractions were concentrated, adjusted to 50% glycerol, and stored at –20°C.
Topoisomerase Activity. Samples were serially diluted 10-fold into otherwise complete reaction mixture (50 mM Na-Hepes, pH 7.5/100 mM KCl/10 mM MgCl2/0.5 mM DTT/0.5 mM Na2EDTA/20 μg/ml acetylated BSA/15 μg/ml supercoiled pBKCMV DNA, final volume 20 μl), incubated for 15 min at 37°C, quenched, separated by gel electrophoresis, stained with ethidium bromide, and visualized by UV transillumination (18). Under these conditions one unit of activity is the amount of enzyme required to relax one-half the supercoiled substrate. Positively supercoiled pBKCMV was prepared by topoisomerase I relaxation at 0°C (19).
Covalent Complex Between Small Subunit and Radiolabeled DNA. Reactions were initiated with 1,000 units of T. brucei (HA Ultragel peak fraction) or calf thymus topoisomerase I added to assay buffer containing 4 ng of 32P-labeled DNA (random prime labeling of a 2.5-kb BsgI–Psp1460I fragment containing the large subunit gene), and either 180 μM camptothecin (Aldrich) or 0.6% vol/vol dimethyl sulfoxide control (50 μl final volume, 15 min, 37°C). Reactions were quenched with KOH, neutralized with HCl then Na-Hepes, dialyzed (50 mM Na-Hepes, pH 7.6/10 mM MgCl2; 30 min, room temperature), treated with 10 units DNase I (bovine pancreas FPLC pure, Amersham Pharmacia; 1h,37°C), and quenched with EDTA. Proteins were precipitated and separated by SDS/PAGE. The gel was dried and exposed to a phosphorimaging screen (Fuji), and band intensities were quantitated (IMAGE GAUGE v3.45).
Small Subunit Protein Microsequence. Proteins in Superdex peak fraction were separated by SDS/PAGE, stained with Coomassie blue, and bands at 37 and 35 kDa (63 ng or 1.8 pmol estimated protein) were excised, equilibrated in 50% acetonitrile, and stored at –20°C. Sequence analysis was performed at the Harvard Microchemistry Facility by tandem mass spectrometry on a Finnigan LCQ Quadrupole Ion Trap Mass Spectrometer.
Sequence of Small Subunit Gene. We used CSWANENNVPISR from the 35-kDa protein to identify a clone in the EMBL sheared genomic DNA database (www.ebi.ac.uk/parasites/parasite-genome.html) whose 5′ end contained a 267-bp region encoding two of the peptide sequences (DVENMLK, CSWANENNVPISR), a topoisomerase IB active site motif, and a stop codon. The entire coding region was amplified from cDNA (ProSTAR kit, Stratagene) by using 5′ miniexon (CGCTATTTATTAGAACAGTTTCTGTACTATATTG) and 3′ coding nucleotides 791–819 as primers. A single product of 857 bp was isolated, sequenced, and used to identify a 639-bp fragment in a sheared DNA library (www.tigr.org/tdb/parasites/), containing the 5′-most 143 bp of the coding region and 496 bp upstream of this. To confirm the coding region, both strands were sequenced from the single 1,436-bp PCR product amplified from genomic DNA by using 5′-primer (bp –356 to –331) and 3′ primer (bp +230 to +258). GeneDB assigns this gene to chromosome 9.
Northern Blots. Total RNA (Trizol, Invitrogen) or mRNA [Poly(A) Quick, Stratagene] were separated by gel electrophoresis, transferred, and probed with either a 1,164-bp SalI–SalI fragment of the large subunit, bp 1–819 (full length) small subunit gene, or bp 1–650 of T. brucei α-tubulin, all radiolabeled by the random prime method.
Results
Topoisomerase IB Gene Sequence Appears Incomplete. An ORF was identified in genomic DNA and both ends were confirmed in cDNA. The 2,088-bp gene encodes a 79-kDa predicted protein (Fig. 1 A and B), comparable to the size of native topoisomerase I reported for kinetoplastids (8–10). It has a pI of 9.47 and multiple bipartite nuclear localization signals (starting at amino acids 27, 165, 626, 636, and 643). The protein sequence aligns with high probability to topoisomerase IB domains from several databases (smart00435, 8e–111; pfam1028, 1e–70; pfam02919, 3e–70; and COG3569, 6e–33). The highly conserved DNA-binding domain is flanked by two nonconserved hydrophilic regions, similar to those seen for other type IB sequences (Fig. 1 A). However, the T. brucei gene appears prematurely terminated at its 3′ end: none of the characteristic catalytic domain motifs appear within the ORF or 650 bp of its 3′ flanking region.
A single 3.6-kb transcript appears in Northern blots (Fig. 1C Left), of similar intensity in insect and bloodstream forms (when adjusted for loading differences), suggesting there is no developmental regulation at the level of transcription. Efforts to obtain catalytically active recombinant enzyme were not successful, from bacteria, yeast, or the baculovirus–insect cell system.
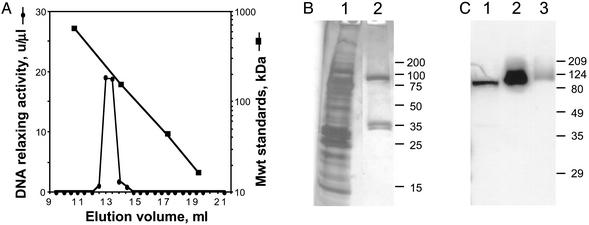
Purification of Catalytically Active Native Topoisomerase IB. To identify the catalytic moiety in enzymatically active protein, topoisomerase IB was isolated from T. brucei. Plasmid DNA relaxing activity was enriched >450-fold by a series of conventional chromatography steps (Table 1). Fluorescence microscopy of the ``nuclear pellet'' showed both intact nuclei and compact mitochondrial kinetoplast DNA, indicating the activity may originate from either or both compartments. Four large-scale preps (each starting with ≈1011 cells) all provided comparable purification and yield, even when phenylsepharose and hydroxyapatite steps were reversed. The final gel filtration step in all four preps also yielded a similar finding: activity elutes at a volume expected for a 207-kDa protein (Fig. 2A), which is substantially larger than the 80 kDa predicted by the gene and difficult to reconcile on the basis of Stokes radius considerations (20). Importantly, SDS/PAGE analysis of the peak Superdex fraction reveals three bands, at 90, 37, and 35 kDa (Fig. 2B, lane 2; sizes assigned on the basis of many experiments). To determine the relationship of these to the topoisomerase gene, polyclonal antibodies against recombinant large subunit were used in an immunoblot analysis. A single protein of 90 kDa is recognized in lysate, Superdex load, and Superdex peak fraction (Fig. 2C, lanes 1–3, respectively). This finding indicates that the large protein in the Superdex peak is encoded by the topoisomerase I gene, and that the smaller proteins are not simply degradation products. Collectively these findings suggest that the native T. brucei topoisomerase IB may be comprised of dissimilar subunits, and that one or both of the small proteins may contain the missing catalytic domain.
Table 1. Purification of native T. brucei topoisomerase IB activity.
| Topoisomerase IB
|
|||||
|---|---|---|---|---|---|
| Fraction | Total protein, mg | Total activity, 106 units | Specific activity, 103 units/mg pro | Purification, -fold | Yield, % |
| Lysate | 1,100 | 7.2 | 6.5 | 1.0 | 100 |
| Nuclear extract | 420 | 4.3 | 10 | 1.5 | 60 |
| DEAE | 340 | 8.4 | 25 | 3.8 | 120 |
| Phosphocellulose | 21 | 1.4 | 67 | 10 | 19 |
| Phenylsepharose | 11 | 1.1 | 100 | 15 | 15 |
| Hydroxyapatite | 1.8 | 0.72 | 400 | 62 | 10 |
| Superdex | 0.06 | 0.18 | 3,000 | 460 | 2.5 |
Fig. 2.
T. brucei topoisomerase IB is heteromultimeric. (A) Proteins in hydroxyapatite fractions were separated by Superdex gel exclusion chromatography and assayed for DNA relaxing activity. In multiple experiments with different flow rates, salt conditions, and sample volumes, peak activity reproducibly eluted at 207 ± 27 kDa (mean ± SD; n = 9), well ahead of most contaminating proteins. (B) Proteins in the Superdex load (lane 1) and peak fraction (lane 2) were separated by SDS/PAGE and silver stained (13% and 2.6% of the total volumes, respectively, were applied to the gel). (C) Proteins in lysate (lane 1), hydroxyapatite fraction (lane 2), and Superdex fraction (lane 3) were immunoblotted with mouse anti-large subunit polyclonal serum. Apparent size changes may reflect different buffer conditions in samples. Size markers are given on the right.
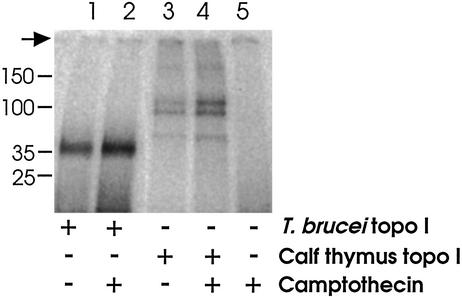
Validation of Small Subunit As Catalytic. To identify the native T. brucei topoisomerase IB catalytic moiety, enzyme was combined with 32P-labeled DNA in the presence or absence of camptothecin. The mixtures were treated with KOH to capture covalent protein–DNA complexes, then neutralized, digested with protease-free DNase and separated by SDS/PAGE. This procedure yields a single broad band migrating at ≈35 kDa (Fig. 3, lane 1), thus localizing the catalytic domain to the small protein(s). Camptothecin enhances the formation of covalent complexes (Fig. 3, lane 2), consistent with our previous studies of cleavable complex formation in trypanosomes (13). Under these conditions, enhancement of cleavable complex formation by camptothecin is comparable for T. brucei and human enzymes (1.7-fold and 2.1-fold, respectively). Although the 37 and 35 kDa proteins in this experiment are discernable by Coomassie staining (data not shown), they are not resolved on the autoradiograph. To determine whether just one or both is labeled, several regions were excised from the gel and counted. Counts in the small bands were 3 to 4 times those of background (or of the 90-kDa band), indicating either small protein can provide catalytic function.
Fig. 3.
Radiolabeling the catalytic domain. Enzyme samples were incubated with 32P-labeled DNA and either camptothecin or dimethyl sulfoxide, denatured, digested with DNase I, separated by SDS/PAGE, and visualized by autoradiography. T. brucei enzyme yields a single broad band at 35 kDa (lane 1), enhanced by camptothecin (lane 2). The calf thymus topoisomerase IB control yields multiple bands [intact enzyme at 90 kDa and two smaller enzymatically active breakdown products (18); lane 3], all enhanced by camptothecin (lane 4). In a mock reaction, the dense signal from radiolabeled DNA (not shown) is nearly eliminated by DNase I treatment (lane 5). Size markers are given at left, the arrow indicates origin.
Sequence for Small Subunit Gene. The entire yield of 35 kDa protein in the Superdex peak from one enzyme prep was submitted for de novo sequence analysis of tryptic peptides by liquid chromatography/tandem mass spectrometry. Four sequences were obtained: (i) REDSDDALLESSAPSK, (ii) SGLMNDIK, (iii) DVENMLK, and (iv) CSWANENNVPISR. Of considerable interest, peptide i was also a prominent product of the 37-kDa protein (peptides ii–iv were not sought), and serine 4 in peptide i (underlined above) was phosphorylated in the 35-kDa, but not the 37-kDa, protein. Remaining sequence for the single-copy small subunit gene was obtained from genomic DNA and confirmed from cDNA. The 819-bp ORF encodes a 30-kDa predicted protein (Fig. 1 A and B), consistent with the ≈36-kDa native proteins, and containing all four tryptic peptide sequences (Fig. 1B, boxes). In contrast to the large subunit, this protein has a low pI of 5.21 and no nuclear localization signals. The C-terminal 69 aa contain residues diagnostic of a topoisomerase IB catalytic domain (smart00435), including the active site Tyr233S (Fig. 1B, asterisk). The long N-terminal region contains no recognized functional domains, but Ser70S (phosphorylated in the 35-kDa protein) lies within a 100-aa region (50S–150S, Fig. 1B) in which intense phosphorylation is predicted. Northern blot analysis reveals a single band at 2.0 kb, present in both lifecycle forms (Fig. 1C Center).
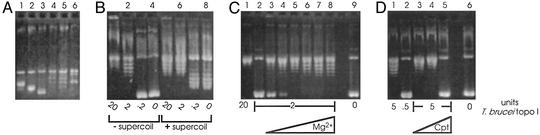
Characteristics of Native T. brucei Topoisomerase IB Activity. Hydroxyapatite peak fractions were used to characterize catalytic properties. An isolated partially relaxed single topoisomer of pBKCMV (Fig. 4A, lane 3) was further relaxed in steps of one by the T. brucei enzyme, in keeping with its type I classification (Fig. 4A, lane 4). Controls demonstrate relaxation in steps of one with calf thymus topoisomerase IB (Fig. 4A, lane 5), and in steps of two by a type II enzyme (Fig. 4A, lane 6). In all other characteristics examined, the T. brucei topoisomerase behaves like a IB enzyme. It relaxes positive and negative supercoils (Fig. 4B, lanes 1–3 and 5–7, respectively). Although detectable in its absence (Fig. 4C, lanes 1 and 2), Mg2+ enhances activity (Fig. 4C, lanes 3–8). Camptothecin inhibits relaxing activity (Fig. 4D) at concentrations comparable to those that affect calf thymus topoisomerase I (ref. 21 and data not shown). Although catalytic activity of T. brucei enzyme in the presence of 10 mM MgCl2 is unaffected by KCl up to 150 mM, it is 50% inhibited by 200 mM KCl, and diminishes rapidly at higher salt concentrations (data not shown). This differs from the behavior of monomeric human topoisomerase IB (whose activity peaks at 200 mM KCl), but mimics that of catalytically active complexes reconstituted from separately expressed human core and catalytic domains (22).
Fig. 4.
Characterization of T. brucei topoisomerase IB catalytic activity. Assays monitored the conversion of supercoiled pBKCMV into more relaxed and slower migrating isomers. Numbers immediately below panels indicate enzyme units. (A) Relaxation in steps of one. Individual topoisomers were gel-isolated (lanes 1–3) and the fastest migrating isomer (lane 3) was used in reactions with T. brucei topoisomerase (2 units; lane 4), calf thymus type IB topoisomerase (2 units; lane 5), or phage T4 type II topoisomerase (5 units; lane 6). (B) Relaxation of negative and positive supercoils. Serial dilutions of T. brucei topoisomerase IB were reacted with native pBKCMV (lanes 1–3) or slightly positively supercoiled pBKCMV (lanes 5–7). No enzyme, lanes 4 and 8. (C)Mg2+-independent relaxing activity. T. brucei topoisomerase IB was assayed in the presence of the following mM concentrations of MgCl2: 0 (lanes 1, 2), 0.5 (lane 3), 1 (lane 4), 2.5 (lane 5), 5 (lane 6), 10 (lane 7), or 20 (lane 8). No enzyme, lane 9. (D) Camptothecin inhibits catalytic activity. T. brucei topoisomerase IB activity was assayed in the presence of the followingμM concentrations of camptothecin: 0 (lanes 1 and 2), 25 (lane 3), 50 (lane 4), and 100 (lane 5). No enzyme, lane 6; all reactions contained 0.5% dimethyl sulfoxide.
Discussion
A wealth of information is available on the type IB topoisomerases (5–7). In particular, their monomeric nature is well documented in dozens of gene sequences (from higher eukaryotes, bacteria, and viruses) and by the characterization of purified native enzyme from numerous organisms. However, we find multiple lines of evidence to indicate that the type IB topoisomerase activity in T. brucei, though conventional in terms of catalytic activity and inhibitor susceptibility (Fig. 4), is highly unusual in its structure. Native proteins and their genes each provide unambiguous evidence that the T. brucei enzyme is a multimer comprised of two dissimilar subunits. The 90- and ≈36-kDa proteins seen in purified fractions (Fig. 2B, lane 2) are translated from distinct mRNAs (Fig. 1C), obtained in turn from independent single-copy genes (Fig. 1 A and B) that are located on separate chromosomes. The composite sequence of the two genes is clearly homologous with conventional type IB enzymes (Fig. 1B), with a core DNA-binding domain predicted in the 90-kDa subunit and a catalytic domain with active site tyrosine predicted in the 36-kDa subunit. This multimeric structure provides a satisfying explanation for the L. donovani topoisomerase-like gene (23), which encodes a large subunit homolog and for which an active site serine was proposed (24). BLAST search of the TIGR databases (which have become available since this work began) now indicates that T. cruzi and several Leishmania species have separate genes for their topoisomerase IB core and catalytic domains (data not shown), and a very recent report shows relaxation of negative supercoils obtained when both subunits of L. donovani topoisomerase IB are coexpressed in yeast (15). These findings suggest that the multisubunit structure is a common feature in kinetoplastids, and indicate that the small subunit was overlooked in early studies of native type IB activity.
The T. brucei topoisomerase IB sequences have a few notable features. First, each subunit contributes essential and highly conserved catalytic residues that characterize both the type IB topoisomerases and the tyrosine recombinases (T. brucei Arg377L, Lys415L, Arg471L, His514L and Tyr233S; ref. 25). Like other IBs, the T. brucei topoisomerase has a Lys at 468L, not the His seen in recombinases (26). Also conserved are some (Gly252L, Asp416L, Asn232S), but not all (e.g., Asp535L and Val239S), of the residues important for camptothecin sensitivity (27). Perhaps the latter contribute to the observed differences in susceptibility to camptothecin analogs (14), which originally prompted these studies. Second, the overall charge difference (pIs of 9.47 and 5.21 for the large and small subunits, respectively) in conjunction with unusual salt sensitivity of the T. brucei enzyme suggests that ionic interactions may be important in holding the subunits together. Third, in terms of subcellular localization, previous studies in kinetoplastids predict that type IB activity should be found in the nucleus and the mitochondrion (9, 13). In higher eukaryotes, such dual distribution of a single protein may be accomplished by the presence of both nuclear and mitochondrial targeting sequences (28). Multiple classic bipartate nuclear localization signals are found in the large but not the small subunit sequence, so the complex likely assembles in the cytosol before nuclear importation. Neither subunit contains a detectable mitochondrial targeting sequence, although in kinetoplastids these may be short and sometimes not recognizable (29). Finally, the purified activity in all four preps yields two small subunits on SDS/PAGE, both of which form covalent complexes with DNA. All available evidence indicates there is only one gene and one transcript for the small subunit. The 35-kDa protein may be a proteolysis product, though protease inhibitors were continuously present during purification. More likely is that the 35- and 37-kDa proteins vary in degree of phosphorylation. Extensive phosphorylation is predicted in the N-terminal nonconserved region, phosphorylation would account for slower than expected migration in SDS/PAGE, and a difference in phosphorylation was evident in peptide i from the two proteins. Trypanosomes have vigorous phosphatase activity (30) and phosphatase inhibitors were not used during purification, hence phosphorylation differences may be artifactual. Alternatively, they may reflect a mechanism for intracellular regulation, in keeping with topoisomerases in other systems (5).
Why in T. brucei and related kinetoplastids are the DNA-binding core domain and the catalytic domain on separate genes? These may have arisen over time from fission of a single progenitor gene; alternatively, they may represent the persistence of discrete genes. There is some phylogenetic evidence to support the latter. As noted above, the tyrosine recombinases and type IB topoisomerases share mechanistic, structural and (to a lesser extent) sequence similarities (25). This has led to the concept that these two enzyme classes share a common ancestral catalytic domain that fused over time with different N-terminal domains (31). In this scenario, kinetoplastids may provide a missing link in evolution between the hypothesized ancient independent catalytic domain and the contemporary fused constructs.
Regardless of phylogenetic origin the persistence of separate subunits suggests they impart some advantage to kinetoplastids. This is unlikely to be in terms of catalytic efficiency. Separation of the core and catalytic domains of human topoisomerase IB by proteolytic cleavage within the unconserved linker does not markedly affect catalysis (32). Separate domains may permit the subunits to function independently, perhaps in conjunction with other partners (a possibility that we should explore with RNA interference studies). An obvious option provided by separate subunits is the possibility of exchanging or replacing domains. Thus for example, enzymatic activity could be restored by replacing a damaged large subunit with a new one. Such a mechanism would be particularly useful in reversing (or preventing) the protein-bound breaks in DNA that occur in vivo when topoisomerase activity is interrupted (12).
A curious and highly reproducible finding in this work is that the native enzyme migrates through a gel filtration column as if it has a mass of 207 kDa (Fig. 2 A). This is substantially larger than the combined sizes seen on SDS/PAGE analysis (90 and 36 kDa; Fig. 2B, lane 2), and may be the result of a nonglobular shape (20). Alternatively, based on the sequence-predicted molecular masses of 80 and 30 kDa, the catalytically active entity may be an A2B2 tetramer. (Additional variations on this could arise from small subunit phosphorylation differences.) Although confirmation of this finding will have to await the availability of recombinant proteins, it is interesting to speculate on the possible consequences or advantages of a tetrameric arrangement. The observed relaxation in steps of one (Fig. 4A) would indicate that close proximity of two single strand breaking units does not lead to type II-like activity. However, the availability of two complete enzymatic units would certainly facilitate binding at helical crossover points, a propensity that may underpin the IB enzymes' clear preference for a supercoiled substrate (6). A final intriguing possibility afforded by an A2B2 structure is the opportunity for domain swapping to support recombinase activity. As noted above, there are close similarities between these classes of enzymes, indeed most recombinases have topoisomerase activity and conversely type IB enzymes may resolve Holiday structures (25, 33).
The multimeric structure of topoisomerase IB in African trypanosomes and related kinetoplastids provides yet another vivid example of the unusual nucleic acid metabolism in these pathogens. The protein–protein interactions essential for the function of this enzyme, in addition to its sequence-mediated structural differences, offer a new and selective target for the design of much-needed antiparasitic chemotherapy.
Acknowledgments
We thank Sara Melville for providing P1 filters, Bill Lane for peptide sequencing, Ken Kreuzer for T4 topo II, and Scott Kaufmann and Jonathan Pevsner for helpful discussions. The generosity of Paul Englund and his laboratory in providing reagents and pleasant scientific discussion are gratefully acknowledged. We thank Barbara Sollner-Webb, Jane Scocca, and Tom Kulikowicz for critically reading the manuscript. This work was supported by National Institutes of Health Grant AI 28855, the Burroughs Wellcome Fund, the Swiss National Research Foundation, the Ciba Jubilaeums Foundation, and the Theodor Engelmann Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY308070 and AY308071).
References
- 1.Moore, A. C. (2002) N. Engl. J. Med. 346, 2069–2076. [DOI] [PubMed] [Google Scholar]
- 2.El-Sayed, N. M., Hegde, P., Quackenbush, J., Melville, S. E. & Donelson, J. E. (2000) Int. J. Parasitol. 30, 329–345. [DOI] [PubMed] [Google Scholar]
- 3.Shapiro, T. A. & Englund, P. T. (1995) Annu. Rev. Microbiol. 49, 117–143. [DOI] [PubMed] [Google Scholar]
- 4.Morris, J. C., Drew, M. E., Klingbeil, M. M., Motyka, S. A., Saxowsky, T. T., Wang, Z. & Englund, P. T. (2001) Int. J. Parasitol. 31, 453–458. [DOI] [PubMed] [Google Scholar]
- 5.Wang, J. C. (1996) Annu. Rev. Biochem. 65, 635–692. [DOI] [PubMed] [Google Scholar]
- 6.Champoux, J. J. (2001) Annu. Rev. Biochem. 70, 369–413. [DOI] [PubMed] [Google Scholar]
- 7.Wang, J. C. (2002) Nat. Rev. Mol. Cell Biol. 3, 430–440. [DOI] [PubMed] [Google Scholar]
- 8.Riou, G. F., Gabillot, M., Douc-Rasy, S., Kayser, A. & Barrois, M. (1983) Eur. J. Biochem. 134, 479–484. [DOI] [PubMed] [Google Scholar]
- 9.Melendy, T. & Ray, D. S. (1987) Mol. Biochem. Parasitol. 24, 215–225. [DOI] [PubMed] [Google Scholar]
- 10.Chakraborty, A. K., Gupta, A. & Majumder, H. K. (1993) Indian J. Biochem. Biophys. 30, 257–263. [PubMed] [Google Scholar]
- 11.Nenortas, E. C., Bodley, A. L. & Shapiro, T. A. (1998) Biochim. Biophys. Acta 1400, 349–354. [DOI] [PubMed] [Google Scholar]
- 12.Li, T.-K. & Liu, L. F. (2001) Annu. Rev. Pharmacol. Toxicol. 41, 53–77. [DOI] [PubMed] [Google Scholar]
- 13.Bodley, A. L. & Shapiro, T. A. (1995) Proc. Natl. Acad. Sci. USA 92, 3726–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bodley, A. L., Wani, M. C., Wall, M. E. & Shapiro, T. A. (1995) Biochem. Pharmacol. 50, 937–942. [DOI] [PubMed] [Google Scholar]
- 15.Villa, H., Otero Marcos, A. R., Reguera, R. M., Balaña-Fouce, R., García-Estrada, C., Pérez-Pertejo, Y., Tekwani, B. L., Myler, P. J., Stuart, K. D., Bjornsti, M.-A. & Ordóñez, D. (2003) J. Biol. Chem. 278, 3521–3526. [DOI] [PubMed] [Google Scholar]
- 16.Brun, R. & Schönenberger, M. (1979) Acta Trop. 36, 289–292. [PubMed] [Google Scholar]
- 17.Carruthers, V. B. & Cross, G. A. M. (1992) Proc. Natl. Acad. Sci. USA 89, 8818–8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, L. F. & Miller, K. G. (1981) Proc. Natl. Acad. Sci. USA 78, 3487–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goto, T. & Wang, J. C. (1982) J. Biol. Chem. 257, 5866–5872. [PubMed] [Google Scholar]
- 20.Unger, K. (1984) Methods Enzymol. 104, 154–169. [DOI] [PubMed] [Google Scholar]
- 21.Hsiang, Y.-H., Hertzberg, R., Hecht, S. & Liu, L. F. (1985) J. Biol. Chem. 260, 14873–14878. [PubMed] [Google Scholar]
- 22.Stewart, L., Ireton, G. C. & Champoux, J. J. (1997) J. Mol. Biol. 269, 355–372. [DOI] [PubMed] [Google Scholar]
- 23.Broccoli, S., Marquis, J. F., Papadopoulou, B., Olivier, M. & Drolet, M. (1999) Nucleic Acids Res. 27, 2745–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das, A., Dasgupta, A., Sharma, S., Ghosh, M., Sengupta, T., Bandopadhyay, S. & Majumder, H. K. (2001) Nucleic Acids Res. 29, 1844–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng, C., Kussie, P., Pavletich, N. & Shuman, S. (1998) Cell 92, 841–850. [DOI] [PubMed] [Google Scholar]
- 26.Nunes-Düby, S. E., Kwon, H. J., Tirumalai, R. S., Ellenberger, T. & Landy, A. (1998) Nucleic Acids Res. 26, 391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staker, B. L., Hjerrild, K., Feese, M. D., Behnke, C. A., Burgin, A. B. & Stewart, L. (2002) Proc. Natl. Acad. Sci. USA 99, 15387–15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin, N. C. & Hopper, A. K. (1994) Biochimie 76, 1161–1167. [DOI] [PubMed] [Google Scholar]
- 29.Maslov, D. A., Zíková, A., Kyselová, I. & Lukes, J. (2002) Mol. Biochem. Parasitol. 125, 113–125. [DOI] [PubMed] [Google Scholar]
- 30.Bodley, A. L., McGarry, M. W. & Shapiro, T. A. (1995) J. Infect. Dis. 172, 1157–1159. [DOI] [PubMed] [Google Scholar]
- 31.Krogh, B. O. & Shuman, S. (2002) Proc. Natl. Acad. Sci. USA 99, 1853–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart, L., Ireton, G. C. & Champoux, J. J. (1996) J. Biol. Chem. 271, 7602–7608. [DOI] [PubMed] [Google Scholar]
- 33.Sherratt, D. J. & Wigley, D. B. (1998) Cell 93, 149–152. [DOI] [PubMed] [Google Scholar]