Abstract
The evolutionarily conserved Mdm20 protein (Mdm20p) plays an important role in tropomyosin–F-actin interactions that generate actin filaments and cables in budding yeast. However, Mdm20p is not a structural component of actin filaments or cables, and its exact function in cable stability has remained a mystery. Here, we show that cells lacking Mdm20p fail to N-terminally acetylate Tpm1p, an abundant form of tropomyosin that binds and stabilizes actin filaments and cables. The F-actin-binding activity of unacetylated Tpm1p is reduced severely relative to the acetylated form. These results are complemented by the recent report that Mdm20p copurifies with one of three acetyltransferases in yeast, the NatB complex. We present genetic evidence that Mdm20p functions cooperatively with Nat3p, the catalytic subunit of the NatB acetyltransferase. These combined results strongly suggest that Mdm20p-dependent, N-terminal acetylation of Tpm1p by the NatB complex is required for Tpm1p association with, and stabilization of, actin filaments and cables.
Tropomyosin is an actin-binding protein found in virtually all eukaryotic cells (1). In sarcomeric muscle it acts together with troponin to regulate myosin access to the actin filament (2). In nonmuscle cells, which lack troponin, its function is believed to consist primarily of filament stabilization (3, 4). Tropomyosin exists in the cell as a parallel coiled-coil dimer that binds longitudinally to the actin filament and spans an integral number of actin monomers (four to seven, depending on cell type) (1, 5). In addition, the protein interacts with itself in head-to-tail fashion, forming a spiraling polymer (6).
Although tropomyosin appears to have a number of quasi-equivalent actin-binding sites along its length, in actuality, the N- and C-terminal regions are especially important for determining the strength of the tropomyosin–actin interaction (6). Early attempts to overexpress vertebrate muscle tropomyosin in bacteria led to the observation that native tropomyosin is acetylated at its N terminus (7). Without this modification, the actin-binding and polymerization capabilities of the protein are compromised severely (7, 8). Analysis of the three-dimensional structure of the tropomyosin N terminus indicates that acetylation allows the α-helical conformation of the protein to extend to the initial Met-1 residue (9). In contrast, the structure of an unacetylated tropomyosin fragment shows the first two residues of the N terminus in a nonhelical conformation that disrupts the coiled-coil domain at the protein extremity (10).
The budding yeast Saccharomyces cerevisiae contains two tropomyosin genes, TPM1 and TPM2 (5, 11). Both the Tpm1 protein (Tpm1p) and the less abundant Tpm2 protein (Tpm2p) have been shown to stabilize actin filaments that subsequently are bundled together to form actin cables (4). Although tpm2 mutations have little effect on gross actin cable structure in yeast cells, tpm1 mutants lack visible actin cables (5, 11). Recent work established that yeast Tpm1p is also acetylated N-terminally, and that this acetylation is critical for normal binding and stabilization of filamentous actin (12). Unlike acetylated Tpm1p, unacetylated Tpm1p exhibits a drastically reduced affinity for F-actin and is incapable of restoring filament formation of polymerization-defective mutant actin (4, 12).
The addition of extra amino acids to the N terminus of muscle and nonmuscle tropomyosins can mimic the effect of N-terminal acetylation (13, 14). Three mutant forms of yeast Tpm1p containing different N-terminal extensions (2–5 aa) exhibited F-actin-binding affinities and binding cooperativities similar to the WT protein (12). The most significant difference between the three mutant forms was their preferred conformational state on F-actin (open or closed), which controls access to the myosin motor-binding site on the actin filament (12).
Although N-terminal acetylation is a common cotranslational modification of cytoplasmic proteins in eukaryotes (15, 16), the functional significance of this modification has been determined in only a small number of cases. In budding yeast, the three N-terminal acetyltransferases are multiprotein complexes known as NatA, NatB, and NatC (15, 16, 34, 35). Three sequence-related proteins, Ard1p, Nat3p, and Mak3p, appear to be the catalytic subunits of the three enzymes, respectively, although other proteins also are required for function. Analysis of the yeast N-terminal acetyltransferases, using knockout strains and an extensive set of substrate proteins, revealed that their substrate specificity is largely dependent on the amino acid identity of the second (sometimes in combination with the third) residue of a target polypeptide (15, 17). Based on these observations, Tpm1p, which has an aspartic acid in the second position, is predicted to be acetylated by Nat3p within the NatB complex.
Over the past decade, evidence has accumulated steadily that mitochondrial inheritance in budding yeast is accomplished by transporting the organelle along actin filaments and cables that extend from the mother cell into the growing bud (18). Yeast cells lacking Tpm1p lose visible actin cables and exhibit significant mitochondrial inheritance defects (19). Additional support for this model comes from studies of the MDM20 gene (MDM, mitochondrial distribution and morphology). Genetic analysis indicates that MDM20 is required for both actin organization and mitochondrial inheritance; mdm20 mutants lack visible actin cables and exhibit a severe temperature-sensitive defect in mitochondrial inheritance and growth (20). These and other studies (21, 22) suggest that MDM20's impact on mitochondrial inheritance is secondary and that its primary cellular role is to establish or maintain the stability of actin filaments and cables. In support of this idea, a variety of dominant mutations in TPM1 or ACT1 (encoding yeast actin) partially restore actin cables and suppress temperature-sensitive mitochondrial inheritance defects and growth defects in an mdm20 mutant (23). However, Mdm20p does not appear to be a structural component of actin filaments or cables and, instead, behaves like a soluble cytoplasmic protein in localization and biochemical fractionation experiments (refs. 20 and 24; J. M. Singer, unpublished data). Although the identification of Mdm20p orthologs in humans, flies, worms, plants, and fission yeast suggests that the function of this protein has been conserved during evolution (J. M. Singer, unpublished data), the mechanism by which it regulates actin cable stability remains unclear.
The recent report that Mdm20p is a component of the NatB acetyltransferase complex (ref. 16; B. Polevoda, T. S. Cardillo, T. C. Doyle, G. S. Bedi, and F. Sherman, personal communication) raised the possibility that Mdm20p was required for the N-terminal acetylation of one or more proteins that contribute to the stabilization of actin filaments and cables. In this study, we show that yeast Mdm20p is required for the N-terminal acetylation of Tpm1p and that normal binding of Tpm1p to F-actin requires this cotranslational modification. We also present genetic evidence that Mdm20p functions cooperatively with Nat3p, the catalytic subunit of the NatB acetyltransferase. Our results identify a new molecular target of the NatB acetyltransferase complex and provide direct evidence that Mdm20p-dependent, N-terminal acetylation of Tpm1p is essential for actin filament and cable stability in yeast.
Materials and Methods
Yeast Strains, Media, and Genetic Techniques. S. cerevisiae strains were derived from the FY strain background (25). Standard yeast genetic techniques were used (26). Rich medium (yeast extract/peptone/dextrose) and synthetic medium (SD = 2% dextrose, SGal = 3% galactose, SG/Lac = 3% glycerol/2% lactate) were prepared as described (26). Isolation and identification of the mdm20-suppressing alleles, TPM1-5 and ACT1-203, were described previously (23). The nat3Δ::HIS3 allele, a precise deletion of the entire NAT3 ORF, was created in a diploid strain, JSY1832, by using the technique of Baudin et al. (27). The disruption was verified by PCR in the resulting heterozygote, JSY4632, which was sporulated to obtain the nat3Δ::HIS3 haploid strain, JSY4649. Strains used included: JSY999, MATα his3Δ200 leu2Δ1 ura3-52; JSY1065, MATα his3Δ200 leu2Δ1 ura3-52 mdm20Δ::LEU2; JSY1832, MATa/MATα his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 lys2Δ202/lys2Δ202 trp1Δ63/trp1Δ63 ura3-52/ura3-52; JSY3094, MATa his3Δ200 leu2Δ1 ura3-52 mdm20Δ::LEU2; JSY3244, MATa his3Δ200 leu2Δ1 lys2Δ202 ura3-52 mdm20Δ::LEU2 TPM1-5; JSY3315, MATa leu2Δ1 ura3-52 TPM1-5; JSY4131, MATα his3Δ200 leu2Δ1 ura3-52 tpm2::HIS3; JSY4625, MATa his3Δ200 leu2Δ1 ura3-52 mdm20Δ::LEU2 tpm2::HIS3; JSY4632, MATa/MATα his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 lys2Δ202/lys2Δ202 trp1Δ63/trp1Δ63 ura3-52/ura3-52 nat3Δ::HIS3/+; JSY4649, MATα his3Δ200 leu2Δ1 lys2Δ202 trp1Δ63 ura3-52 nat3Δ::HIS3; JSY4650, MATα his3Δ200 leu2Δ1 lys2Δ202 trp1Δ63 ura3-52; JSY4922, MATa his3Δ200 leu2Δ1 trp1Δ63 ura3-52 nat3Δ::HIS3; JSY4923, MATa his3Δ200 leu2Δ1 lys2Δ202 trp1Δ63 ura3-52 nat3Δ::HIS3 ACT1-203; JSY4924, MATa his3Δ200 leu2Δ1 lys2Δ202 ura3-52 nat3Δ::HIS3 TPM1-5; JSY4925, MATa his3Δ200 leu2Δ1 lys2Δ202 trp1Δ63 ura3-52 nat3Δ::HIS3 TPM1-5; JSY4984, MATa his3Δ200 leu2Δ1 lys2Δ202 ura3-52 mdm20Δ::LEU2; JSY4985, MATα his3Δ200 leu2Δ1 trp1Δ63 ura3-52; JSY4986, MATa his3Δ200 leu2Δ1 trp1Δ63 ura3-52 mdm20Δ::LEU2 nat3Δ::HIS3; JSY4987, MATα his3Δ200 leu2Δ1 lys2Δ202 ura3-52 nat3Δ::HIS3.
Analysis of Actin Organization and Mitochondrial Inheritance Phenotypes. The actin cytoskeleton was visualized with 0.5 μM Alexa Fluor 568-conjugated phalloidin (Molecular Probes) as described (23). Because actin cables are best visualized in cells with small to medium buds, only cells at this stage were scored for the presence or absence of cable(-like) structures.
Mitochondrial inheritance was quantified in strains harboring the pDO12 plasmid (28) expressing a mitochondrial matrix-targeted COX4-GFP fusion protein. Overnight cultures were grown at 25°C in synthetic medium, diluted to an OD600 of ≤0.5 in fresh medium, and grown for an additional3hat25°Cor37°C before quantification. Because mitochondrial membranes are not always transported immediately into small buds (even in WT strains), mitochondrial inheritance was scored only in medium- and large-budded cells. Digital microscopic images of cells were acquired as described (28).
Biochemical Characterization of Tpm1-5p and Tpm1p. Whole-cell protein lysates of JSY999 and JSY3315 were separated by electrophoresis on an SDS/12% polyacrylamide gel, transferred to nitrocellulose, and processed for Western blotting by using standard methods (20). Tpm1-5p, Tpm1p, and Tpm2p were recognized by using a rabbit polyclonal antiserum, B43 (1:3,000) (5, 29), with cross-reactivity to all three proteins (provided by A. Bretscher, Cornell University, Ithaca, NY). Detection was carried out by using a monoclonal horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:50,000) (Jackson ImmunoResearch) and the enhanced chemiluminescence kit (New England Nuclear).
Purification of Tpm1p from MDM20 and mdm20 yeast strains was performed by established methods (5, 30) from strains JSY4131 and JSY4625 containing a galactose-inducible overexpression vector, pBD100 (ref. 5; provided by A. Bretscher). Because the purification scheme used isolates Tpm2p as well as Tpm1p, a tpm2Δ allele was introduced into the background of both Tpm1p overexpression strains.
The mass of purified Tpm1 proteins was determined by electrospray ionization mass spectrometry. Lyophilized samples first were redissolved in 0.1% SDS. The percentage of SDS subsequently was reduced to a tolerable level for electrospray ionization (≤0.01%) by passage of the samples through a PD-10 desalting column (Amersham Biosciences). Protein solutions were concentrated by using Centricon YM-30 centrifuge filter devices (Millipore). Immediately before injection into the Quattro II triple quadrupole mass spectrometer (Micromass, Manchester, U.K.), final protein samples (200 μl) were prepared in 50% acetonitrile/0.1% formic acid. Proteins were scanned over an m/z range of 1,000–3,000, and both positive and negative ion spectra were detected. Mass spectral data were processed by using the Micromass masslynx operating system and the maxent 1 software tool.
Sedimentation Assays. WT yeast actin (a generous gift from P. Rubenstein, University of Iowa, Iowa City) was purified by a DNase I affinity chromatography protocol as described (31, 32). F-actin sedimentation assays were performed as described (33). Briefly, F-actin was diluted to 10 μM in F-buffer (10 mM Tris·HCl, pH 7.5/0.2 mM CaCl2/0.2 mM ATP/2 mM MgCl2/50 mM KCl/0.5 mM DTT), combined with increasing concentrations of yeast Tpm1p (0–3.2 μM, derived from WT or mdm20 mutant cells), and incubated for 30 min at room temperature. After sedimentation for 45 min in a Beckman Coulter TL-100 rotor at 75,000 rpm, supernatant and pellet fractions were analyzed by SDS/PAGE. Under these conditions, Tpm1p (0.5 μM) derived from WT or mdm20 cells did not sediment in the absence of F-actin. Digitally scanned Coomassie blue-stained gels were analyzed by using nih image 1.62 software. The data obtained were used to calculate the amount of F-actin-bound Tpm1p (derived from WT or mdm20 mutant cells) in the pellet fractions.
Results
TPM1-5 Encodes an N-Terminally Extended Form of Tpm1p That Suppresses Actin Cable Defects in mdm20. In a previous study, we described the isolation of 12 dominant mdm20-suppressing mutations (23). Each of these mutations fell in one of two genes, ACT1 or TPM1, that encode the only conventional actin in budding yeast and the predominant form of tropomyosin. The finding that mdm20 phenotypes are suppressed by dominant alleles of ACT1 and TPM1 suggested that Mdm20p functions to regulate actin–tropomyosin interactions. One allele, in particular, TPM1-5, provided significant insight into the mechanism of Mdm20p function.
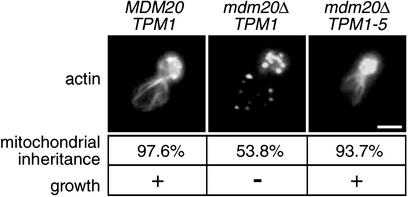
As with all other mdm20-suppressing alleles, TPM1-5 restores visible actin cables and suppresses temperature-sensitive growth defects when combined with an mdm20-null mutation (mdm20Δ; ref. 23; Fig. 1). Although cables in the TPM1-5 mdm20Δ mutant strain are less robust than those observed in WT cells, they are easily visible in the majority of cells in the population (96.8%, n = 253). In contrast, visible cables are almost entirely absent in mdm20Δ cells lacking the TPM1-5 suppressor (refs. 20 and 23; Fig. 1). In the mdm20Δ strain, long cables extending to the base of the mother cell are never observed, and short faint cables restricted to the bud neck are visible in only 15.9% of the cells (ref. 23; Table 1; Fig. 1).
Fig. 1.
The TPM1-5 allele partially restores actin cables and suppresses mitochondrial inheritance defects in an mdm20Δ strain. Alexa Fluor 568-phalloidin-labeled actin cables and/or patches are shown in budding MDM20 TPM1, mdm20Δ TPM1, and mdm20Δ TPM1-5 cells grown at 25°C (buds are in the upper right portion of each image). Representative cells are shown. The percentage of large-budded cells containing GFP-labeled mitochondria in the bud at 37°C is indicated. The ability (+) or failure (–) of each strain to grow on dextrose-containing medium at 37°C also is indicated. (Bar = 5 μm.)
Table 1. Actin cable and mitochondrial inheritance phenotypes.
| Mitochondria in large-budded cells
|
||||||
|---|---|---|---|---|---|---|
| Actin cables in budding cells
|
At 25°C
|
At 37°C
|
||||
| Genotype | % | n | % | n | % | n |
| WT | 99.4 | 718 | 99.2 | 649 | 98.1 | 701 |
| nat3 | 31.6 | 653 | 94.2 | 800 | 71.5 | 800 |
| nat3 TPM1-5 | 96.8 | 339 | 99.4 | 509 | 98.7 | 600 |
| nat3 ACT1-203 | 95.8 | 189 | 98.8 | 250 | 99.0 | 300 |
| nat3 mdm20 | 16.1 | 217 | 74.5 | 200 | 64.0 | 200 |
| mdm20 | 15.9 | 566 | 76.9 | 347 | 54.9 | 375 |
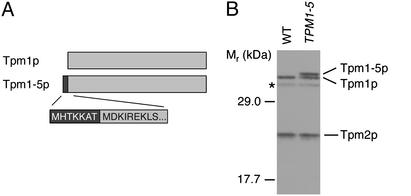
Because of the creation of a premature initiation codon 19 bp upstream of the normal translation start site, the TPM1-5 mutation (ATA → ATG) results in a 7-aa extension at the N terminus of the otherwise WT Tpm1-5 protein (Fig. 2A). The slower-migrating Tpm1-5p can be visualized by Western blotting (Fig. 2B), although the persistence of the WT Tpm1p in the same lane suggests that translation may be initiated at both the old and new start sites in TPM1-5 strains. As reported previously for Tpm1p (11), the apparent molecular masses of both Tpm1p and Tpm1-5p are larger than predicted from their respective amino acid sequences. Production of the elongated Tpm1-5p was verified by N-terminal protein sequencing (23). A mixture of WT and mutant protein was not detected by the latter method, presumably because WT Tpm1p is acetylated N-terminally and, therefore, refractory to sequencing by Edman degradation.
Fig. 2.
Tpm1-5p contains an N-terminal extension. (A) A schematic diagram of Tpm1p and Tpm1-5p. Creation of a premature translation initiation codon by the TPM1-5 mutation results in a 7-aa extension (black) at the N terminus of Tpm1-5p, relative to WT Tpm1p (gray). The identity of the seven amino acids shown in white text was verified by protein sequencing (23). (B) Western blot analysis of whole-cell lysates from WT and TPM1-5 cells by using an antibody that recognizes all yeast tropomyosins. The WT Tpm1p (33 kDa), elongated Tpm1-5p (34 kDa), and Tpm2p (25 kDa) are marked (the apparent molecular masses of all three proteins are larger than predicted). The asterisk marks a background band also present in extracts prepared from tpm1Δ control strains.
Cells Lacking Mdm20p Fail to N-Terminally Acetylate Tpm1p. The following four observations strongly suggested that Mdm20p functions in the N-terminal acetylation of Tpm1p: (i) TPM1-5 can suppress mdm20 phenotypes, (ii) N-terminal acetylation is essential for the normal function of WT Tpm1p, (iii) short peptides fused to Tpm1p's N terminus can restore function in the absence of acetylation, and (iv) Mdm20p and Nat3p copurify as components of the NatB N-terminal acetylase complex. To test this hypothesis, we overexpressed WT Tpm1p in yeast strains containing the WT MDM20 gene (JSY4131, MDM20) or a chromosomal disruption of the MDM20 coding region (JSY4625, mdm20Δ). Tpm1p purified from each strain then was analyzed by electrospray mass spectrometry (Fig. 3).
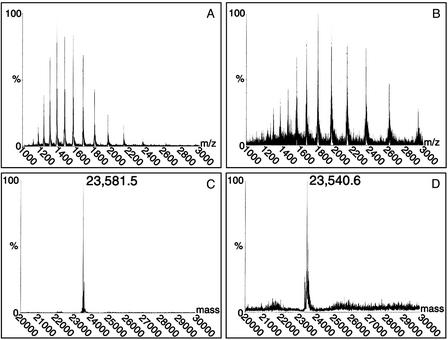
Fig. 3.
Mdm20p is required for Tpm1p acetylation. (A and C) Negative ion electrospray mass spectrometry data for Tpm1p derived from MDM20 cells (A) and the corresponding deconvoluted mass spectrum (C) show the molecular mass (in Da) of Tpm1p produced in a WT genetic background. The determined value (23,581.5 Da) is very close to that expected for an acetylated form of the protein. (B and D) The electrospray mass spectrum (B) and deconvoluted spectrum (D) for Tpm1p derived from mdm20 mutant cells. The mass estimate (23,540.6 Da) is 40.9 Da smaller than the sample in A and C and corresponds to the predicted mass for the unacetylated form of Tpm1p, indicating that an acetyl group is no longer present. A more detailed version of this figure is available as Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org.
As shown in Fig. 3, the determined molecular mass of Tpm1p expressed in a WT MDM20 background was 23,581.5 Da, whereas the mass of Tpm1p expressed in an mdm20Δ background was 23,540.6 Da. The mass difference between the two samples, 40.9 Da, is consistent with the expected mass difference from the presence or absence of an acetyl group (42 Da). In addition, the masses we observed for the two proteins correspond well to the predicted masses for acetylated and unacetylated Tpm1p (23,583 and 23,541 Da, respectively) and to the previously determined masses for these forms of the yeast protein (12). Edman degradation sequencing revealed that the N terminus of Tpm1p isolated from WT cells was blocked, consistent with the presence of an N-terminal acetyl group. In contrast, the N-terminal amino acid sequence of Tpm1p isolated from mdm20 mutant cells was readily obtained (data not shown). Together, these results indicate that Mdm20p is required for N-terminal acetylation of Tpm1p.
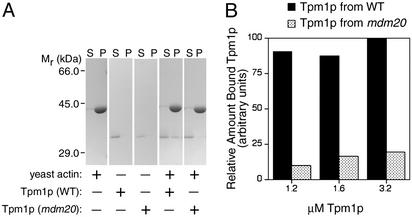
N-Terminal Acetylation of Yeast Tpm1p Is Necessary for Efficient Actin Filament Binding. The importance of Mdm20p-dependent, Tpm1p N-terminal acetylation for binding to yeast actin was evaluated in sedimentation assays. At a concentration of 10 μM, yeast actin spontaneously assembles into filaments (F-actin) that sediment upon ultracentrifugation. In contrast, Tpm1p isolated from either WT or mdm20 cells failed to sediment in the absence of F-actin (Fig. 4A). Although N-terminally acetylated Tpm1p (isolated from WT cells) binds F-actin and can be recovered in the pellet fraction, unacetylated Tpm1p (isolated from mdm20 cells) remains in the supernatant (Fig. 4A). When present at equivalent concentrations (1.2–3.2 μM), the amount of unacetylated Tpm1p bound to F-actin was reduced significantly relative to the acetylated form (Fig. 4B). These results directly demonstrate that Mdm20p-mediated N-terminal acetylation of Tpm1p is important for actin binding.
Fig. 4.
N-terminal acetylation of Tpm1p is required for efficient actin binding. (A) Yeast actin alone, Tpm1p alone (isolated from WT or mdm20 cells), or both yeast actin and Tpm1p were incubated in F-buffer (10 mM Tris·HCl, pH 7.5/0.2 mM ATP/0.2 mM CaCl2/0.5 mM DTT/2.0 mM MgCl2/50 mM KCl). After sedimenting at 75,000 rpm in a TL-100 rotor, supernatant (S) and pellet (P) fractions were analyzed by SDS/PAGE and Coomassie blue staining. Only Tpm1p bound to F-actin is found in the pellet fraction in this assay. (B) Quantitative scanning of Coomassie blue-stained gels was used to determine the relative amount of bound Tpm1p in reaction mixtures containing 10 mM yeast actin and increasing concentrations of WT or mdm20-derived Tpm1p (1.2–3.2 μM). Tpm1p isolated from WT cells (filled bars) consistently bound better to F-actin than did Tpm1p isolated from mdm20 mutant cells (stippled bars).
Mdm20p and the Nat3p Acetyltransferase Act in the Same Cellular Pathway. The idea that Mdm20p acts in conjunction with Nat3p as a part of the NatB acetyltransferase complex leads to several testable predictions. The first prediction is that loss of Nat3p function will induce a set of phenotypes that include, but may not be limited to, those caused by the loss of Mdm20p function. A second, related prediction is that mdm20-like phenotypes resulting from loss of Nat3p function will be suppressed by mutations that suppress mdm20.
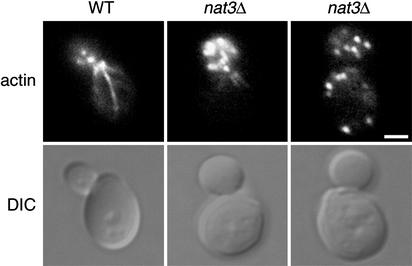
To test these predictions, we disrupted the NAT3 gene in a WT diploid strain and sporulated and dissected this strain to obtain haploid nat3Δ progeny. Consistent with previous reports, we found that nat3Δ mutants were viable but grew very slowly on fermentable as well as nonfermentable carbon sources and at temperatures ranging from 25°C to 37°C (ref. 17; J. M. Singer, unpublished data). In this respect, nat3Δ strains differ from mdm20Δ strains, which grow more quickly at 25°C and do not grow at all at 37°C (20, 23). As with mdm20Δ mutants, however, nat3Δ cells exhibited defects in both actin organization and mitochondrial inheritance. Visible actin cables were absent in 68.4% of nat3Δ mutant cells, a phenotype also observed in mdm20Δ mutants (84.1% lacking cables) (Fig. 5 and Table 1). In the remaining nat3Δ cells, only short, faint cables were detected emanating from the bud neck. Also, like mdm20Δ mutants, nat3Δ mutant cells often had abnormally large bud necks at 37°C, a phenotype related to defects in actin organization (ref. 20; Fig. 6, differential interference contrast microscopy image of nat3Δ cell).
Fig. 5.
Deletion of the NAT3 gene results in the loss of actin cables. Differential interference contrast (DIC) microscopy images and Alexa Fluor 568-phalloidin actin labeling of WT and nat3Δ cells grown at 25°C are shown. Representative cells are shown. (Bar = 2.5 μm.)
Fig. 6.
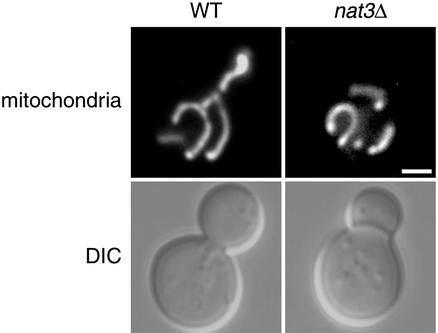
nat3 mutants exhibit a mitochondrial inheritance defect. Differential interference contrast (DIC) microscopy images and mitochondrial-GFP labeling of representative WT and nat3Δ cells grown at 25°C and shifted to 37°C for 3 h are shown. (Bar = 2.5 μm.)
With respect to mitochondrial inheritance, the nat3Δ mutant again behaved like mdm20. The phenotypic similarity was all the more striking given that defective mitochondrial inheritance is temperature-dependent in both mutants. Although only 5.8% of large nat3Δ buds lacked mitochondria at 25°C, the inheritance defect increased to 28.5% at 37°C (Fig. 6 and Table 1). By comparison, mdm20 mutants displayed mitochondrial inheritance defects in 23.1% of buds formed at 25°C and 45.1% of buds formed at 37°C (Table 1 and ref. 23). In WT cells, mitochondria are inherited by >98% of the buds formed at both temperatures (Fig. 6 and Table 1).
The nat3Δ mdm20Δ double mutant exhibited a combination of nat3Δ and mdm20Δ phenotypes. Like nat3Δ, the double mutant grew slowly on all carbon sources (J. M. Singer, unpublished data). However, the double mutant displayed actin cable and mitochondrial inheritance defects similar to those of mdm20Δ cells (Table 1). Combined with the single-mutant analysis, this result suggests that Nat3p acetylates a variety of cellular substrates in vivo and that disruption of Mdm20p function affects the acetylation of only a subset of these substrates.
To determine whether suppressors of mdm20 also could suppress nat3, we introduced two mdm20-suppressing mutations, ACT1-203 (23) and TPM1-5, into a nat3Δ background. Neither ACT1-203 nor TPM1-5 could suppress the slow-growth phenotype associated with nat3Δ (J. M. Singer, unpublished data). Conversely, both mutations were able to restore actin cables and suppress mitochondrial inheritance defects to near-WT levels in nat3Δ mutants (Table 1). Together, the genetic data presented here indicate that the NAT3 and MDM20 genes function in the same pathway, providing additional support for the idea that Mdm20p acts together with Nat3p, as a part of the NatB acetyltransferase complex, to acetylate Tpm1p.
Discussion
Our previous studies indicated that Mdm20p function is important for tropomyosin–F-actin interactions that stabilize actin cables in budding yeast (23). In this article, we demonstrate that Mdm20p functions as an essential component of the machinery responsible for Tpm1p N-terminal acetylation. N-terminal acetylation is required for proper Tpm1p function, and Tpm1p function is required for the stabilization of long actin filaments that are bundled together to form actin cables. Because mitochondria are transported inefficiently into yeast buds lacking actin cables (especially at elevated temperatures), evaluation of mitochondrial inheritance provides us with an additional, sensitive assay to monitor Mdm20p function and actin cable stability in vivo.
The discovery that Mdm20p is required for efficient Tpm1p function is consistent with our previous characterization of Mdm20p. Initial studies indicated that the effects of mdm20 mutations on actin organization were limited to actin cables (20). Correspondingly, Tpm1p subcellular localization is restricted to actin cables (11). As noted above, mdm20 mutant phenotypes are suppressed by certain TPM1 gain-of-function alleles, but they also are suppressed partially by the overexpression of WT Tpm1p (20). However, because Tpm1p acts downstream of Mdm20p, overexpression of Mdm20p cannot compensate for the complete absence of Tpm1p (20).
A recent in vivo study showed that Mdm20p function is required for the association of Tpm1p with actin (21). In otherwise WT cells, an activated form of the yeast formin Bni1p (Bni1ΔRBD) caused the accumulation of actin filaments in the bud cortex. Because Tpm1p colocalized to these filaments, they were presumed to be cable-like in structure. Bud cortical actin still was produced when Bni1ΔRBD was introduced into cells lacking Mdm20p; however, Tpm1p failed to associate with this actin. Together with the data reported here, these results indicate that Tpm1p must be acetylated by the Mdm20p-containing NatB complex before it can associate with actin filaments and cables in vivo.
We observed that nat3 mutants have similar, but consistently less severe, phenotypes than mdm20 mutants with respect to actin cable loss and mitochondrial inheritance. The simplest explanation for this observation is that a small proportion of Tpm1p is acetylated even in the absence of Nat3p. However, promiscuous acetylation of Tpm1p by the NatA or NatC acetyltransferase complexes is not supported by previous work. Biochemical studies showed that N-terminal acetylation of NatA-, NatB-, or NatC-specific substrates was abolished in cells lacking the relevant acetyltransferase activity (17). An alternative explanation is that Nat3p (independent of Mdm20p) is required for acetylation of a non-Tpm1p substrate whose function inhibits or suppresses actin cable formation and mitochondrial inheritance. Consistent with this idea, mdm20 nat3 double mutants display less severe phenotypes than cells mutant for mdm20 alone (Table 1). Further identification and analysis of Nat3p substrates and their activities will help to elucidate the molecular basis for the phenotypic differences between mdm20 and nat3 cells.
The nat3 slow-growth phenotype at 25°C is much more severe than that of the mdm20 mutant and is unaffected by mdm20-suppressing mutations, suggesting that Nat3p is required for acetylation of a wider range of substrates than Mdm20p. This difference might arise if inclusion of Mdm20p in the NatB complex is cell cycle-regulated. An argument against this model is that actin cables can be observed at all cell cycle stages in WT cells but are almost never visualized in mdm20 mutants, indicating that some level of Mdm20p function is required throughout the cell cycle. Two alternative scenarios to account for Mdm20p-independent Nat3p functions are that (i) Mdm20p is present but functionally neutral as NatB acetylates substrates other than Tpm1p or (ii) a fraction of cellular NatB complexes contain Mdm20p, whereas others replace Mdm20p with an alternate protein(s) that generates different substrate specificity. If Mdm20p were a permanent component of the NatB complex that could be activated or inactivated depending on the substrate target, we would have expected mdm20Δ and nat3Δ cells to exhibit a nearly identical range of phenotypes in our genetic studies. Conversely, if Mdm20p occupies a position in the complex in which multiple proteins can fit to provide alternate substrate specificities, the absence of Mdm20p would have little effect on Mdm20p-independent functions. This latter explanation is consistent with the observation that the NatB complex remains at least partially functional in the absence of Mdm20 protein.
Act1p is a known substrate of the NatB complex (17). Whether Mdm20p also is necessary for the acetylation of Act1p has not been determined. Nonetheless, it appears that Mdm20p is required for the acetylation of at least one substrate in addition to Tpm1p because a tpm1 mutant alone is viable, whereas the combination of tpm1 and mdm20 mutations results in synthetic lethality (20). The most likely candidate for a second Mdm20-dependent acetylation substrate is Tpm2p, the minor form of tropomyosin in budding yeast. Combinations of tpm1 and tpm2 mutations are also synthetically lethal in yeast (5). Thus, a tpm1 tpm2 genotype may effectively mimic the situation in which a tpm1 mdm20 cell finds itself, having no Tpm1p and only unacetylated, crippled Tpm2p.
Mdm20p-dependent Tpm1p acetylation now can be incorporated into a model of actin cable formation proposed by Evangelista et al. (21). First, the polymerization of actin filaments is initiated in the bud by the formins Bni1p/Bnr1p (36). The growing filament extends toward the bud neck and the mother cell. Independently, the NatB acetyltransferase complex cotranslationally acetylates the N terminus of Tpm1p, with Nat3p providing the catalytic function and Mdm20p aiding in the identification of the target. Acetylated Tpm1p then binds to individual actin filaments, stabilizing them and allowing them to become very long. Finally, Sac6p (fimbrin), perhaps aided by Abp140, bundles the filaments together into a cable (37, 38).
Once formed, actin cables are used as tracks to transport a variety of membrane-bound organelles, including mitochondria (18–20), vacuoles (39), peroxisomes (40), late Golgi elements (41), and secretory vesicles (29) from the mother cell into the bud. In the case of the latter four organelles, this transport is mediated by Myo2p, an actin-dependent, class V myosin motor. In the case of mitochondria, a subset of the organelles in the mother cell moves along actin cables toward the bud, propelled by either an unidentified motor protein or localized bursts of secondary actin polymerization initiated by Arp2/3 (42, 43). In muscle cells, access of myosin to binding sites on the actin filament is regulated by proteins that change the conformation of bound tropomyosin (2). In nonmuscle cells, the role of tropomyosins, like Tpm1p, in masking/exposing motor-binding sites is less clear. In the future, it will be important to identify the machinery that controls Tpm1p conformational changes and regulates binding of Myo2p and/or other organelle motors to the actin filament.
Supplementary Material
Acknowledgments
We thank Anthony Bretscher and David Pruyne for reagents and discussions, Elliot Rachlin of the University of Utah Chemistry Department Mass Spectrometry Facility and Peter Anderson for help with mass spectrometry experiments, Peter Rubenstein for providing yeast actin, and members of the Shaw laboratory for helpful discussions. This work was supported by National Institutes of Health Grant GM-53466 (to J. M. Shaw) and National Institutes of Health Predoctoral Genetics Training Grant T32-GM07464 (to J. M. Singer). Purchase of the Micromass Quattro II mass spectrometer was funded by National Science Foundation Grant CHE-9708413 and the University of Utah Institutional Funds Committee. The University of Utah DNA and Peptide Facility is supported in part by a grant from the National Cancer Institute (CA42014).
References
- 1.Lees-Miller, J. P. & Helfman, D. M. (1991) BioEssays 13, 429–437. [DOI] [PubMed] [Google Scholar]
- 2.Gordon, A. M., Homsher, E. & Regnier, M. (2000) Physiol. Rev. 80, 853–924. [DOI] [PubMed] [Google Scholar]
- 3.Lin, J. J., Warren, K. S., Wamboldt, D. D., Wang, T. & Lin, J. L. (1997) Int. Rev. Cytol. 170, 1–38. [DOI] [PubMed] [Google Scholar]
- 4.Wen, K. K., Kuang, B. & Rubenstein, P. A. (2000) J. Biol. Chem. 275, 40594–40600. [DOI] [PubMed] [Google Scholar]
- 5.Drees, B., Brown, C., Barrell, B. G. & Bretscher, A. (1995) J. Cell Biol. 128, 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hitchcock-DeGregori, S. E. (1994) Adv. Exp. Med. Biol. 358, 85–96. [DOI] [PubMed] [Google Scholar]
- 7.Hitchcock-DeGregori, S. E. & Heald, R. W. (1987) J. Biol. Chem. 262, 9730–9735. [PubMed] [Google Scholar]
- 8.Cho, Y. J., Liu, J. & Hitchcock-DeGregori, S. E. (1990) J. Biol. Chem. 265, 538–545. [PubMed] [Google Scholar]
- 9.Greenfield, N. J., Montelione, G. T., Farid, R. S. & Hitchcock-DeGregori, S. E. (1998) Biochemistry 37, 7834–7843. [DOI] [PubMed] [Google Scholar]
- 10.Brown, J. H., Kim, K. H., Jun, G., Greenfield, N. J., Dominguez, R., Volkmann, N., Hitchcock-DeGregori, S. E. & Cohen, C. (2001) Proc. Natl. Acad. Sci. USA 98, 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, H. & Bretscher, A. (1989) Cell 57, 233–242. [DOI] [PubMed] [Google Scholar]
- 12.Maytum, R., Geeves, M. A. & Konrad, M. (2000) Biochemistry 39, 11913–11920. [DOI] [PubMed] [Google Scholar]
- 13.Monteiro, P. B., Lataro, R. C., Ferro, J. A. & Reinach, F. d. C. (1994) J. Biol. Chem. 269, 10461–10466. [PubMed] [Google Scholar]
- 14.Urbancikova, M. & Hitchcock-DeGregori, S. E. (1994) J. Biol. Chem. 269, 24310–24315. [PubMed] [Google Scholar]
- 15.Polevoda, B. & Sherman, F. (2000) J. Biol. Chem. 275, 36479–36482. [DOI] [PubMed] [Google Scholar]
- 16.Polevoda, B. & Sherman, F. (2003) J. Mol. Biol. 325, 595–622. [DOI] [PubMed] [Google Scholar]
- 17.Polevoda, B., Norbeck, J., Takakura, H., Blomberg, A. & Sherman, F. (1999) EMBO J. 18, 6155–6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boldogh, I. R., Yang, H. C. & Pon, L. A. (2001) Traffic 2, 368–374. [DOI] [PubMed] [Google Scholar]
- 19.Simon, V. R., Karmon, S. L. & Pon, L. A. (1997) Cell Motil. Cytoskeleton 37, 199–210. [DOI] [PubMed] [Google Scholar]
- 20.Hermann, G. J., King, E. J. & Shaw, J. M. (1997) J. Cell Biol. 137, 141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evangelista, M., Pruyne, D., Amberg, D. C., Boone, C. & Bretscher, A. (2002) Nat. Cell Biol. 4, 32–41. [DOI] [PubMed] [Google Scholar]
- 22.Belmont, L. D. & Drubin, D. G. (1998) J. Cell Biol. 142, 1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singer, J. M., Hermann, G. J. & Shaw, J. M. (2000) Genetics 156, 523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hermann, G. J. (1998) Ph.D. thesis (Univ. of Utah, Salt Lake City).
- 25.Winston, F., Dollard, C. & Ricupero-Hovasse, S. L. (1995) Yeast 11, 53–55. [DOI] [PubMed] [Google Scholar]
- 26.Sherman, F., Fink, G. R. & Hicks, J. B. (1986) Methods in Yeast Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 27.Baudin, A., Ozier-Kalogeropoulos, O., Denouel, A., Lacroute, F. & Cullin, C. (1993) Nucleic Acids Res. 21, 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otsuga, D., Keegan, B. R., Brisch, E., Thatcher, J. W., Hermann, G. J., Bleazard, W. & Shaw, J. M. (1998) J. Cell Biol. 143, 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pruyne, D. W., Schott, D. H. & Bretscher, A. (1998) J. Cell Biol. 143, 1931–1945. [DOI] [PubMed] [Google Scholar]
- 30.Liu, H. & Bretscher, A. (1989) Proc. Natl. Acad. Sci. USA 86, 90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuang, B. & Rubenstein, P. A. (1997) J. Biol. Chem. 272, 4412–4418. [DOI] [PubMed] [Google Scholar]
- 32.Cook, R. K., Root, D., Miller, C., Reisler, E. & Rubenstein, P. A. (1993) J. Biol. Chem. 268, 2410–2415. [PubMed] [Google Scholar]
- 33.Cheng, D., Marner, J. & Rubenstein, P. A. (1999) J. Biol. Chem. 274, 35873–35880. [DOI] [PubMed] [Google Scholar]
- 34.Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M. & Seraphin, B. (1999) Nat. Biotechnol. 17, 1030–1032. [DOI] [PubMed] [Google Scholar]
- 35.Polevoda, B. & Sherman, F. (2001) J. Biol. Chem. 276, 20154–20159. [DOI] [PubMed] [Google Scholar]
- 36.Pruyne, D., Evangelista, M., Yang, C., Bi, E., Zigmond, S., Bretscher, A. & Boone, C. (2002) Science 297, 612–615. [DOI] [PubMed] [Google Scholar]
- 37.Adams, A. E. M., Botstein, D. & Drubin, D. G. (1991) Nature 354, 404–408. [DOI] [PubMed] [Google Scholar]
- 38.Asakura, T., Sasaki, T., Nagano, F., Satoh, A., Obaishi, H., Nishioka, H., Imamura, H., Hotta, K., Tanaka, K., Nakanishi, H., et al. (1998) Oncogene 16, 121–130. [DOI] [PubMed] [Google Scholar]
- 39.Hill, K. L., Catlett, N. L. & Weisman, L. S. (1996) J. Cell Biol. 135, 1535–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoepfner, D., van den Berg, M., Philippsen, P., Tabak, H. F. & Hettema, E. H. (2001) J. Cell Biol. 155, 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossanese, O. W., Reinke, C. A., Bevis, B. J., Hammond, A. T., Sears, I. B., O'Connor, J. & Glick, B. S. (2001) J. Cell Biol. 153, 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boldogh, I. R., Yang, H. C., Nowakowski, W. D., Karmon, S. L., Hays, L. G., Yates, J. R., III, & Pon, L. A. (2001) Proc. Natl. Acad. Sci. USA 98, 3162–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon, V. R., Swayne, T. C. & Pon, L. A. (1995) J. Cell Biol. 130, 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.