Abstract
In an attempt to better understand and control the processes that regulate stem cell fate, we have set out to identify small molecules that induce neuronal differentiation in embryonic stem cells (ESCs). A high-throughput phenotypic cell-based screen of kinase-directed combinatorial libraries led to the discovery of TWS119, a 4,6-disubstituted pyrrolopyrimidine that can induce neurogenesis in murine ESCs. The target of TWS119 was shown to be glycogen synthase kinase-3β (GSK-3β) by both affinity-based and biochemical methods. This study provides evidence that GSK-3β is involved in the induction of mammalian neurogenesis in ESCs. This and such other molecules are likely to provide insights into the molecular mechanisms that control stem cell fate, and may ultimately be useful to in vivo stem cell biology and therapy.
Although stem cells hold considerable promise for the treatment of a number of degenerative diseases, including Parkinson's disease and diabetes (1–3), obstacles such as host rejection, control of stem cell fate, and availability of stem cells must be overcome before their therapeutic potential can be realized. These challenges require a better understanding of the signaling pathways that control stem cell fate and an improved ability to manipulate stem cell proliferation and differentiation. Stem cell differentiation is controlled by both intrinsic regulators and the extracellular environment, and is typically controlled ex vivo by cell culture manipulation with ``cocktails'' of growth factors, signaling molecules, and/or by genetic manipulation. Cell-permeable small molecules such as retinoic acid (RA) have also proven extremely useful tools in inducing differentiation of various progenitor cells such as embryonic stem cells (ESCs), neural stem cells (NSCs), and mesenchymal stem cells (MSCs) (4–6). However, the action of RA is pleiotropic. Clearly the identification of additional small molecules that permit precise regulation of stem cell self-renewal and differentiation, as well as the dedifferentiation of lineage-committed cells, would be useful in understanding the underlying molecular mechanisms of these processes and ultimately could facilitate therapeutic applications of stem cells (7, 8).
We therefore set out to identify small molecules that can induce the selective differentiation of stem cells to neurons. Previous reports have shown that mouse and human ESCs (9, 10) have the capacity to differentiate in vitro into neural progenitor cells, which can further mature into neurons and glial cells. Because kinases are likely to play an important role in these processes, given their involvement in many cell cycle and developmental events, combinatorial libraries of heterocyclic compounds designed around a large number of kinase-directed molecular scaffolds (ref. 11 and references therein) were screened. Herein, we report the discovery of small molecules from these libraries that induce neuronal differentiation in pluripotent murine embryonal carcinoma (EC) cells and ESCs, as well as studies that have begun to define a mechanism for their action.
Materials and Methods
Plasmid Construction. To construct the pTα1-Luc vector, 1.1 kb of the 5′-flanking region of the rat Tα1 gene was amplified by PCR by using the 5′-primer 5′-GGGACCTCGAGGTATCCAGACTCACTCCTTTC-3′ and the 3′-primer 5′-GGCAGAAGCTTGTTTGCTCACCATGGTTGCT-3′. The PCR product and pGL3-Basic Vector (Promega) were digested with XhoI and HindIII and ligated together to generate pTα1-Luc vector. The pTOPFLASH luciferase vector was obtained from J. Liu (The Scripps Research Institute).
Cell Culture. P19 cells [American Type Culture Collection (ATCC) CRL-1825] were cultured in minimum essential medium α (MEMα; GIBCO) with 7.5% calf serum and 2.5% FBS (HyClone; P19 growth medium) at 37°C in 5% CO2. To induce differentiation, P19 cells were grown to a density of 106/100-mm Petri dish (Fisher) in MEMα with 5% FBS and 0.5 μM all-trans RA (neural induction medium). After 2 days, the P19 aggregates were transferred to fresh neural induction medium and incubated for another 2 days. After 4 days, the P19 aggregates were dissociated by trypsinization and plated in a tissue culture plate. The undifferentiated ESC line D3 (ATCC CRL-1934) was cultured on a feeder layer of irradiated mouse fibroblasts with ESC growth medium (knockout-DMEM, GIBCO) supplemented with 1× penicillin/streptomycin, 1× nonessential amino acids, 1× nucleosides (Specialty Media), 0.1 mM 2-mercaptoethanol (Sigma), 2 mM l-glutamine, 15% FBS (HyClone), and 1,000 units/ml leukemia inhibitory factor (LIF; Chemicon). Before in vitro differentiation, ESCs were cultured for an additional two passages on fresh gelatin-coated tissue culture dish with ESC growth medium with FBS substituted by 15% serum replacement (GIBCO).
Transfection, Reporter Assay, and Compound Screens. Transfections were performed by using FuGENE6 (Roche) as directed by the manufacturer. For a 35-mm dish, 2.0 μg of pTα1-Luc vector and 0.1 μg of pcDNA3.1 (Invitrogen) were used. After 2 days, 700 μg/ml G418 (GIBCO) was added, and stable single clones were obtained by using cloning cylinders; these clones were expanded in P19 growth medium with G418 (600 μg/ml). Single stable clones of P19 transfectant with pTα1-Luc were grown in Petri dishes in MEMα with 5% FBS and treated under three conditions: (i) with 0.5 μM all-trans RA; (ii) with 1% DMSO; and (iii) with no added drug. After 4 days (2 + 2) in suspension culture, cell aggregates were dispersed into single cell suspension and plated into tissue culture plates. Cells were then lysed and tested for luciferase activities at different time points after replating. The clone (P19Ta1Luc-17), which afforded a high luciferase signal upon neuronal differentiation and low luciferase signal under the other two control conditions, was selected for screening. For the small molecule screen, undifferentiated P19 cells (clone P19Ta1Luc-17) were plated into white 384-well tissue culture plates (Greiner) at a density of 2,000 cells per well in MEMα with 5% FBS. Compounds were added at a final concentration of 5 μM 12 h after plating. Cells were treated with compounds for 4 days and tested for luciferase activity as directed by the manufacturer (Promega). To assay β-catenin-induced T cell factor/lymphoid enhancer factor (TCF/LEF) reporter activity in P19 cells treated with TWS analogs and RA, P19 cells in a 100-mm tissue culture dish were cotransfected with 6 μg of pTOPFLASH (containing four consensus LEF-1/TCF-1 binding sites, a minimal promoter, and a firefly luciferase reporter) and 3 μg of Renilla luciferase control reporter (Promega) by using FuGENE6 (Roche) as directed by the manufacturer. After 24 h, cells were trypsinized and replated into a 96-well tissue culture plate and treated with TWS analogs, or plated into Petri dishes and treated with RA in MEMα with 5% FBS 12 h postplating. Thirty-six hours later, cells were lysed, and protein extracts were assayed for luciferase activity as directed by the manufacturer. The increase in firefly luciferase activity was normalized to Renilla luciferase activity and represents the average of three experiments.
Immunocytochemistry and Antibodies. Cells were fixed for 20 min with 4% paraformaldehyde in PBS (GIBCO). Immunostaining was carried out with standard protocols. Primary antibodies were used at the following dilutions: βIII-tubulin (TuJ1) mouse monoclonal (Babco, Berkeley, CA, 1:500) and rabbit polyclonal (Babco, 1:2,000), mouse monoclonal microtubule-associated protein (Map) 2(a+b) (Sigma, 1:1,000), mouse monoclonal neurofilament M (Chemicon, 1:1,000), mouse monoclonal NeuN (Chemicon, 1:100), mouse monoclonal nestin (Becton Dickinson, 1:1,000), rabbit polyclonal synapsin I (Calbiochem, 1:1,000), and rabbit polyclonal glutamate (Chemicon, 1:300). Secondary antibodies were Cy2- or Cy3-conjugated anti-rabbit or anti-mouse (Jackson ImmunoResearch, 1:500). Cells were imaged by using a Nikon Eclipse TE2000 microscope with ×200 magnification. Double-label images were assembled in metamorph.
Affinity Chromatography. The synthesis of TWS101, -102, -113, -119, and -121, their linker derivatives, and the affinity supports will be described elsewhere. Cells were lysed with PY buffer (pH 7.2/60 mM β-glycerophosphate/15 mM nitrophenylphosphate/25 mM Mops/15 mM EGTA/15 mM MgCl2/2 mM DTT/1 mM sodium vanadate/1 mM sodium fluoride/1 mM sodium phenylphosphate/100 μM benzamidine) plus 0.4% Nonidet P-40 for 10 min on ice, followed by centrifugation at 14,000 rpm at 4°C for 10 min (Eppendorf). The clear supernatant was collected and the pellet was discarded. The total protein concentration in the supernatant was determined by using a BCA protein quantification kit (Pierce). Forty microliters of affinity support (suspension in PBS, Vaffinity-support/VPBS = 1/2) was washed two times in a microcentrifuge tube with bead buffer (pH 7.4/50 mM Tris A/5 mM sodium fluoride/250 mM sodium chloride/5 mM EDTA/5 mM EGTA/0.1% Nonidet P-40/100 μM benzamidine). Protein extracts (300 μg) were added to each affinity support, and the volume was made up to 900 μl with bead buffer. After agitation at 4°C for 1 h, the affinity supports were centrifuged at 6,000 rpm for 1 min at 4°C (Eppendorf). The supernatant was removed, and the affinity supports were washed three times with chilled bead buffer (in the competition assay, bead buffer containing 50 μM small molecule was used as the incubation buffer and the wash buffer.) After the final wash, the supernatant was removed, and 18 μl of SDS sample buffer was added to the resin and heated at 95°C for 5 min. Samples were loaded and separated on 10% Tris-glycine SDS-polyacrylamide gel (Invitrogen). For silver staining, Silver Stain Plus Kit (Bio-Rad) was used. For Western blot analysis, after electroblotting on nitrocellulose membrane, the membrane was blocked for 1 h at room temperature with 5% nonfat milk powder and immunoblotted overnight at 4°C with rabbit anti-glycogen synthase kinase-3β (anti-GSK-3β) polyclonal antibody (1:1,000, Cell Signaling, Beverly, MA). The membrane was washed three times with PBS/0.05% Tween 20, incubated with anti-rabbit peroxidase-conjugated affinity-purified secondary antibody (1:5,000, Sigma) for 1 h at room temperature, washed again three times, and developed by using SuperSignal chemiluminescence (Pierce).
Surface Plasmon Resonance (SPR). SPR measurements were performed on a BIAcore S51 instrument (Uppsala) at 25°C in PBS buffer. Relative light units (11,000–17,000) of a GSK-3β–GST fusion protein were immobilized on S series CM5 biosensor chips by EDC [1-ethyl-3-(3-dimethylaminopropyl)carbodiimide] coupling procedures. Briefly, GSK-3β was diluted into mildly acidic buffer (10 mM KAc, pH 5.0) and injected for 7 min at concentrations of 20–200 μg/ml onto a sensor chip previously activated by a 7-min incubation with a 1:1 mixture of EDC (0.2 M) and N-hydroxysuccinimide (NHS, 0.05 M). Finally, unreacted sites of the biosensor matrix were blocked by a 7-min injection of ethanolamine (1 M, pH 8.5). Subsequently, TWS119 was injected for 90 s at increasing concentrations (0, 2, 4, 8, 16, 32, 62.5, 125, 250, 500, and 1,000 nM), and dissociation of GSK-3β–TWS119 complexes was followed for 300 s. Evaluation of SPR data was performed by using the S51 evaluation software package (BIA-core, Piscataway, NJ). Data from two independent titration experiments performed on four different surfaces were averaged. A 1:1 binding model was assumed for the determination of thermodynamic and kinetic binding constants.
Results and Discussion
Small Molecule Screens of Neurogenesis in EC Cells. Mouse P19 EC cells were initially chosen for the primary screen because, like ESCs, they are pluripotent with a broad range of embryonic developmental potentials; P19 cells have been differentiated into neuronal, glial, cardiac, and skeletal muscle cells under proper conditions (ref. 12; ref. 13 and references therein). Moreover, P19 cells have a low frequency of spontaneous neuronal differentiation and are easy to culture, possess a normal karyotype, and are amenable to genetic manipulation. To screen large numbers (>105) of discrete compounds for those molecules that selectively induce neuronal differentiation, a luciferase reporter was constructed by inserting 1.1 kb of the regulatory region of neuronal Tα1 tubulin (14, 15), a specific neuronal marker, upstream of the luciferase gene. A single stable P19 clone transfected with the pTα1-Luc reporter afforded a 10- to 15-fold increase in the luciferase signal upon standard neuronal differentiation induced with all-trans RA. Whereas aggregation of P19 cells to form embryoid bodies (EB) is required for neuronal differentiation using RA, here, cells were screened in a P19 monolayer model based on the observation that activation of neuronal basic helix–loop–helix (bHLH) transcription factors such as NeuroD is sufficient to convert monolayers of P19 cells to neurons (16).
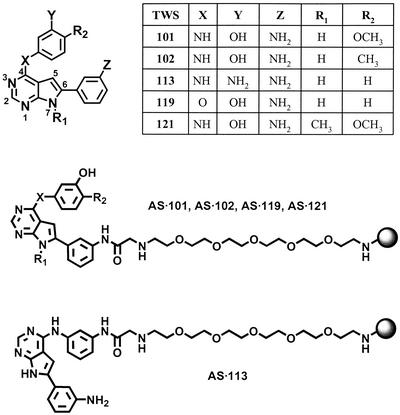
To carry out the primary screen, P19 cells were treated with 5 μM (final concentration) compounds in MEMα with 5% FBS 12 h after plating. Cells were treated for 4 days and lysed to determine luciferase activity. Primary hits were confirmed by direct immunostaining with βIII-tubulin/TuJ1 and observation of the characteristic neuronal morphology. A number of molecules were found to induce neuronal differentiation in P19 cell monolayers; the 4,6-disubstituted-pyrrolopyrimidine analog (Fig. 1) was chosen for further studies. A preliminary structural-activity relationship (SAR) analysis of the primary screen data revealed that the N7-H is critical (methylation completely abolishes activity); X can be nitrogen or oxygen (although oxygen substitution increases activity); Y and Z are OH or NH2 in all active analogs; and small non-polar groups such as methyl and methoxy are favored at R2. Based on this analysis, a focused library of pyrrolopyrimidines was synthesized and screened, and a molecule with improved activity (TWS119) was identified (Fig. 1).
Fig. 1.
Structures of 4,6-disubstituted pyrrolopyrimidines and affinity matrices.
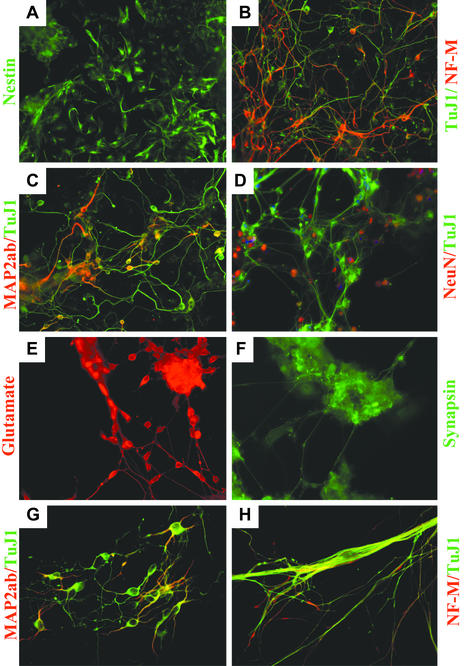
Phenotypic Characterization of Ligands. Treatment of a monolayer of P19 cells with 1 μM TWS119 caused 30–40% cells to differentiate specifically into neuronal lineages based on counting of TuJ1 positive cells (Fig. 2B) with correct neuronal morphology (up to 60% neuronal differentiation occurred through the standard EB formation protocol with concomitant TWS119 treatment). Most of the remaining TuJ1-negative cells immunostain positive for nestin (Fig. 2 A), indicating they are neural progenitor cells. Glial fibrillary acidic protein (GFAP; a glial marker) positive cells and MF20 (a muscle marker) positive cells were not detected under these conditions. Transient compound treatment for 2 days followed by incubation for an additional 2 days in compound-free medium resulted in a higher percentage of neurons (40–60%), suggesting that extended exposure of neural progenitors to early differentiation signals is unfavorable for late-stage neuronal maturation. Within hours of TWS119 treatment, cells began to grow as adherent ``aggregates,'' mimicking EB formation. As expected, the TuJ1-positive cells have a neuronal morphology with a round soma body and asymmetric multiple processes. Longer incubation times (up to 2 wk in B27-supplemented neurobasal medium) led to positive staining for neurofilament-M, Map2(a+b), NeuN, glutamate, and synapsin 1 (Fig. 2 B–F). The ability of TWS119 to induce neuronal differentiation is not limited to P19 EC cells, as treatment of primary mouse ESCs with 400 nM compound under similar conditions also effectively induces neuronal differentiation (≈50–60%) as indicated by immunostaining with TuJ1, Map2(a+b), and neurofilament-M (Fig. 2 G–H). Because neuronal differentiation can be achieved without EB formation and RA treatment, TWS119 may act by a novel mechanism on early processes involved in determining cell fate.
Fig. 2.
Induction of neurogenesis by TWS119. P19 cells were treated with 1–5 μM TWS119 for 2 days and cultured in compound-free MEMα supplemented with 2% FBS for an additional 2–14 days (B27-supplemented neurobasal medium was used for culturing longer than 10 days) before fixing and immunostaining with various neuronal specific antibodies. (A–F) Immunofluorescence staining for nestin, neurofilament-M/TuJ1, Map2(a+b)/TuJ1, NeuN/TuJ1, glutamate, and synapsin 1. (G–H) Under similar conditions, TWS119 can also induce murine ESCs (D3 cell line) to differentiate into neurons as indicated by immunofluorescence staining for neurofilament-M/TuJ1 and Map2(a+b)/TuJ1.
Target Identification. To further investigate the mechanism by which TWS119 induces neuronal differentiation, we attempted to identify the cellular targets of this compound by affinity chromatography (17). Structure–activity relationship data suggested that derivatization of the anilino group of TWS119 (Z = NHAc) does not adversely affect its ability to induce neurogenesis. Consequently, TWS119 was linked through this position to an agarose affinity matrix (Fig. 1). To distinguish specific binding to TWS119 vs. nonspecific interactions with the affinity matrix, a series of five affinity supports was used (Fig. 1). Three of the affinity supports were derived from TWS119 (AS·119) and its active analogs, TWS101 (AS·101) and TWS102 (AS·102); one (AS·121) was synthesized from an inactive analog TWS121 as negative control; and one (AS·113) contained TWS113 linked through a nonpermissive site (Fig. 1).
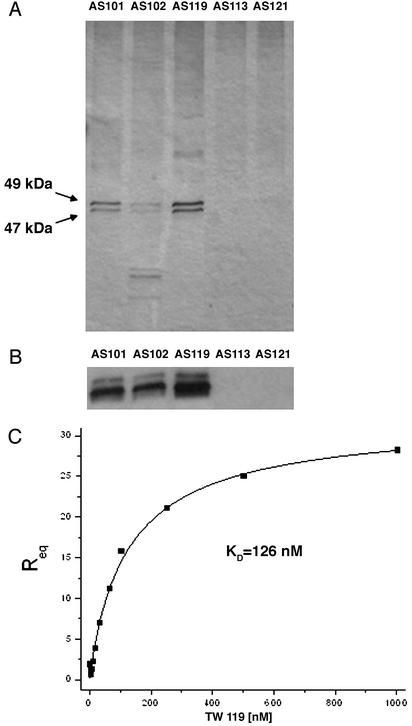
The proteins isolated with these affinity matrices were analyzed by SDS/PAGE. Two bands at ≈47 and 49 kDa were bound by the affinity matrices derived from the active TWS analogs, but were not retained on the underivatized matrix or matrices derivatized with the inactive analogues (Fig. 3A). Moreover, the addition of 50 μM TWS119 to the lysates during affinity chromatography effectively blocked binding of the 47- and 49-kDa proteins to the affinity matrices, further demonstrating specificity for TWS119. These two bands were identified to be GSK-3β by LC/MS, and possibly represent two different splicing isoforms (18) or posttranslationally modified products. More specifically, six individual tryptic-digest peptide fragments of GSK-3β were identified with estimated false positive rates of <1% (19). This result was confirmed independently by GSK-3β Western blot analysis (Fig. 3B). Moreover, recombinant GSK-3β bound selectively to the affinity supports derivatized with (and only with) the active analogue. This tight binding of TWS119 to GSK-3β (KD = 126 nM, Fig. 3C) was quantified by SPR and further demonstrated by its potent GSK-3β kinase inhibitory activity (IC50 = 30 nM).
Fig. 3.
Silver staining and Western blot analysis of proteins retained by affinity supports and SPR-based binding assay. (A) Total protein extracts from P19 cells lysed with PY buffer plus 0.4% Nonidet P-40 were loaded onto affinity supports AS·101, AS·102, AS·119, AS·113, and AS·121. After three washes with bead buffer, bound proteins were resolved by SDS/PAGE and revealed by silver staining. (B) Proteins retained by AS·101, AS·102, and AS·119 affinity supports were confirmed to be GSK-3β by Western blotting with anti-GSK-3β antibody. (C) SPR-based interaction analysis of TWS119 with recombinant GSK-3β. A GST-GSK-3β fusion protein was immobilized on an S series CM5 biosensor chip (≈17,000 relative light units) via EDC coupling reactions. Ligand binding with increasing concentrations of TWS119 (2–1,000 nM) were analyzed on a BIAcore S51 instrument. Analysis of the equilibrium binding responses was performed assuming a 1:1 binding model. The average affinity for TWS119 binding to GSK-3β was determined at KD = 126 ± 11 nM in four independent experiments.
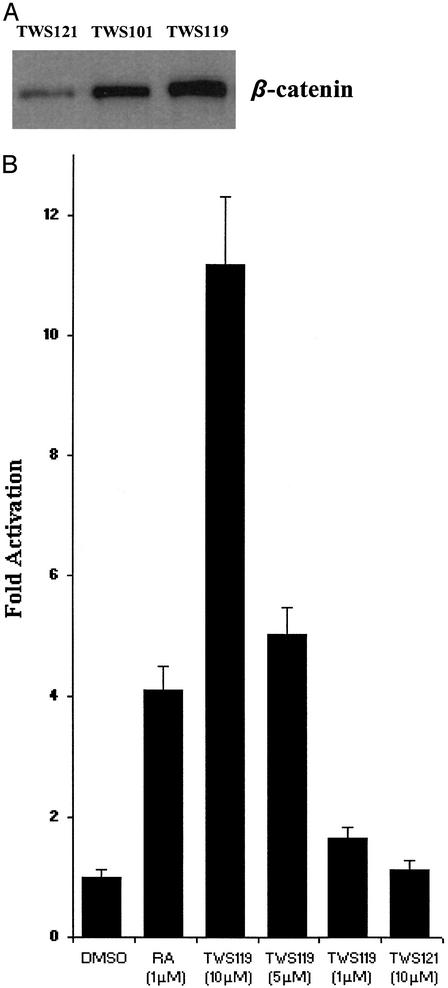
Mechanism of Action. To determine the effects of GSK-3β inhibition in P19 cells treated with TWS analogues, lysates were analyzed by Western blots, revealing that treatment with active analogs increased the level of β-catenin, a downstream substrate of GSK-3β in the Wnt signaling pathway, relative to the control treatment with TWS121 (Fig. 4A). Under normal cellular conditions, β-catenin is phosphorylated by GSK-3β, leading to ubiquitin-mediated degradation of the protein. On activation of the Wnt pathway, GSK-3β is inhibited, resulting in the accumulation and nuclear translocation of β-catenin, interaction with the TCF/LEF DNA binding proteins and regulation of transcription (20, 21). To confirm that GSK-3/β-catenin signaling is modulated in P19 cells by treatment with active TWS analogs, a TCF/LEF reporter assay (containing four consensus LEF-1/TCF-1 binding sites, a minimal promoter, and a luciferase reporter) (22) was carried out. TWS119 caused an activation of β-catenin-induced TCF/LEF reporter activity in a dose-dependent manner with an 11-fold increase at 10 μM after 36 h treatment (Fig. 4B). GSK-3β phosphorylates β-catenin in a tight complex containing axin and APC proteins. Hence, it is entirely possible that TWS119 binds to GSK-3β and modulates the activity of the complex, triggering downstream transcriptional events that lead the neuronal induction.
Fig. 4.
Effects on β-catenin-dependent signaling on treatment of P19 cells with TWS119. (A) β-catenin levels were analyzed by Western blot with anti-β-catenin antibody in P19 cells treated with TWS121 (24 h, 10 μM), TWS101 (24 h, 5 μM), and TWS119 (24 h, 3 μM). (B) pTOPFLASH luciferase reporter assay in a P19 monolayer treated with TWS analogs and P19 aggregates treated with RA: P19 cells were cotransfected with pTOPFLASH (containing four consensus LEF-1/TCF-1 binding sites, a minimal promoter, and a firefly luciferase reporter) and Renilla luciferase control reporter (Promega). After 24 h, cells were trypsinized and replated into a 96-well tissue culture plate and treated with TWS analogs, or plated into Petri dishes and treated with RA in MEMα with 5% FBS 12 h postplating. Thirty-six hours later, cells were lysed, and protein extracts were assayed for luciferase activity. The increase in firefly luciferase activity was normalized to Renilla luciferase activity and represents the average of three experiments.
GSK-3β is a multifunctional serine/threonine kinase involved in pattern formation during embryonic development, cell fate determination, transcriptional control, metabolism, oncogenesis, and neurological diseases. Unlike most protein kinases, GSK-3β is generally active and is primarily regulated by inactivation through various signaling pathways. Canonical Wnt signaling entails inactivation of GSK-3β, followed by nuclear translocation of β-catenin, which activates a LEF/TCF-mediated transcription (20, 21). Wnt signaling has recently been implicated in neural induction from pluripotent ES or EC cells. Ectopic expression of Wnt-1 inhibits neural differentiation in response to RA treatment (23). Removal of this inhibition by Sfrp2, a Wnt antagonist, has neural-inducing effects in mouse ES cells. In another report, however, overexpression of Wnt-1 induces neuronal differentiation of P19 aggregates in the absence of RA (24).
GSK-3β has also emerged as a crucial player in controlling bHLH transcription factors responsible for retinogenesis. In Xenopus, GSK-3β inhibits the bHLH transcription factor XNeu-roD through phosphorylation; a dominant negative GSK-3β resulted in neuronal differentiation of early retinal cells (25). Interestingly, the consensus sequence for GSK-3β phosphorylation is absent in mouse NeuroD sequence, raising the possibility that there might be other bHLH factors that are under the control of GSK-3β. Finally, there is evidence that shows Wnt is involved in regulating BMP signaling, and inhibition of BMP signaling is necessary to produce a neural fate (ref. 26 and references therein).
Alternatively, TWS119 might promote neural induction or survival of neuronal progenitors via novel mechanisms other than the canonical Wnt signaling pathway. Such mechanisms might include the inhibition of one or more kinases that were not apparent in the affinity experiments (possibly due to low abundance or other factors), or other proteins involved in controlling stem cell fate. Clearly, the precise details by which TWS119 functions will require additional structural, biochemical and cellular studies.
Summary. In summary, we have identified a small molecule from a phenotypic cellular screen that induces differentiation of pluripotent EC and ES cells to neurons. This molecule seems to act by modulation of GSK-3β functions. This and other molecules are likely to provide new insights into the molecular mechanisms that control stem cell fate, and may ultimately be useful to in vivo stem cell biology and therapy.
Acknowledgments
We thank Dr. David Anderson and Dr. Anne Bang for helpful discussions. Funding was provided by the Skaggs Institute for Chemical Biology (to P.G.S.), the Novartis Research Foundation (to N.S.G.), a predoctoral fellowship from the Howard Hughes Medical Institute (to S.D.), and a postdoctoral fellowship from the Humboldt Foundation (to A.B.).
Abbreviations: ESC, embryonic stem cell; EC, embryonal carcinoma; Map, microtubule-associated protein; SPR, surface plasmon resonance; GSK-3β, glycogen synthase kinase-3β; RA, retinoic acid; TCF, T cell factor; LEF, lymphoid enhancer factor; EDC, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide; EB, embryoid bodies; bHLH, basic helix–loop–helix.
References
- 1.Kim, J.-H., Auerbach, J. M., Rodríguez-Gómez, J. A., Velasco, I., Gavin, D., Lumelsky, N., Lee, S.-H., Nguyen, J., Sánchez-Pernaute, R., Bankiewicz, K. & McKay, R. (2002) Nature 418, 50–56. [DOI] [PubMed] [Google Scholar]
- 2.Lumelsky, N., Blondel, O., Laeng, P., Velasco, I., Ravin, R. & McKay, R. (2001) Science 292, 1389–1394. [DOI] [PubMed] [Google Scholar]
- 3.Tsai, R. Y. L., Kittappa, R. & McKay, R. (2002) Dev. Cell 2, 707–712. [DOI] [PubMed] [Google Scholar]
- 4.Wichterle, H., Lieberam, I., Porter, J. & Jessell, T. (2002) Cell 110, 385–397. [DOI] [PubMed] [Google Scholar]
- 5.Palmer, T. D., Takahashi, J. & Gage, F. H. (1997) Mol. Cell. Neurosci. 8, 389–404. [DOI] [PubMed] [Google Scholar]
- 6.Skillington, J., Choy, L. & Derynck, R. (2002) J. Cell Biol. 159, 135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosania, G. R., Chang, Y. T., Perez, O., Sutherlin, D., Dong, H., Lockhart, D. J. & Schultz, P. G. (2000) Nat. Biotechnol. 18, 304–308. [DOI] [PubMed] [Google Scholar]
- 8.Wu, X., Ding, S., Ding, Q., Gray, N. S. & Schultz, P. G. (2002) J. Am. Chem. Soc. 124, 14520–14521. [DOI] [PubMed] [Google Scholar]
- 9.Lee, S. H., Lumelsky, N., Auerbach, J. M. & McKay, R. D. (2000) Nat. Biotechnol. 18, 675–679. [DOI] [PubMed] [Google Scholar]
- 10.Zhang, S.-C., Wernig, M., Duncan, I. D., Brüstle, O. & Thomson, J. A. (2001) Nat. Biotechnol. 19, 1129–1133. [DOI] [PubMed] [Google Scholar]
- 11.Ding, S., Gray, N. S., Wu, X., Ding, Q. & Schultz, P. G. (2002) J. Am. Chem. Soc. 124, 1594–1596. [DOI] [PubMed] [Google Scholar]
- 12.Bain, G., Kitchens, D., Yao, M., Huettner, J. E. & Gottlieb, D. I. (1995) Dev. Biol. 168, 342–357. [DOI] [PubMed] [Google Scholar]
- 13.McBurney, M. W. (1993) Int. J. Dev. Biol. 37, 135–140. [PubMed] [Google Scholar]
- 14.Wang, S., Wu, H., Jiang, J., Delohery, T. M., Isdell, F. & Goldman, S. A. (1998) Nat. Biotechnol. 16, 196–201. [DOI] [PubMed] [Google Scholar]
- 15.Roy, N. S., Wang, S., Jiang, L., Kang, J., Benraiss, A., Harrison-Restelli, C., Fraser, R. A. R., Couldwell, W. T., Kawaguchi, A., Okano, H., et al. (2000) Nat. Med. 6, 271–277. [DOI] [PubMed] [Google Scholar]
- 16.Farah, M. H., Olson, J. M., Sucic, H. B., Hume, R. I., Tapscott, S. J. & Turner, D. L. (2000) Development 127, 693–702. [DOI] [PubMed] [Google Scholar]
- 17.Knockaert, M., Gray, N., Damiens, E., Chang, Y.-T., Grellier, P., Grant, K., Fergusson, D., Mottram, J., Soete, M., Dubremetz, J.-F., et al. (2000) Chem. Biol. 7, 411–422. [DOI] [PubMed] [Google Scholar]
- 18.Mukai, F., Ishiguro, K., Sano, Y. & Fujita, S. C. (2002) J. Neurochem. 81, 1073–1083. [DOI] [PubMed] [Google Scholar]
- 19.Peng, J., Elias, J. E., Thoreen, C. C., Licklider, L. J. & Gygi, S. P. (2003) J. Proteome Res. 2, 43–50. [DOI] [PubMed] [Google Scholar]
- 20.Li, L., Yuan, H., Weaver, C. D., Mao, J., Farr, G. H., III, Sussman, D. J., Jonkers, J., Kimelman, D. & Wu, D. (1999) EMBO J. 18, 4233–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moon, R. T., Bowerman, B., Boutros, M. & Perrimon, N. (2002) Science 296, 1644–1646. [DOI] [PubMed] [Google Scholar]
- 22.van de Wetering, M., Cavallo, R., Dooijes, D., van Beest, M., van Es, J., Loureiro, J., Ypma, A., Hursh, D., Jones, T., Bejsovec, A., et al. (1997) Cell 88, 789–799. [DOI] [PubMed] [Google Scholar]
- 23.Aubert, J., Dunstan, H., Chambers, I. & Smith, A. (2002) Nat. Biotechnol. 20, 1240–1245. [DOI] [PubMed] [Google Scholar]
- 24.Tang, K., Yang, J., Gao, X., Wang, C., Liu, L., Kitani, H., Atsumi, T. & Jing, N. (2002) Biochem. Biophys. Res. Commun. 293, 167–173. [DOI] [PubMed] [Google Scholar]
- 25.Moore, K. B., Schneider, M. L. & Vetter, M. L. (2002) Neuron 34, 183–195. [DOI] [PubMed] [Google Scholar]
- 26.Muñoz-Sanjuán, I. & Brivanlou, A. H. (2002) Nat. Rev. Neurosci. 3, 271–280. [DOI] [PubMed] [Google Scholar]