Abstract
Notch signaling is involved in numerous cell fate decisions in invertebrates and vertebrates. The Notch receptor is a type I transmembrane (TM) protein that undergoes two proteolytic steps after ligand binding, first by an ADAM (a distintegrin and metalloprotease) in the extracellular region, followed by γ-secretase-mediated cleavage inside the TM domain. We demonstrate here that the murine ligand Delta1 (Dll1) undergoes the same sequence of cleavages, in an apparently signal-independent manner. Identification of the ADAM-mediated shedding site localized 10 aa N-terminal to the TM domain has enabled us to generate a noncleavable mutant. Kuzbanian/ADAM10 is involved in this processing event, but other proteases can probably substitute for it. We then show that Dll1 is part of a high-molecular-weight complex containing presenilin1 and undergoes further cleavage by a γ-secretase-like activity, therefore releasing the intracellular domain that localizes in part to the nucleus. Using the shedding-resistant mutant, we demonstrate that this γ-secretase cleavage depends on prior ectodomain shedding. Therefore Dll1 is a substrate for regulated intramembrane proteolysis, and its intracellular region possibly fulfills a specific function in the nucleus.
The Notch receptor is part of an evolutionarily conserved signaling pathway involved in cell fate decisions through local cell–cell interactions (1). It is a large single-pass transmembrane (TM) receptor, matured in the secretory pathway by a convertase of the furin family (at a site called S1) (2), and presented at the cell surface as a heterodimeric molecule (2, 3). Notch ligands are divided into two subclasses, the Delta and the Serrate/Jagged families (4, 5). These ligands are TM proteins with a small intracellular (IC) domain, a large extracellular (EC) region comprising epidermal growth factor-like repeats, and an amino-terminal region DSL for Delta Serrat Log-2 that is specific to this class of proteins. Mammalian ligands include two members of the Serrate family, Jagged 1 and 2, and three members of the Delta family (Delta1, Delta3, and Delta4, also known as Dll1, Dll3, and Dll4). These molecules interact via their DSL domains with a specific region in the EC domain of Notch, causing a proteolytic cleavage event by a protease of the ADAM (a distintegrin and metalloprotease) family, TACE, at a site N-terminal to the TM domain (S2 site) (6, 7). The remaining membrane-tethered Notch fragment is then cleaved within its TM domain at the S3 site by a γ-secretase-like activity (8–10), leading to the release of the IC domain, which translocates into the nucleus, where it participates in transcriptional activation of target genes together with the CSL and the Mastermind gene products (11, 12).
Although the Notch pathway seems to be activated mainly through cell–cell interactions, proteolytic cleavage of both receptors and ligands has been shown to be important for signaling. Although the role of Notch processing seems to be adequately documented, that of ligand processing is a bit more difficult to fit into the current understanding of this signaling cascade. Genetic studies have implicated the gene Kuzbanian (Kuz) encoding a membrane metalloprotease of the ADAM family (also known as ADAM10, and closely related to TACE/ADAM17) in the Notch pathway (13–16). The precise role of Kuz in this pathway remains controversial, but its activity seems to be required for signaling. Recent data suggest that Drosophila Kuz can cleave Notch at the S2 site (17), whereas other evidence indicates that it is required for Delta processing, at least in Drosophila (18–20). Delta cleavage results in the shedding of its EC region, raising a controversial question about the function of this soluble EC region (see discussion in ref. 19). One way to address this question would be to identify the cleavage site and mutate it to generate a noncleavable form of the molecule.
Struhl and Adachi (21) have proposed that presenilin can mediate the cleavage of any type I TM protein, provided that the EC domain is short enough. Based on these data and the similarities in processing with other proteins such as β-amyloid precursor protein (APP) or Notch, which undergo an intramembranous cleavage after ectodomain shedding, we hypothesized that Dll1 could be a new substrate for presenilin-mediated intramembrane cleavage.
In the study presented here we have characterized in detail the processing event(s) that affect the murine Notch ligand Dll1. We have confirmed that it is constitutively cleaved in the apparent absence of signal, and that this cleavage is strongly diminished in Kuz–/– cells. We have identified the cleavage site and generated a noncleavable form of Dll1. Finally, we have demonstrated that Dll1 associates with presenilins and undergoes a γ-secretase-like cleavage, resulting in the release of its IC region, which then localizes in part to the nucleus. A mutation that blocks ADAM-mediated cleavage prevents the generation of the γ-secretase cleavage product, indicating that the former is required for the latter to take place. These results suggest that Dll1 is a substrate for regulated intramembrane proteolysis (22) and undergoes the same succession of proteolytic events that affect Notch during signaling, raising the issue of the possible role that the IC fragment of Dll1 might play in the nucleus.
Materials and Methods
Dll1 Constructs. A vesicular stomatitis virus (VSV) and a Flag tag were cloned into the SacII site and the MfeI site of mDll1, respectively, giving rise to the VSV-Flag-Dll1 (V-F-Dll1)/pcDNA3 construct. V-F-Dll1 was cloned into the BglII/XhoI sites of the murine stem cell virus-internal ribosomal entry site-GFP (MIG) vector (23). The V-F-Dll1-Apa construct was generated by deletion of 48 bp between two ApaI sites. The V-F-Dll1-D8 construct was generated by substitution of 8 aa to aspartate (SERHMESQ → D8). The Dll1IC-V5 construct was generated by cloning Dll1IC (Val-569 to Val-722) into pcDNA3.1-V5-His. The Dll1-Myc6 construct was a kind gift of J. Nye (Amersham Biosciences) (24). Further details on the constructs will be provided on request.
Cell Culture and Transfection. Kuz+/+ and Kuz–/– cells are mouse embryonic fibroblasts (MEFs) immortalized with simian virus 40 (6). Presenilin1 (PS1)+/+ and PS1–/– cells are MEF cells immortalized with large T antigen (25). Human embryonic kidney (HEK) 293T, HeLa, Plat-E, and MEF cells were cultured in DMEM supplemented with 10% FCS and blasticidin and puromycin for the Plat-E cell line. HEK293T cells were transiently transfected by using the calcium phosphate coprecipitation procedure and harvested 24 h later. HeLa and Plat-E cells were transfected by using Fugene (Roche Molecular Biochemicals).
Transduction of MEF Cells, Flow Cytometry, and Cell Sorting. High titers of empty (MIG) or recombinant (V-F-Dll1) viruses were obtained after transfection of the Plat-E ecotropic packaging cell line (26). Retroviruses containing supernatant were collected 48 h after transfection and added to 5 × 105 MEF cells. Retrovirally transduced MEF cells were collected 48 h later and analyzed for GFP expression by flow cytometry. GFP-positive cells were enriched by sorting on a MoFlo cytometer (DAKO), giving rise to a ≥98% pure population as determined by postsort analysis.
Preparation of Cell Extracts, Immunoprecipitation, and Immunoblotting. Whole-cell extracts, immunoprecipitations, and immunoblottings were carried out as described (2). For cellular subfractionation, cells were resuspended in an hypotonic buffer (20 mM Tris, pH 7.4/10 mM KCl/0.1 mM EDTA/protease inhibitors mixture). After 10 min, Nonidet P-40 was added to 0.15% and cell lysates were centrifuged at 800 × g for 5 min. The supernatant was ultracentrifuged at 105,000 × g for 1 h and constituted the cytosolic and membrane fractions. The nuclear pellet was resuspended in an extraction buffer containing 400 mM NaCl. After 30 min, the nuclear extract was recovered by centrifugation at 10,000 × g for 20 min. When mentioned, cells were treated with 5 μM lactacystin or 70 μM MW167.
Metabolic Labeling and Immunofluorescence. Pulse–chase experiments were performed as described (2). HeLa cells were stained as described (27), and images were captured by using an Axio-plan2 fluorescent microscope and an Axiocam digital camera (Zeiss).
Antibodies. For Dll1 antiserum, a peptide encoding amino acids 676–696 was coupled to keyhole limpet hemocyanin and injected into rabbits. This serum was diluted 1/4,000 for immunoblotting. The rabbit anti-PS1 antibody (a kind gift of L. Buée, Institut National de la Santé et de la Recherche Médicale, Lille, France) and anti-VSV (P5D4), anti-Myc (9E10), and anti-Flag (M2, Sigma) were diluted 1/200 for immunoprecipitations. Anti-Flag, anti-β-tubulin (Sigma), anti-GFP (Oncogene Science), and anti-CBF1/Su(H)/Log 1 (CSL) (7) were diluted 1/2,000 for immunoblotting. The anti-V5 antibody (Invitrogen) was diluted 1/200 for immunofluorescence.
Radiosequencing of Dll1TMIC. HEK293T cells were transfected with Dll1-Myc6 and radioactively labeled with [35S]Met (200 μCi/ml, 1,000 Ci/mmol) or [3H]Leu (50 μCi/ml, 161 Ci/mmol) for 4 h. After immunoprecipitation with the anti-Myc antibody, proteins were separated by SDS/PAGE and blotted on a poly(vinylidene difluoride) membrane. After autoradiography, the Dll1TMIC band was excised and subjected to radio sequencing on an Applied Biosystems 473A sequencer.
Analytical Gel Filtration. Whole extracts from HEK293T cells (1.5 × 107 cells) transfected with Dll1 were loaded on a Superose 6 column (Amersham Biosciences) preequilibrated with buffer (20 mM Tris, pH 8/0.3 M NaCl/5 mM MgCl2/0.3% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate) and calibrated with dimeric BSA (134 kDa) and ferritin (440 kDa). Fractions were collected and analyzed by immunoblotting with the Dll1 antiserum.
Results
Characterization of Dll1 Shedding. To investigate the mechanism of action of Notch ligands in mammals we undertook the analysis of murine Dll1 processing. We performed a pulse–chase analysis in HEK293T cells transiently expressing Dll1 with a VSV tag in the EC part and a Flag tag in the IC part (Fig. 1A). By immunoprecipitation with an anti-VSV antibody, we observed the progressive disappearance of the 85-kDa, full-length Dll1 that became undetectable at 6 h, in parallel with the appearance of a 55-kDa Dll1-soluble form in the medium (Dll1EC; Fig. 1B). A similar kinetics was observed for the appearance of a 30-kDa Dll1-derived cell-associated form (Dll1TMIC; Fig. 1C), revealed by anti-Flag immunoprecipitation. A progressive upshift of this form was noted and could be explained by some posttranslational modification: 32P labeling experiments indicate that Dll1TMIC is phosphorylated (data not shown). Therefore Dll1 undergoes an apparently constitutive processing, giving rise to a shed form Dll1EC and a membrane-associated fragment Dll1TMIC, as shown by biotinylation experiments (Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). This processing was also observed in a stable cell line expressing Dll1 (Fig. 2D, lane 2).
Fig. 1.
Murine Dll1 undergoes a constitutive ectodomain shedding. (A) Schematic map of the tagged Dll1 molecule used in B and C. (B and C) Pulse– chase analysis of Dll1. HEK293T cells were transfected with V-F-Dll1. After 24 h the cells were pulsed with [35S]Met (t0) for 20 min and chased for 0.5, 1, 2, 4, and 6 h. Cell extracts were immunoprecipitated with anti-VSV (B) or anti-Flag (C) antibodies. Culture media were immunoprecipitated with anti-VSV antibody (B Lower). Dll1 indicates full-length Dll1, Dll1TMIC is the membrane-associated processing product, and Dll1EC is the shedding product. Molecular mass markers are indicated on the left.
Fig. 2.
Identification of the EC cleavage site, generation of a Dll1 mutant for this cleavage, and processing of Dll1 in Kuz–/– cells. (A) Alignment of mDll1 with Dll1-D8 and Dll1-Apa mutants and Delta orthologs, in the juxtamembrane region (amino acids 516–545 of mDll1). The two cleavage sites identified by Mishra-Gorur et al. (19) in Drosophila Delta and the identified cleavage site of Dll1 are indicated. (B) Identification of the cleavage site. HEK293T cells were transfected with Delta-Myc6. After 24 h, cells were labeled with [3H]Leu or [35S]Met. Whole-cell extracts were immunoprecipitated with anti-Myc antibody. By radio sequencing of  a 35S peak was detected at cycle 1 and a 3H peak at cycle 19. The corresponding amino acid sequence is shown (Lower). (C) Analysis of the noncleavable Dll1-D8 and Dll1–Apa mutants. HEK293T cells were transfected with WT Dll1 (lane 2), Dll1-D8 (lane 3), or Dll1-Apa (lane 4), and whole-cell extracts were blotted with Dll1 antiserum. (D) Proteolytic cleavage of Dll1 is reduced in Kuz–/– cells. Kuz +/+ (lanes 1 and 2) and –/– (lanes 3 and 4) MEFs were infected with a retrovirus encoding GFP alone (MIG) or V-F-Dll1-internal ribosomal entry site-GFP (V-F-Dll1). GFP-enriched pools of cells were analyzed by immunoblotting of whole-cell extracts by using anti-Flag antibody. Equal protein loading was controlled by using an anti-GFP antibody (the level of GFP being correlated with the level of Dll1 expression).
a 35S peak was detected at cycle 1 and a 3H peak at cycle 19. The corresponding amino acid sequence is shown (Lower). (C) Analysis of the noncleavable Dll1-D8 and Dll1–Apa mutants. HEK293T cells were transfected with WT Dll1 (lane 2), Dll1-D8 (lane 3), or Dll1-Apa (lane 4), and whole-cell extracts were blotted with Dll1 antiserum. (D) Proteolytic cleavage of Dll1 is reduced in Kuz–/– cells. Kuz +/+ (lanes 1 and 2) and –/– (lanes 3 and 4) MEFs were infected with a retrovirus encoding GFP alone (MIG) or V-F-Dll1-internal ribosomal entry site-GFP (V-F-Dll1). GFP-enriched pools of cells were analyzed by immunoblotting of whole-cell extracts by using anti-Flag antibody. Equal protein loading was controlled by using an anti-GFP antibody (the level of GFP being correlated with the level of Dll1 expression).
Identification of Dll1 Cleavage Site and Generation of a Noncleavable Mutant. To isolate sufficient amounts of Dll1TMIC for microsequencing we took advantage of a 6×Myc-tagged Dll1 construct (24). HEK293T cells expressing Dll1-Myc6 were labeled with [35S]Met or [3H]Leu. After immunoprecipitation, the radiolabeled Dll1TMIC was subjected to automated Edman degradation. The major peaks of 35S and 3H radioactivity were in fractions 1 and 19, respectively. This process allowed us to conclude that Dll1 is cleaved 10 aa N-terminal to the TM domain, between His-535 and Met-536 (Fig. 2 A and B), a juxtamembrane localization consistent with known shedding sites of other type I TM proteins.
To generate a noncleavable mutant we undertook the mutagenesis of this region. Because point mutations at or around the cleavage site did not affect processing, we generated a mutant with a substitution of 8 aa to aspartate, Dll1-D8, and a mutant deleted of 16 aa in the region covering the cleavage site, Dll1-Apa (Fig. 2 A). These mutants were expressed in HEK293T cells and their processing was monitored with the Dll1 antiserum. The appearance of the Dll1TMIC form was almost completely abolished (Fig. 2C). The proper membrane localization of these mutants was confirmed by immunofluorescence analysis (data not shown) and biotinylation experiments for Dll1-Apa (Fig. 6).
Kuz/ADAM10 Is Involved in Dll1 Processing. Kuz/ADAM10 has been suggested to mediate Delta processing in Drosophila. To assess the implication of Kuz in murine Dll1 processing we used Kuz–/– and Kuz+/+ MEFs (6). These cell lines were infected with MIG retrovirus expressing V-F-Dll1 or with a control MIG retrovirus. GFP-expressing cells were enriched by flow cytometry, and Dll1 processing was monitored by using the anti-Flag antibody. In Kuz–/– cells production of Dll1TMIC was reduced by at least 50% in comparison to Kuz+/+ cells (Fig. 2D), while the full-length Dll1 accumulated. This experiment demonstrates that ADAM10 is partially responsible for Dll1 processing and that another metalloprotease probably accounts for the remaining processing in Kuz–/– cells. One good candidate could be ADAM17 (TACE), the closest relative to ADAM10 (28).
Dll1 Is Part of a Macromolecular Complex Containing PS1. To get further insight into the physiology of this processing event, we investigated the state of oligomerization and the distribution of Dll1 and its membrane-associated processing product on a sizing column. By coimmunoprecipitation experiments, we first demonstrated that full-length Dll1 can form homodimers (Fig. 7, which is published as supporting information on the PNAS web site). We then analyzed the full-length and Dll1TMIC forms of Dll1 by gel filtration on a Sepharose 6 column. Analysis of the column fractions with the Dll1 antiserum revealed two distinct elution profiles. The first one containing full-length Dll1 peaked around an apparent molecular mass of 300–400 kDa (fraction 17), whereas the second one containing Dll1TMIC peaked ≈130 kDa (fraction 23) (Fig. 3A Upper). This finding suggests that Dll1 and Dll1TMIC are part of multimolecular complexes. Because there is accumulating evidence that shed TM proteins are often substrates for presenilin-dependent γ-secretase cleavage, we tested the presence of PS1 in the different fractions. Immunoblotting with PS1 antiserum revealed a profile overlapping the distribution of the Dll1 full-length protein (Fig. 3A Lower). To confirm that Dll1 and PS1 can be found in the same complex, we performed coimmunoprecipitation experiments using PS1 antiserum on extracts of cells cotransfected with Dll1 and PS1. Immunoblotting with the Dll1 antiserum revealed that full-length Dll1 coprecipitated with PS1, but not the endoproteolytic fragment Dll1TMIC (Fig. 3B, lane 3). The reciprocal experiment allowed us to show that endogenous PS1 was associated with transfected V-F-Dll1 after anti-Flag immunoprecipitation (data not shown). Taken together, these results demonstrate that full-length Dll1 and PS1 can be found in the same high-molecular-weight complex, and that Dll1TMIC is part of a distinct smaller complex (see Discussion).
Fig. 3.
Full-length Dll1 and its proteolytic fragment Dll1TMIC are present in distinct complexes. (A) Gel filtration analysis of extracts from 293T cells transfected with Dll1. Whole-cell extracts were fractionated through a Superose 6 column. Fractions were analyzed by immunoblotting, using Dll1 antiserum (Upper), followed by PS1 antiserum (Lower) after membrane stripping. Positions of full-length Dll1, Dll1TMIC, and the N-terminal processing product of PS1 (PS1 NTF) are indicated on the right. (B) Dll1 interacts with PS1. HEK293T cells were transfected with plasmids encoding V-F-Dll1 and/or PS1 as indicated. Whole-cell extracts (WE; lane 1) and anti-PS1 immunoprecipitates (IP anti-PS1; lanes 2–4) were analyzed on SDS/PAGE and immunoblotted with an anti-Flag antibody. The position of Igs is indicated on the right.
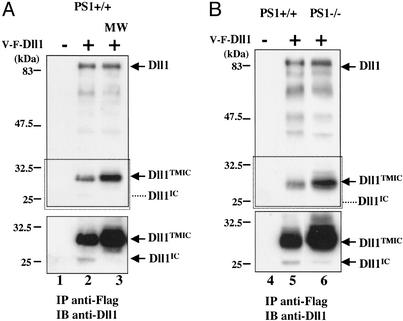
Dll1 Is a Substrate for Presenilin-Dependent γ-Secretase Activity. To address the question of a possible PS1-dependent cleavage of Dll1, we established stable cell lines expressing V-F-Dll1-internal ribosomal entry site-GFP or a control MIG construct in PS1–/– and PS1+/+ MEFs (25). Immunoprecipitation with an anti-Flag antibody, followed by immunoblotting with Dll1 antiserum revealed the existence of a Dll1IC form migrating ≈25 kDa in PS1+/+ cells (Fig. 4A, lane 2). This Dll1IC form was absent in cells treated with the specific γ-secretase inhibitor MW167, whereas the Dll1TMIC form was stabilized (Fig. 4A, compare lanes 2 and 3). Moreover in PS1–/– cells this Dll1IC form was hardly detectable, whereas the level of Dll1TMIC was increased (Fig. 4B, compare lanes 5 and 6). This experiment demonstrates that Dll1 undergoes a presenilin-dependent γ-secretase cleavage that releases a Dll1IC form. To determine the subcellular localization of this Dll1IC form we prepared membrane, cytosolic, and nuclear fractions from HEK293T cells transiently transfected with V-F-Dll1 and treated with lactacystin, a proteasome inhibitor. Analysis of these fractions with an anti-Flag antibody revealed the presence of the DllIC form in the soluble fractions (cytosol and nucleus) (Fig. 5A, lanes 4 and 7). Moreover, immunofluorescence analysis of HeLa cells transfected with a Dll1IC-V5 construction demonstrated that Dll1IC was localized mainly to the nucleus (Fig. 5B).
Fig. 4.
Dll1 is cleaved by a presenilin-dependent γ-secretase activity. PS1 +/+ and –/– MEFs were infected by a retrovirus encoding GFP alone (–) or V-F-Dll1-internal ribosomal entry site-GFP (+). (A) Dll1IC release is inhibited by MW167. GFP-enriched pools of PS1+/+ cells were analyzed by immunoprecipitation using the anti-Flag antibody followed by immunoblotting with the Dll1 antiserum. (Upper) Short exposure. (Lower, corresponding to the boxed region) Long exposure. Dll1IC indicates the position of the soluble IC fragment. In lane 3, PS1+/+ cells were treated for 15 h in the presence of the γ-secretase inhibitor MW167 (MW) before lysis. (B) Absence of Dll1IC release in PS1–/– cells. A similar experiment to lanes 1 and 2 of A was performed in parallel in PS1+/+ (lanes 4 and 5) and PS1–/– (lane 6) cells.
Fig. 5.
Generation of Dll1TMIC is a preliminary requirement for γ-secretase cleavage to occur. (A) Dll1-Apa and Dll1-D8 do not undergo γ-secretase cleavage. HEK293T cells were transfected with V-F-Dll1 (lanes 1, 4, and 7), V-F-Dll1-Apa (lanes 2, 5, and 8), and V-F-Dll1-D8 (lanes 3, 6, and 9) and treated with lactacystin for 3 h. Membrane, cytosolic, and nuclear extracts were prepared and analyzed by immunoblotting using the anti-Flag antibody, followed by immunoblotting with anti-β-tubulin (a cytosolic marker) and anti-CSL (a nuclear marker) antibodies as a control for the purity of the fractions and equal protein loading. (B) Dll1IC localized to the nucleus. HeLa cells were transfected with Dll1IC-V5 and treated with lactacystin for 3 h. Cells were stained with the anti-V5 antibody (Left) and Hoechst for nuclear staining (Center). Light blue in the merged image (Right) indicates colocalization.
We then demonstrated that this cleavage did not occur in the shedding-resistant mutants Dll1-Apa and Dll1-D8 (Fig. 5A, compare lanes 4 and 7 with 5 and 8 and 6 and 9). Therefore, the generation of the Dll1TMIC form is a preliminary requirement for γ-secretase cleavage. From these data we can conclude that Dll1 undergoes two consecutive processing events: a shedding event that generates a soluble EC form and an IC membrane-anchored form, followed by a γ-secretase cleavage releasing an IC fragment that localizes in part to the nucleus.
Discussion
The results presented here indicate that the Notch ligand Dll1 is constitutively cleaved, first by an ADAM (Kuz being involved in this cleavage) at a site located 10 aa N-terminal to the TM region, and then by a γ-secretase-like activity that releases the IC part of the molecule (Dll1IC), which then translocates, at least in part, to the nucleus. Impairment of ADAM cleavage prevents the appearance of Dll1IC, indicating that ectodomain shedding is required before the γ-secretase cleavage step.
The exact function of Notch ligands ectodomain shedding is unclear. Recent data suggest that ligand processing results in its inactivation, this event being required for the establishment of a distinction between receiving and signaling cells, a prerequisite for effective Notch signaling (19). The cleavage of mammalian Dll1 in the EC region has been observed, but not characterized in detail (24). Genetic data suggest that Kuz is required for Notch signaling, both in the emitting and the receiving cell (13–16). However, as published data suggest that Kuz is involved in the cleavage of both Notch and its ligands, it is impossible to conclude that ligand cleavage is absolutely required for Notch signaling.
In a recent paper, Mishra-Gorur et al. (19) have identified two cleavage sites of Drosophila Delta introduced into S2 cells, respectively, 2 and 14 aa N-terminal to the TM domain. No sequence similarity can be found between the sites used for Drosophila and murine Delta, but this is not very surprising as ADAMs seem to recognize a structure more than a primary sequence; in addition, very little sequence conservation can be found between the juxtamembrane regions of Delta orthologs (see Fig. 2 A). This lack of sequence specificity has already been reported for other ADAM's substrates such as Ephrin (29) and is confirmed by the multiple unsuccessful point mutations we tested before we could isolate a noncleavable mutant.
The question remains as to which member of the ADAM family is responsible for Dll1 cleavage. Our experiments using Kuz–/– cells suggest that this metalloprotease is involved, but is not the exclusive one. The possibility exists that depending on the cell type, either Kuz/ADAM10 or TACE/ADAM17 can cleave Dll1 (and possibly other ligands). The same probably applies to Notch or APP (30).
One important point is whether the ADAM cleavage observed is constitutive or inducible. At the moment it is difficult to answer this question as multiple soluble or membrane-associated ``ligands'' might be expressed by HEK293T cells. The same applies to APP, which seems to be processed by α-secretase in the absence of external stimulus. Of course, it would be interesting to determine whether the EC region of Notch, which has been postulated to be transendocytosed by Delta-expressing cells during signaling in Drosophila (31), somehow modulates Delta processing.
Having characterized in detail this cleavage event, we decided to push the analogy with Notch further and determine whether Dll1 cleavage by an ADAM might be followed by an intramembrane γ-secretase processing event involving presenilins. Several type I TM proteins have already been shown to undergo ectodomain shedding by an ADAM and subsequent cleavage by γ-secretase, including Notch, APP, ErbB4, CD44, low-density lipoprotein receptor-related protein, and E-cadherin (32–35). We indeed observed that a fragment migrating faster than Dll1TMIC could be detected in a stable cell line expressing Dll1, that the generation of this processing product was completely abolished by the γ-secretase inhibitor MW167, and that this product was hardly detectable in PS1–/– cells. The weak remaining cleavage in the latter can be explained by the presence of PS2. We conclude from these data that Dll1 is a substrate for presenilin-dependent γ-secretase cleavage. In addition, Dll1 was found to be associated with PS1. The lack of PS1 association with Dll1TMIC is a bit unexpected. A possible explanation is that after ADAM cleavage, PS1/Dll1TMIC association is only transient and disappears when γ-secretase cleavage is completed, whereas the pool of Dll1TMIC we detect is not directed toward further cleavage.
The mutation that abolishes the ADAM cleavage of Dll1 was shown to prevent the appearance of this fast-migrating species, strongly suggesting that the ADAM cleavage is required for the subsequent cleavage step to occur. We also demonstrated through subcellular fractionation that the released Dll1IC was present in the cytosolic and nuclear fraction, which was confirmed by the predominant nuclear localization observed by immunofluorescence. It can be noted in this latter experiment that a small amount of Dll1IC localized to the plasma membrane, possibly as a consequence of the presence of a highly conserved PDZ-binding domain in the very C-terminal part of Dll1.
The localization of the γ-secretase cleavage site has not been precisely determined, but it is interesting to notice that a conserved Val residue is present 4 aa N-terminal to the end of the TM region of Delta orthologs, a position identical to the Val residue recognized by γ-secretase during Notch cleavage.
An obvious question relates to the role of the γ-secretase cleavage in Dll1 metabolism. In the case of Notch and possibly also of APP, the signaling role of the IC region has been convincingly established. Therefore the possibility exists that after processing, the IC region of Dll1 plays some specific role in the nucleus, whether transcriptional or otherwise. Interestingly a certain number of experiments have suggested that Notch ligands might play a role in cis in the cells where they are expressed, in connection with the presence of the PDZ-binding domain mentioned above (36), although this role might be more connected to a structural function at the plasma membrane, through modulation of cell adhesion (37).
A putative transcriptional role of the IC region of Dll1 could be assayed by gene profiling experiments, allowing us to determine whether the putative Dll1 target genes are also Notch target genes or whether a different set of genes are modulated.
While this manuscript was under revision, two groups reported that Drosophila Delta and murine Dll1 are substrates for γ-secretase cleavage (38, 39).
In conclusion, we have shown that the Notch ligand Dll1 is subjected to an apparently constitutive series of processing events that lead to the nuclear translocation of the IC region of the molecule. Future studies will have to determine whether these events are indeed constitutive or regulated by some soluble or membrane-associated ligand (such as Notch), whether they modulate Notch signaling, and which role is played by the IC region of Dll1 once in the nucleus.
Supplementary Material
Acknowledgments
We are grateful to P. Saftig and B. De Strooper for the kind gift of PS1–/– MEF cells, D. J. Pan for the kind gift of the Kuz–/– MEF cells, and T. Kitamura for providing the Plat-E cell line. We thank L. Pradier for the PS1 construct, L. Buée for the anti-PS1 antiserum, S. Goffinont for assistance with the gel filtration analysis, and the Institut Pasteur flow cytometry team for cell sorting. This work was supported in part by a grant from the Association pour la Recherche sur le Cancer (to A.I.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ADAM, a distintegrin and metalloprotease; APP, β-amyloid precursor protein; Dll1, Delta1; TM, transmembrane; IC, intracellular; EC, extracellular; MEF, mouse embryonic fibroblast; MIG, murine stem cell virus-internal ribosomal entry site-GFP; PS1, presenilin1; Kuz, Kuzbanian; VSV, vesicular stomatitis virus; V-F-Dll1, VSV-Flag-Dll1; HEK, human embryonic kidney; CSL, CBF1/SU(H)/Log 1.
References
- 1.Artavanis-Tsakonas, S., Rand, M. D. & Lake, R. J. (1999) Science 284, 770–776. [DOI] [PubMed] [Google Scholar]
- 2.Logeat, F., Bessia, C., Brou, C., Lebail, O., Jarriault, S., Seidah, N. G. & Israël, A. (1998) Proc. Natl. Acad. Sci. USA 95, 8108–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaumueller, C. M., Qi, H. L., Zagouras, P. & Artavanis-Tsakonas, S. (1997) Cell 90, 281–291. [DOI] [PubMed] [Google Scholar]
- 4.Fleming, R. J., Purcell, K. & Artavanis-Tsakonas, S. (1997) Trends Cell Biol. 7, 437–441. [DOI] [PubMed] [Google Scholar]
- 5.Lendahl, U. (1998) BioEssays 20, 103–107. [DOI] [PubMed] [Google Scholar]
- 6.Mumm, J. S., Schroeter, E. H., Saxena, M. T., Griesemer, A., Tian, X., Pan, D. J., Ray, W. J. & Kopan, R. (2000) Mol. Cell 5, 197–206. [DOI] [PubMed] [Google Scholar]
- 7.Brou, C., Logeat, F., Gupta, N., Bessia, C., LeBail, O., Doedens, J. R., Cumano, A., Roux, P., Black, R. A. & Israël, A. (2000) Mol. Cell 5, 207–216. [DOI] [PubMed] [Google Scholar]
- 8.Song, W. H., Nadeau, P., Yuan, M. L., Yang, X. D., Shen, J. & Yankner, B. A. (1999) Proc. Natl. Acad. Sci. USA 96, 6959–6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Struhl, G. & Greenwald, I. (1999) Nature 398, 522–525. [DOI] [PubMed] [Google Scholar]
- 10.De Strooper, B., Annaert, W., Cupers, P., Saftig, P., Craessaerts, K., Mumm, J. S., Schroeter, E. H., Schrijvers, V., Wolfe, M. S., Ray, W. J., et al. (1999) Nature 398, 518–522. [DOI] [PubMed] [Google Scholar]
- 11.Jarriault, S., Brou, C., Logeat, F., Schroeter, E., Kopan, R. & Israël, A. (1995) Nature 377, 355–358. [DOI] [PubMed] [Google Scholar]
- 12.Wu, L. Z., Aster, J. C., Blacklow, S. C., Lake, R., Artavanis-Tsakonas, S. & Griffin, J. D. (2000) Nat. Genet. 26, 484–489. [DOI] [PubMed] [Google Scholar]
- 13.Hartmann, D., de Strooper, B., Serneels, L., Craessaerts, K., Herreman, A., Annaert, W., Umans, L., Lubke, T., Lena Illert, A., von Figura, K., et al. (2002) Hum. Mol. Genet. 11, 2615–2624. [DOI] [PubMed] [Google Scholar]
- 14.Rooke, J., Pan, D., Xu, T. & Rubin, G. M. (1996) Science 273, 1227–1231. [DOI] [PubMed] [Google Scholar]
- 15.Wen, C. H., Metzstein, M. M. & Greenwald, I. (1997) Development (Cambridge, U.K.) 124, 4759–4767. [DOI] [PubMed] [Google Scholar]
- 16.Sotillos, S., Roch, F. & Campuzano, S. (1997) Development (Cambridge, U.K.) 124, 4769–4779. [DOI] [PubMed] [Google Scholar]
- 17.Lieber, T., Kidd, S. & Young, M. W. (2002) Genes Dev. 16, 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi, H. L., Rand, M. D., Wu, X. H., Sestan, N., Wang, W. Y., Rakic, P., Xu, T. & Artavanis-Tsakonas, S. (1999) Science 283, 91–94. [DOI] [PubMed] [Google Scholar]
- 19.Mishra-Gorur, K., Rand, M. D., Perez-Villamil, B. & Artavanis-Tsakonas, S. (2002) J. Cell Biol. 159, 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klueg, K. M., Parody, T. R. & Muskavitch, M. A. (1998) Mol. Biol. Cell 9, 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Struhl, G. & Adachi, A. (2000) Mol. Cell 6, 625–636. [DOI] [PubMed] [Google Scholar]
- 22.Brown, M. S., Ye, J., Rawson, R. B. & Goldstein, J. L. (2000) Cell 100, 391–398. [DOI] [PubMed] [Google Scholar]
- 23.Van Parijs, L., Refaeli, Y., Lord, J. D., Nelson, B. H., Abbas, A. K. & Baltimore, D. (1999) Immunity 11, 281–288. [DOI] [PubMed] [Google Scholar]
- 24.Franklin, J. L., Berechid, B. E., Cutting, F. B., Presente, A., Chambers, C. B., Foltz, D. R., Ferreira, A. & Nye, J. S. (1999) Curr. Biol. 9, 1448–1457. [DOI] [PubMed] [Google Scholar]
- 25.De Strooper, B., Saftig, P., Craessaerts, K., Vanderstichele, H., Guhde, G., Annaert, W., Von Figura, K. & Van Leuven, F. (1998) Nature 391, 387–390. [DOI] [PubMed] [Google Scholar]
- 26.Morita, S., Kojima, T. & Kitamura, T. (2000) Gene Ther. 7, 1063–1066. [DOI] [PubMed] [Google Scholar]
- 27.Jarriault, S., Le Bail, O., Hirsinger, E., Pourquié, O., Logeat, F., Strong, C. F., Brou, C., Seidah, N. G. & Israël, A. (1998) Mol. Cell. Biol. 18, 7423–7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seals, D. F. & Courtneidge, S. A. (2003) Genes Dev. 17, 7–30. [DOI] [PubMed] [Google Scholar]
- 29.Hattori, M., Osterfield, M. & Flanagan, J. G. (2000) Science 289, 1360–1365. [DOI] [PubMed] [Google Scholar]
- 30.Asai, M., Hattori, C., Szabo, B., Sasagawa, N., Maruyama, K., Tanuma, S. & Ishiura, S. (2003) Biochem. Biophys. Res. Commun. 301, 231–235. [DOI] [PubMed] [Google Scholar]
- 31.Parks, A. L., Klueg, K. M., Stout, J. R. & Muskavitch, M. A. (2000) Development (Cambridge, U.K.) 127, 1373–1385. [DOI] [PubMed] [Google Scholar]
- 32.Marambaud, P., Shioi, J., Serban, G., Georgakopoulos, A., Sarner, S., Nagy, V., Baki, L., Wen, P., Efthimiopoulos, S., Shao, Z., et al. (2002) EMBO J. 21, 1948–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lammich, S., Okochi, M., Takeda, M., Kaether, C., Capell, A., Zimmer, A. K., Edbauer, D., Walter, J., Steiner, H. & Haass, C. (2002) J. Biol. Chem. 277, 44754–44759. [DOI] [PubMed] [Google Scholar]
- 34.May, P., Reddy, Y. K. & Herz, J. (2002) J. Biol. Chem. 277, 18736–18743. [DOI] [PubMed] [Google Scholar]
- 35.Ni, C. Y., Murphy, M. P., Golde, T. E. & Carpenter, G. (2001) Science 294, 2179–2181. [DOI] [PubMed] [Google Scholar]
- 36.Ascano, J. M., Beverly, L. J. & Capobianco, A. J. (2003) J. Biol. Chem. 278, 8771–8779. [DOI] [PubMed] [Google Scholar]
- 37.Lowell, S. & Watt, F. M. (2001) Mech. Dev. 107, 133–140. [DOI] [PubMed] [Google Scholar]
- 38.Ikeuchi, T. & Sisodia, S. S. (2003) J. Biol. Chem. 278, 7751–7754. [DOI] [PubMed] [Google Scholar]
- 39.Bland, C. E., Kimberly, P. & Rand, D. (2003) J. Biol. Chem. 278, 13607–13610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.