Abstract
Glucose homeostasis is controlled by insulin in part through the translocation of intracellular glucose transporter 4 to the plasma membrane in muscle and fat cells. Akt/protein kinase B downstream of phosphatidylinositol 3-kinase has been implicated in this insulin-signaling pathway, but results with a variety of reagents including Akt1–/– and Akt2–/– mice have been equivocal. Here we report the application of small interfering RNA-directed gene silencing to deplete both Akt1 and Akt2 in cultured 3T3-L1 adipocytes. Loss of Akt1 alone slightly impaired insulin-mediated hexose transport activity but had no detectable effect on glycogen synthase kinase (GSK)-3 phosphorylation. In contrast, depletion of Akt2 alone by 70% inhibited approximately half of the insulin responsiveness. Combined depletions of Akt1 plus Akt2 in these cells even more markedly attenuated insulin action on glucose transporter 4 movements, hexose transport activity, and GSK-3 phosphorylation. These data demonstrate a primary role of Akt2 in insulin signaling, significant functional redundancy of Akt1 and Akt2 isoforms in this pathway, and an absolute requirement of Akt protein kinases for regulation of glucose transport and GSK-3 in cultured adipocytes.
Insulin is a key regulator of glucose homeostasis, and its absence is lethal in humans. The ability of insulin to lower blood glucose stems in part from its actions in muscle and fat to enhance sugar uptake through regulation of glucose transporter 4 (GLUT4) (for a recent review, see ref. 1). These transporters are sequestered in perinuclear membranes in unstimulated cells and are rapidly induced to recycle to the plasma membrane in response to insulin (2). Insulin signaling through its receptor tyrosine kinase mediates a phosphatidylinositol 3-kinase-dependent pathway that produces the phosphatidylinositol (3,4,5) trisphosphate required for this effect (for review, see refs. 3–5). Among the downstream effectors strongly implicated in linking this signaling pathway to components that regulate GLUT4 trafficking is the protein kinase Akt (also known as protein kinase B) based on studies with cultured cells and a gene knockout mouse model (6–10). However, this important issue has been controversial (11), and recent data from mice lacking the Akt1 isoform showed no compromise in insulin sensitivity (12). Skeletal muscle from mice deficient in Akt2, the predominant isoform expressed in muscle and fat, is only modestly less sensitive to low levels of insulin and actually responds normally to maximal doses of the hormone (10). The diabetes observed in these mice may be largely due to the dramatic attenuation of insulin action on liver gluconeogenesis (10). Although gene-specific knockout mice provide an excellent tool to study acute actions of signaling molecules, it is possible that phenotypes observed in mice may result from unrelated changes in gene expression and developmental regulation. No data on insulin signaling in isolated primary or cultured adipocytes lacking Akt1 or Akt2 have been published.
Here we have addressed this issue by selectively inhibiting the expression of Akt protein kinases in intact cultured adipocytes through the use of interference RNA (for reviews see refs. 13 and 14). This powerful approach overcomes the problems encountered in mouse gene knockouts where loss of both Akt1 and Akt2 genes is lethal. First discovered in Caenorhabditis elegans (15), this gene-silencing technique uses double-stranded RNA to activate nuclease-containing protein complexes (RNA-induced silencing complex) to target a specific mRNA species, which is then degraded (16, 17). Before insertion into protein-silencing complexes, processing of double-stranded RNA into small interfering RNA (siRNA) duplexes of 21–23 nt occurs by enzymes known as Dicers (18–20). Extension of the technique to mammalian cells has involved the use of synthetic siRNA duplexes of 21-base lengths transfected directly into cultured cells (21), where decreased levels of selected proteins can be observed in response to siRNA after 24–72 h (21). We report here the details of a method that can be used successfully to silence genes in insulin-sensitive cultured adipocytes and show that virtual complete ablation of Akt1 and ≈70% depletion of Akt2 can be achieved. Combined depletion of these Akt isoforms largely attenuates insulin signaling to both GLUT4 glucose transporters and glycogen synthase kinase (GSK)-3, demonstrating an obligatory role of Akt protein kinases in these insulin-signaling cascades.
Materials and Methods
Materials. Human insulin was obtained from Eli Lilly. Goat polyclonal anti-Akt1 Ab (antigen human Akt1 peptide near C terminus, sc-7126), horseradish peroxidase-conjugated donkey anti-goat IgG, mouse monoclonal anti-lamin A/C (sc7293), monoclonal anti-GSK-3α/β, and polyclonal anti-protein kinase C (PKC)λ/ζ (C-20, sc216) were from Santa Cruz Biotechnology. Rabbit polyclonal anti-Akt2 Ab (antigen peptide at C-terminal of human Akt2) was kindly provided by Morris J. Birnbaum (University of Pennsylvania, Philadelphia). Rabbit polyclonal Ab against adipocyte complement-related protein of 30 kDa (Acrp30) was from Affinity BioReagents (Golden, CO) and Ab against nonmuscle myosin IIB was from Covance (Richmond, CA). Polyclonal Abs against phospho-Akt Thr-308/309, phospho-GSK-3α/β (Ser-21/9), and phospho-Erk-1/2 were from Cell Signaling Technology (Beverly, MA). Monoclonal phosphotyrosine Ab (4G10) was from Upstate USA (Charlottesville, VA). The FITC-conjugated goat anti-mouse was from Bio-Source International (Camarillo, CA). The plasmid expressing Myc-tagged GLUT4-EGFP was constructed as described (22).
Design and Synthesis of siRNA Duplexes. The 21-mer sense and antisense strands of RNA oligonucleotides were designed as described (21). The RNA oligonucleotides were synthesized in the 2′-ACE(TM) protected form, which enhances overall RNA stability and resistance to nucleases. The complementary sense and antisense strands of RNA oligonucleotides were mixed, 2′-deprotected, annealed, and purified by PAGE. Gel-purified duplexes were subsequently desalted by using reverse-phase column chromatography, followed by washing with 75% ethanol twice to ensure complete salt removal and dried by use of a Speed-Vac. The pellets were resuspended in nuclease-free water before transfection into cultured cells.
Cell Culture and Electroporation of 3T3L1 Adipocytes. The 3T3-L1 fibroblasts were grown in DMEM supplemented with 10% FBS, 50 μg/ml streptomycin, and 50 units/ml penicillin and differentiated into adipocytes as described (23). The 3T3-L1 adipocytes were transfected with siRNA duplexes by electroporation. In brief, the adipocytes at day 5 of differentiation were detached from culture dishes with 0.25% trypsin and 0.5 mg of collagenase/ml in PBS, washed twice, and resuspended in PBS. Approximately 5 million cells (half of the cells from one p150 dish) were then mixed with siRNA duplexes, which were delivered to the cells by a pulse of electroporation with a Bio-Rad gene pulser II system at the setting of 0.18 kV and 960 μF capacitance. After electroporation, cells were immediately mixed with fresh medium for 10 min before reseeding onto multiple-well plates designed for the deoxyglucose uptake assay, Western blotting, and immunofluorescence microscopic analysis.
Immunofluorescence Microscopy. To visualize lamin A/C, cells were fixed with 4% formaldehyde and permeabilized with PBS containing 1% FBS and 0.5% Triton X-100. Cells were then incubated with primary mouse anti-rat lamin A/C Ab for overnight at 4°C. After washing, cells were incubated with FITC-labeled goat anti-mouse IgG for 30 min at room temperature. To analyze GLUT4 translocation in adipocytes, cells were cotransfected with Myc-GLUT4-EGFP plasmid and siRNAs by electroporation. After this, adipocytes were serum-starved, treated as noted in the figure legends, washed, and immunostained by using the procedure described (22). In brief, the cell surface Myc-GLUT4-EGFP was visualized with mouse anti-Myc Ab and rhodamine-labeled anti-mouse secondary Ab. After washing, the coverslips were mounted in 90% glycerol containing 2.5% diazabicyclo[2.2.2]octane. Fluorescence microscopy was carried out with a IX70-inverted microscope (Olympus, Melville, NY) with charge-coupled device camera (Roper Scientific, Trenton, NJ) and metamorph image processing software (Universal Imaging, Downington, PA).
Western Blotting. After experimental treatments, the cells were solubilized as described (22). To detect phosphorylation of Akt Thr-308/309, GSK-3α/β Ser-21/9, and tyrosine phosphorylation of Erk-1/2, 50 μg protein from 3T3-L1 adipocyte lysates were resolved with 8% SDS/PAGE and electrotransferred to nitrocellulose membranes, which were incubated with anti-phospho-specific Abs (1:1,000 dilution) overnight at 4°C and then with horseradish peroxidase-linked anti-rabbit IgG Abs (1:10,000 dilution) for 1 h at room temperature. Tyrosin phosphorylation of insulin receptor substrate (IRS) proteins was detected with monoclonal phosphotyrosine Ab followed by horseradish peroxidase-linked anti-mouse IgG Abs. Akt1 was detected with primary goat polyclonal Ab (1:750 dilution) and secondary horseradish peroxidase-linked donkey anti-goat Ab (1:10,000 dilution). Primary rabbit polyclonal Abs against Akt2 (1:1,000 dilution), nonmuscle myosin IIB (0.1 μg/ml), Acrp30 (0.5 μg/ml), and PKCλ/ζ (1:500 dilution) were used to detect their antigens by using 25 μg of protein from total cell lysates. The membranes were washed with wash buffer (PBS, pH 7.4/0.1% Tween 20) for 1 h at room temperature after incubation with each Ab. Finally, the levels of target proteins or phosphorylated proteins were detected with an enhanced chemiluminescence kit. To use the same nitrocellulose membrane to detect several proteins and phospho-proteins, the blots were incubated with gentle shaking in stripping buffer (62.5 mM Tris·HCl, pH 6.7/100 mM 2-mercaptoethanol/2% SDS) for 30–45 min at 60°C and washed for at least 1 h with wash buffer before reblotting with the Ab designed for the next experiment.
The 2-Deoxyglucose Uptake Assay. Insulin-stimulated glucose transport in 3T3-L1 adipocytes was estimated by measuring 2-deoxyglucose uptake. In brief, siRNA-transfected cells were reseeded on 12-well plates and cultured for 40 h and then washed twice with DMEM before incubation with DMEM containing 0.5% BSA for 4 h at 37°C. Cells were then washed twice with Krebs–Ringer's Hepes buffer (130 mM NaCl/5 mM KCl/1.3 mM CaCl2/1.3 mM MgSO4/25 mM Hepes, pH 7.4) and further starved for 1.5 h with Krebs–Ringer's Hepes buffer supplemented with 0.5% BSA and 2 mM sodium pyruvate. Cells were then stimulated with insulin for 30 min at 37°C. Glucose uptake was initiated by addition of [1,2-3H]2-deoxy-d-glucose to a final assay concentration of 100 μM for 5 min at 37°C. Assays were terminated by four washes with ice-cold Krebs–Ringer's Hepes buffer, and the cells were solubilized with 0.4 ml of 1% Triton X-100, and 3H was determined by scintillation counting. Nonspecific deoxyglucose uptake was measured in the presence of 20 μM cytochalasin B and subtracted from each determination to obtain specific uptake.
Results and Discussion
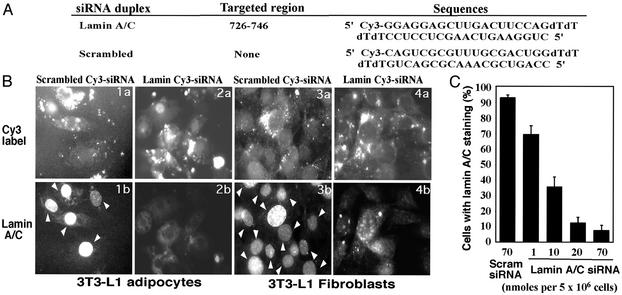
siRNA-Induced Gene Silencing in Cultured Adipocytes. To test whether siRNA can induce gene-specific silencing in adipocytes, we first targeted the gene encoding lamin A/C, previously shown to be sensitive to this technique in other cells. Cy3-tagged 21-nt siRNA duplexes directed against mouse lamin A/C mRNA were designed and synthesized (Fig. 1A). The initial experiments we performed showed that conditions developed for siRNA-mediated gene silencing in other cells types (21) worked well in 3T3-L1 fibroblasts (Fig. 1B Right), but failed to work in 3T3-L1 adipocytes (not shown). We therefore developed alternate methodology to transfect lamin A/C siRNA into 3T3-L1 adipocytes by using electroporation. With this method, Cy3-siRNA was introduced with virtually 100% efficiency into the cultured adipocytes, and by 48 h nearly all cells showed loss of nuclear lamin A/C compared to cells transfected with a scrambled Cy3-tagged siRNA species (Fig. 1B). Quantification of these results showed that adding 20 nmol of siRNA to a suspension of 5 × 106 adipocytes results in loss of lamin A/C in ≈90% of the adipocytes with no detectable toxicity (Fig. 1C). These findings provide the basis for reliable gene silencing in insulin-sensitive cultured adipocytes.
Fig. 1.
Sequence-specific gene silencing by siRNA transfected into 3T3-L1 fibroblasts and 3T3-L1 adipocytes. (A) Sequences of Cy3-labeled lamin A/C and scrambled siRNA duplexes. Lamin A/C siRNA was designed according to the mouse lamin A mRNA sequence obtained from the National Center for Biotechnology Information database (accession no. BC015302) and is unique to mouse lamin A/C according to the mouse EST database. Mouse scrambled siRNA was designed as a random sequence whose sense stand does not show homology to any gene obtained in the mouse EST database or any vertebrate database. (B) The 3T3-L1 adipocytes (day 5) were transfected with Cy3-labeled mouse lamin A/C(2a and 2b) or scrambled siRNA duplexes (1a and 1b) by electroporation at a concentration of 80 nmol per 5 million cells. The 3T3-L1 fibroblasts in six-well plates were transfected with 500 nM Cy3-labeled lamin A/C(4a and 4b) or scrambled siRNAs (3a and 3b) by using Oligofectamine reagent (Invitrogen). After transfection for 48 h, nuclear membrane localization of lamin A/C (1b and 2b for adipocytes and 3a and 4a for fibroblasts) was visualized by immunofluorescence by using Ab against mouse lamin A/C as described in Materials and Methods. Arrows point to nuclear lamin A/C staining in adipocytes (1b) and fibroblasts (3b). (C) Concentration dependence of lamin A/C gene silencing by siRNA in 3T3-L1 adipocytes. Cy3-labeled scrambled siRNA or lamin A/C siRNA at the concentrations indicated were electroporated with differentiated 3T3-L1 adipocytes (day 5). After immunostaining with anti-lamin A/C Ab, cells with lamin A/C staining were counted. Data represent mean ± SD of three independent experiments.
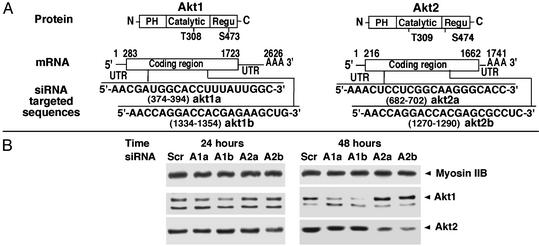
Two siRNA species directed against each of the Akt isoforms Akt1 and Akt2 were then tested for their abilities to inhibit expression of these protein kinases in 3T3-L1 adipocytes (Fig. 2). Each of the Akt1-directed siRNA species inhibited expression of Akt1 protein at both 24 and 48 h after transfection. One of these (akt1b) directed virtually total ablation of Akt1 expression by 48 h, whereas Akt2 expression was unaffected (Figs. 2B and 3 A and B). Similarly, Akt2 expression could be selectively attenuated by ≈70% after transfection of the siRNA species akt2b, whereas the akt2a siRNA was less effective (Figs. 2B and 3 A and B). Interestingly, the akt1b and akt2b siRNA species that show most efficacy are targeted to similar regions of the Akt1 and Akt2 mRNA sequences that encode amino acids 351–357 and 352–358 in Akt1 and Akt2, respectively (Fig. 2 A). Whether the secondary structure of this Akt mRNA region is particularly susceptible to siRNA-directed RNA degradation requires further study. The selectivity of akt1b vs. akt2b siRNAs to silence their respective target mRNA species is apparent even though only 4 of 21 nucleotides are different (Fig. 2 A). Expression of several other unrelated proteins (e.g., myosin IIB shown in Figs. 2, 3, and 5 and adipocyte-specific protein Acrp30 shown in Figs. 3 and 5) were also unaffected by akt1b or akt2b siRNAs.
Fig. 2.
Time-dependent gene silencing of Akt1 and Akt2 by synthetic siRNA. Synthetic siRNA duplexes targeting different regions of Akt1 and Akt2 mRNAs (A) were used for transfection into 3T3-L1 adipocytes by electroporation at 20 nmol per 5 × 106 cells (cells from one-half of p150 dishes). (B) After reseeding for 24 or 48 h, the cells were lysed and 50 μg of protein was used for Akt1 Western blotting and 25 μg of protein was used for Akt2 and nonmuscle myosin IIB Western blotting. Akt1 and Akt2 siRNA duplexes were designed according to mouse Akt1 and Akt2 mRNA sequences obtained from National Center for Biotechnology Information database (accession nos. NM_009652 and U22445, respectively). Sc, scramble; A1a, akt1a siRNA; A1b, akt1b siRNA; A2a, akt2a siRNA; A2b, akt2b siRNA.
Fig. 3.
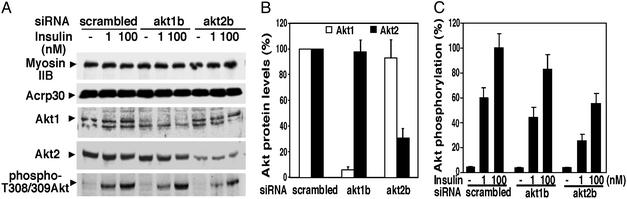
Akt2 accounts for most of the insulin-stimulated Akt protein kinase activity in cultured 3T3-L1 adipocytes. The 3T3-L1 adipocytes were transfected with siRNAs (40 nmol of siRNA duplexes/5 × 106 cells) by electroporation, reseeded for 42 h, and serum-starved for 6 h before the treatment with insulin for 15 min at 37°C. Protein (50 μg) of total cell lysate was used for detection of Akt1 and phospho-Thr-308/309 Akt. (A) Protein (25 μg) was used for detection of Akt2, nonmuscle myosin IIB, and Acrp30. (B) Quantification of Akt1 and Akt2 protein levels, respectively. (C) Quantifications of Akt Thr-308/309 phosphorylation. Data are presented as a mean ± SD of three independent experiments.
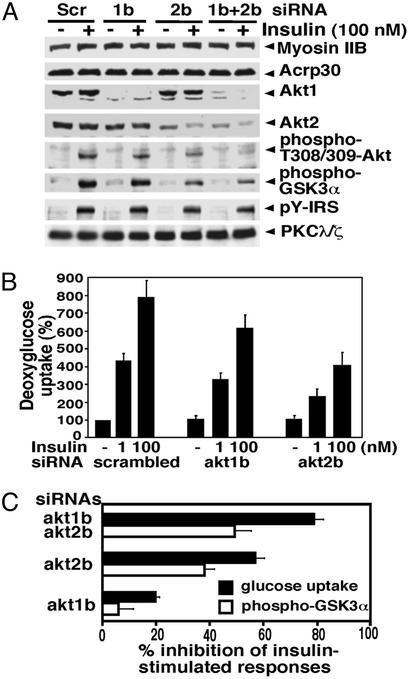
Fig. 5.
Redundancy of Akt1 and Akt2 in the insulin-signaling pathway to hexose transport and GSK-3 phosphorylation in 3T3-L1 adipocytes. Cells were transfected with akt1b or akt2b siRNA separately (40 nmol of siRNA duplexes/5 × 106 cells) or in combination (20 nmol each siRNA duplexes/5 × 106 cells) by electroporation, reseeded, and cultured for 42 h. The serum-starved cells were then stimulated with insulin and deoxyglucose uptake was assayed as described in Materials and Methods. (A) Representative Western blot images for Akt protein levels, phospho-Thr-308/309 levels, phospho-Ser-21-GSK-3α levels, phospho-Tyr in IRS proteins, and PKCλ/ζ protein levels in total lysates. (B) Dose dependence of insulin-stimulated deoxyglucose uptake. Quantitative data are presented as a mean ± SD of four independent experiments. (C) Inhibition of 100 nM insulin-induced glucose uptake and GSK-3α phosphorylation by knockdown Akt1 and Akt2 protein levels. Quantitative data are presented as a mean ± SD of three independent experiments. Scr, scrambled siRNA; 1b, akt1b siRNA; 2b, akt2b siRNA.
Differential Effects of Akt1 and Akt2 Gene Silencing on Insulin Signaling. Akt1 and Akt2 are phosphorylated and activated by the protein kinase PDK1 at Thr-308 or Thr-309, respectively, in the activation T-loop, and further activation occurs through phosphorylation at Ser-473 or Ser-474, respectively (24–26). The effects of selective loss of Akt1 vs. Akt2 proteins on insulin-stimulated phosphorylation of total Thr-308/309 contained in both proteins were assessed by Western blotting with an anti-phospho-Thr-308/309 Ab. Total loss of Akt1 protein resulted in only a 10–20% reduction in total Thr-308/309 phosphorylation of Akt protein kinases in cultured 3T3-L1 adipocytes, consistent with previous results showing Akt1 is much less abundant than Akt2 in adipocytes (7, 27, 28). In contrast, reduction of Akt2 expression by ≈70% caused a marked 55–60% decrease in insulin-stimulated Thr phosphorylation of the Akt protein kinases (Fig. 3). Taken together, these data confirm the predominance of Akt2 over Akt1 in insulin-sensitive cultured adipocytes (7).
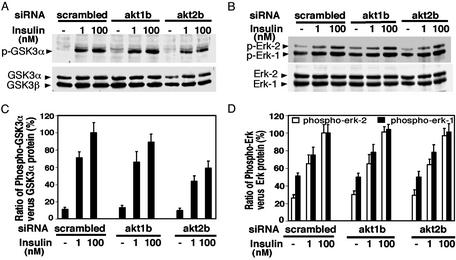
Next, we assessed the consequences of the selective attenuation of Akt1 or Akt2 expression on a downstream target of insulin signaling–GSK-3 (29, 30). GSK-3α is phosphorylated by Akt protein kinases in response to the hormone in a dose-dependent manner (Fig. 4). In three independent experiments, loss of 95% or more of Akt1 directed by the akt1b siRNA caused no significant attenuation of insulin-mediated GSK-3α phosphorylation (Fig. 4 A and B), although a 10–20% effect may go undetected in these studies. In contrast, attenuation of Akt2 expression by 70% caused ≈40% inhibition of insulin-mediated GSK-3α phosphorylation (Figs. 4 A and B and 5 A and C). In control studies, no diminution of insulin signaling to the mitogen-activated protein kinases Erk-1 and Erk-2 was observed when either Akt1 or Akt2 are depleted (Fig. 4 C and D), confirming the specificity of the effect of silencing the Akt protein kinases by this method. These data indicate that insulin action on GSK-3α in cultured adipocytes specifically requires Akt2.
Fig. 4.
Selective gene silencing of Akt2 attenuates insulin-stimulated GSK-3 phosphorylation in 3T3-L1 adipocytes. The nitrocellulose membranes used for Akt1/2 and phospho-Akt analysis (shown in Fig. 2) were stripped and reblotted with phospho-GSK-3 and GSK-3 (A) or phospho-Erk-1/2 and Erk-1/2(B) Abs. (C) Phosphorylation of GSK-3α was quantified by measurement of the ratio of the intensity of phospho-GSK-3α bands vs. the intensity of GSK-3α protein bands. (D) Similarly, phosphorylation of Erk-1/2 was quantified by measurement of the ratio of the intensity of phospo-Erk-1/2 bands vs. the intensity of Erk-1/2 protein bands. Data are presented as a mean ± SD of three independent experiments.
Redundancy of Akt1 and Akt2 in Insulin Signaling. Applying this same approach to hexose transport regulation, we found that ablation of Akt1 expression (Fig. 5A) leads to a small but significant 20–30% decrease in insulin-stimulated 2-deoxyglucose uptake in 3T3-L1 adipocytes (Fig. 5B). Akt2 protein depletion to ≈70% of normal levels dampened the insulin response by 50–58%. These data indicate that both Akt1 and Akt2 contribute to insulin responsiveness of hexose transport in cultured adipocytes roughly in proportion to their contributions to total activated Akt in these cells (see ref. 7 and Fig. 3). We then tested the effects of depleting both Akt1 and Akt2 in 3T3-L1 adipocytes by using the combination of akt1b and akt2b siRNA species (Fig. 5 A and C). This combined treatment virtually completely ablated Akt1 expression and reduced Akt2 expression by >65% whereas insulin-stimulated phosphorylation of total Akt detected by anti-phospho Thr-308/309 Ab decreased by 81% (Fig. 5A). Importantly, insulin-stimulated deoxyglucose uptake was inhibited by nearly 80% under these conditions, compared to ≈58% when only Akt2 was depleted (Fig. 5C). Under these conditions, GLUT4 expression was unchanged (not shown). In control experiments to test whether engagement of the gene silencing process itself nonspecifically inhibits insulin signaling, we observed no significant effect of lamin siRNA on insulin-stimulated glucose transport under conditions where lamin A/C protein levels were markedly reduced (Fig. 1 and data not shown). Taken together, these data demonstrate that although Akt2 is the major protein kinase required in this response, Akt1 can also play a role. Thus under conditions where insulin-stimulated glucose uptake is significantly compromised by partial depletion of Akt2, Akt1 is required for half of the remaining insulin signal (Fig. 5C). GSK-3α phosphorylation in response to insulin was also inhibited to a greater extent when both protein kinases were depleted vs. when only Akt2 was reduced (Fig. 5 A and C).
In additional control experiments, we tested whether expression of other insulin-signaling elements such as IRS proteins (31, 32) or PKCλ/ζ (11, 33) is affected by Akt1 or Akt2 siRNAs. As shown in Fig. 5A, depletion of Akt1 or Akt2 had no significant effect on insulin-stimulated tyrosine phosphorylation of IRS proteins or expression levels of PKCλ/ζ.
The effect of combined depletion of both Akt1 and Akt2 by siRNA on insulin-mediated GLUT4 translocation was next examined in 3T3-L1 adipocytes. Cotransfection of myc-GLUT4-EGFP plasmid DNA with the mixture of akt1b and akt2b siRNAs was performed, and 48 h later reductions of ≈90% and 65% of Akt1 and Akt2 protein levels, respectively, were observed (Fig. 6B). This combined knockdown of Akt1 and Akt2 proteins resulted in the loss of insulin-stimulated cell surface Myc signal (detected by anti-Myc Ab) in ≈70% of adipocytes transfected with the Myc-GLUT4-GFP construct (Fig. 6 A and C). Quantifying the ratio of cell surface Myc rim signal over the total Myc-GLUT4-GFP signal in positive transfected cells revealed that loss of both Akt1 and Akt2 resulted in a 60% decrease in insulin-stimulated Myc-GLUT4-GFP on the cell surface (Fig. 6D). The attenuation of GLUT4 responsiveness is observed at both maximal and submaximal concentrations of insulin (Fig. 6C), whereas no significant effect of Akt depletion on GLUT4 translocation in the absence of insulin is detected.
Fig. 6.
Inhibition of insulin-mediated GLUT4 glucose transporter translocation by combined depletion of Akt1 and Akt2 proteins. The 3T3-L1 adipocytes (5 × 106 cells) were transfected with Myc-GLUT4-EGFP plasmid DNA (40 μg) together with either scrambled siRNA (40 nmol) or the mixture of akt1b and akt2b siRNAs (20 nmol each) by electroporation. After reseeding for 42 h, adipocytes were starved for 5 h before incubation with or without insulin (1 or 100 nM) for 30 min at 37°C. Myc-GLUT4-EGFP translocation to the cell surface in GFP-positive cells was detected with mouse anti-Myc epitope primary Ab and rhodamine-conjugated goat anti-mouse IgG as described in Materials and Methods. (A) Representative images for GFP-positive cells and exofacial Myc staining. The concentration of insulin used was 100 nM. Images for Myc staining and GFP signal were taken at an optical plane near the middle of cells where the Myc signal around the rim of cells was the brightest. (B) Akt protein levels in adipocytes transfected with Myc-GLUT4-EGFP and siRNAs for 48 h. (C) Percentage of the transfected adipocytes showing a Myc-GLUT4-GFP rim on the cell surface. Data are presented as the mean ± SD of three independent experiments with >200 cells counted in each experiment. (D) Quantification of the ratio of cell surface Myc signal over the total GFP signal in adipocytes expressing Myc-GLUT4-GFP by using methods as described (22). Again, the concentration of insulin used was 100 nM. Comparison of data presented as mean ± SD was performed by using the unpaired Student's t test. *, P < 0.001 for difference between cells transfected with scrambled siRNA vs. cells transfected with akt1b/akt2b siRNAs. Scr, scrambled siRNA; 1b, akt1b siRNA; 2b, akt2b siRNA.
The findings presented here show an absolute requirement of Akt protein kinases for normal insulin signaling to glucose transport and GSK-3α and imply direct proportionality of total available Akt1 plus Akt2 to the degree of insulin responsiveness (Fig. 5). This conclusion is in keeping with the similar insulin dose–response relationships observed for activation of total Akt and glucose transport (compare Figs. 3C and 5B). Progressive loss of Akt1, Akt2, or both leads to a correspondingly progressive loss in glucose transport stimulation (Fig. 5). These considerations show that Akt1 can in part replace Akt2 to sustain glucose transport responsiveness, potentially providing an explanation for why skeletal muscle of Akt2–/– mice exhibits only a mild impairment in insulin responsiveness (10). No data on insulin signaling in muscle or fat cells from Akt1–/– mice have been published, but the normal glucose tolerance in these animals (12) is consistent with our data (Fig. 5) that Akt2 alone can sustain most of the insulin-regulated glucose uptake. Although it is possible that other signaling pathways (22, 34, 35) and protein kinases are also involved in insulin action on glucose transport, our results underscore the absolute requirement of Akt for this process.
Acknowledgments
We thank Dr. Morris J. Birnbaum for kindly providing anti-Akt2 Ab and Jane Erickson for assistance with the manuscript. This work was supported by National Institutes of Health Grants DK30648 and DK60837 (to M.P.C.).
Abbreviations: GLUT4, glucose transporter 4; GSK-3, glycogen synthase kinase-3; siRNA, small interfering RNA; Acrp30, adipocyte complement-related protein of 30 kDa; IRS, insulin receptor substrate.
References
- 1.Bryant, N. J., Govers, R. & James, D. E. (2002) Nat. Rev. Mol. Cell Biol. 3, 267–277. [DOI] [PubMed] [Google Scholar]
- 2.Cushman, S. W. & Wardzala, L. J. (1980) J. Biol. Chem. 255, 4758–4762. [PubMed] [Google Scholar]
- 3.Virkamaki, A., Ueki, K. & Kahn, C. R. (1999) J. Clin. Invest. 103, 931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White, M. F. (2002) Am. J. Physiol. 283, E413–E422. [DOI] [PubMed] [Google Scholar]
- 5.Czech, M. P. & Corvera, S. (1999) J. Biol. Chem. 274, 1865–1868. [DOI] [PubMed] [Google Scholar]
- 6.Kohn, A. D., Summers, S. A., Birnbaum, M. P. & Roth, R. A. (1996) J. Biol. Chem. 49, 31372–31378. [DOI] [PubMed] [Google Scholar]
- 7.Hill, M. M., Clark, S. F., Tucker, D. F., Birnbaum, M. J., James, D. E. & Macaulay, S. L. (1999) Mol. Cell. Biol. 19, 7771–7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang, Q., Somwar, R., Bilan, P. J., Liu, Z., Jin, J., Woodgett, J. R. & Klip, A. (1999) Mol. Cell. Biol. 19, 4008–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueki, K., Yamamoto-Honda, R., Kaburagi, Y., Yamauchi, T., Tobe, K., Burgering, B. M., Coffer, P. J., Komuro, I., Akanuma, Y., Yazaki, Y., et al. (1998) J. Biol. Chem. 273, 5315–5322. [DOI] [PubMed] [Google Scholar]
- 10.Cho, H., Mu, J., Kim, J. K., Thorvaldsen, J. L., Chu, Q., Chrenshaw, E. B., Kaestner, K. H., Bartolomei, M. S., Shulman, G. I. & Birnbaum, M. J. (2001) Science 292, 1728–1731. [DOI] [PubMed] [Google Scholar]
- 11.Kotani, K., Ogawa, W., Matsumoto, M., Kitamura, T., Sakaue, H., Hino, Y., Miyake, K., Sano, W., Akimoto, K., Ohno, S. & Kasuga, M. (1998) Mol. Cell. Biol. 18, 6971–6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho, H., Thorvaldsen, J. L., Chu, Q., Feng, F. & Birnbaum, M. J. (2001) J. Biol. Chem. 276, 38349–38352. [DOI] [PubMed] [Google Scholar]
- 13.Hutvagner, G. & Zamore, P. D. (2002) Curr. Opin. Genet. Dev. 12, 225–232. [DOI] [PubMed] [Google Scholar]
- 14.Hannon, G. J. (2002) Nature 418, 244–251. [DOI] [PubMed] [Google Scholar]
- 15.Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 16.Hammond, S. M., Bermstein, E., Beach, D. & Hannon, G. J. (2001) Nature 404, 293–296. [DOI] [PubMed] [Google Scholar]
- 17.Hammond, S., Boettcher, S., Caudy, A., Kobayashi, R. & Hannon, G. J. (2001) Science 293, 1146–1150. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. (2001) Nature 409, 363–366. [DOI] [PubMed] [Google Scholar]
- 19.Zamore, P., Tuschl, T., Sharp, P. & Bartel, D. (2000) Cell 101, 25–33. [DOI] [PubMed] [Google Scholar]
- 20.Elbashir, S. M., Lendeckel, W. & Tuschl, T. (2000) Genes Dev. 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, Z. Y., Chawla, A., Bose, A., Way, M. & Czezh, M. P. (2002) J. Biol. Chem. 277, 509–515. [DOI] [PubMed] [Google Scholar]
- 23.Harrison, S. A., Buxton, J. M., Clancy, B. M. & Czech, M. P. (1990) J. Biol. Chem. 265, 20106–20116. [PubMed] [Google Scholar]
- 24.Alessi, D. R., Deak, M., Casamayor, A., Caudwell, F. B., Morrice, N., Norman, D. G., Gaffney, P., Reese, C. B., MacDougall, C. N., Harbison, D., et al. (1997) Curr. Biol. 7, 776–789. [DOI] [PubMed] [Google Scholar]
- 25.Williams, M. R., Arthur, J. S., Balendran, A., van der Kaay, J., Poli, V., Cohen, P. & Alessi, D. R. (2000) Curr. Biol. 10, 439–448. [DOI] [PubMed] [Google Scholar]
- 26.Brazil, D. P. & Hemmings, B. A. (2001) Trends Biochem. Sci. 26, 657–664. [DOI] [PubMed] [Google Scholar]
- 27.Summers, S. A., Whiteman, E. L., Cho, H., Lipfert, L. & Birnbaum, M. J. (1999) J. Biol. Chem. 274, 23858–23867. [DOI] [PubMed] [Google Scholar]
- 28.Calera, M. R., Martinez, C., Liu, H., El Jack, A. K., Birnbaum, M. J. & Pilch, P. F. (1998) J. Biol. Chem. 273, 7201–7204. [DOI] [PubMed] [Google Scholar]
- 29.Cross, D. A., Alessi, D. R., Cohen, P., Andjelkovich, M. & Hemmings, B. A. (1995) Nature 378, 785–789. [DOI] [PubMed] [Google Scholar]
- 30.Cohen, P. & Frame, S. (2001) Nat. Rev. Mol. Cell Biol. 2, 769–776. [DOI] [PubMed] [Google Scholar]
- 31.Li, J., DeFea, K. & Roth, R. A. (1999) J. Biol. Chem. 274, 9351–9356. [DOI] [PubMed] [Google Scholar]
- 32.Paz, K., Liu, Y.-F., Shorer, H., Hemi, R., LeRoith, D., Quan, M., Kannah, K., Seger, R. & Zick, Y. (1999) J. Biol. Chem. 274, 28816–28822. [DOI] [PubMed] [Google Scholar]
- 33.Standaert, M. L., Galloway, L., Karnam, P., Bandyopadhyay, G., Moscat, J. & Farese, R. V. (1997) J. Biol. Chem. 272, 30075–30082. [DOI] [PubMed] [Google Scholar]
- 34.Chiang, S.-H., Baumann, C. A., Kanzaki, M., Thurmond, D. C., Watson, R. T., Neudauer, C. L., Macara, I. G., Pessin, J. E. & Saltiel, A. R. (2001) Nature 410, 944–948. [DOI] [PubMed] [Google Scholar]
- 35.Wang, Q., Bilan, P. J., Tsakiridis, T., Hinek, A. & Klip, A. (1998) Biochem. J. 331, 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]