Abstract
Chlorarachniophytes are amoeboflagellate algae that acquired photosynthesis secondarily by engulfing a green alga and retaining its plastid (chloroplast). An important consequence of secondary endosymbiosis in chlorarachniophytes is that most of the nuclear genes encoding plastid-targeted proteins have moved from the nucleus of the endosymbiont to the host nucleus. We have sequenced and analyzed 83 cDNAs encoding 78 plastid-targeted proteins from the model chlorarachniophyte Bigelowiella natans (formerly Chlorarachnion sp. CCMP621). Phylogenies inferred from the majority of these genes are consistent with a chlorophyte green algal origin. However, a significant number of genes (≈21%) show signs of having been acquired by lateral gene transfer from numerous other sources: streptophyte algae, red algae (or algae with red algal endosymbionts), as well as bacteria. The chlorarachniophyte plastid proteome may therefore be regarded as a mosaic derived from various organisms in addition to the ancestral chlorophyte plastid. In contrast, the homologous genes from the chlorophyte Chlamydomonas reinhardtii do not show any indications of lateral gene transfer. This difference is likely a reflection of the mixotrophic nature of Bigelowiella (i.e., it is photosynthetic and phagotrophic), whereas Chlamydomonas is strictly autotrophic. These results underscore the importance of lateral gene transfer in contributing foreign proteins to eukaryotic cells and their organelles, and also suggest that its impact can vary from lineage to lineage.
Keywords: molecular evolution, chlorarachniophytes, chloroplast, phylogeny, secondary endosymbiosis
It is now clear that the plastids (chloroplasts) of photosynthetic eukaryotes ultimately arose from a single, ancient endosymbiosis between a heterotrophic eukaryote and a photosynthetic cyanobacterium (1–3). It is also clear that the primary plastids of red and green algae have spread laterally among distantly related organisms through the process of secondary endosymbiosis, in which a heterotrophic eukaryote engulfs a primary plastid-containing alga and retains its photosynthetic apparatus (1, 2, 4).
Secondary endosymbiosis has been a major force in eukaryotic evolution and is responsible for generating an enormous amount of algal biodiversity. Seven distinct eukaryotic groups harbor plastids derived from secondary endosymbiosis: haptophytes, heterokonts (stramenopiles), cryptomonads, dinoflagellates, apicomplexans, euglenids, and chlorarachniophytes (1, 4). The chlorarachniophytes are a relatively small group of mixotrophic marine flagellates and amoeboflagellates with green plastids surrounded by four membranes (5, 6). Together with the cryptomonads, the chlorarachniophytes are distinguished by the retention of a “nucleomorph,” the reduced algal endosymbiont nucleus that resides between the plastid inner and outer membrane pairs (2, 6). The chlorarachniophyte host belongs to a large and diverse lineage referred to as Cercozoa (7–10). The chlorarachniophyte endosymbiont, on the other hand, is derived from a green alga. This finding was first suggested based on plastid pigments (5), and is now supported by numerous molecular phylogenies inferred from plastid- and nucleomorph-encoded genes (11–15). More specifically, these analyses point to the chlorophyte green algae as the probable source of the endosymbiont (12, 13, 15).
All plastid genomes, both primary and secondary, encode only a small fraction of the proteins required for plastid function (16). The vast majority of plastid proteins are encoded by genes in the host nuclear genome, and are posttranslationally targeted to the plastid with N-terminal-targeting peptides (17). Protein targeting to primary plastids is mediated by a transit-peptide, whereas proteins targeted to secondary plastids are first directed to the endomembrane system with a signal peptide, and then to the plastid with a transit peptide (18–20). It is generally believed that many of these nucleus-encoded, plastid-targeted proteins were originally encoded in the cyanobacterial genome, and that the genes were transferred to the host nuclear genome during the integration of the endosymbiont. In secondary plastid-containing algae, most or all of these genes have subsequently moved from the primary algal nucleus to the secondary host nucleus. The protein complement of a plastid should thus reflect a common evolutionary ancestry.
Here, we have characterized 83 genes for plastid-targeted proteins in the chlorarachniophyte Bigelowiella natans. Although the majority of nucleus-encoded, plastid-targeted proteins in chlorarachniophytes are derived from the chlorophyte endosymbiont, a significant number are not. Twenty-one percent of the B. natans genes encoding plastid-targeted proteins show signs of lateral gene transfer from a variety of exogenous sources, including streptophyte algae, red algae and algae with red algal endosymbionts, and bacteria. In stark contrast, however, the homologous plastid proteins in Chlamydomonas show no indications of lateral gene transfer. Lateral gene transfer seems to have played a significant role in shaping the complement of plastid proteins in chlorarachniophytes, and perhaps other mixotrophic secondary plastid-containing algae.
Materials and Methods
Identification of B. natans Plastid-Targeted Proteins. ESTs from B. natans (CCMP621) were analyzed in the course of an ongoing EST sequencing project of a unialgal cDNA library. cDNAs encoding putative plastid-targeted proteins were identified according to three criteria: their phylogenetic relationship to other plastid homologs, their involvement in a process known to be localized in the plastid, and/or their possession of distinctive bipartite plastid-targeting leader sequences consisting of a signal peptide and a transit peptide. cDNAs encoding plastid-targeted proteins were assembled to form contiguous ORFs, and unique clones were completely sequenced by primer walking. The N-terminal regions of conceptual amino acid sequences were analyzed for the presence of signal peptides by using the neural network-based tool SIGNALP (21) and IPSORT (22). IPSORT was also used to detect putative transit peptides.
Dataset Assembly. The nonredundant (NR), EST, and genomic sequence survey (GSS) public databases were searched by blast (23) using B. natans EST sequences as queries. ESTs and GSSs were downloaded and assembled to form contiguous ORFs encoding plastid-targeted proteins from the following organisms: Porphyra yezoensis, Phaeodactylum tricornutum, Chlamydomonas reinhardtii, Dunaliella salina, Marchantia polymorpha, Lycopersicon esculentum, Hordeum vulgare, Glycine max, and Physcomitrella patens. Amino acid sequences inferred from these genes and the B. natans ESTs were aligned with homologs obtained from the NR database using CLUSTALX (24). Alignments were adjusted manually, and ambiguously aligned regions were removed before phylogenetic analysis. Alignments are available in Data Set 1, which is published as supporting information on the PNAS web site, www.pnas.org.
Phylogenetic Analysis. Phylogenetic trees were inferred from amino acid sequence alignments by using maximum likelihood (ML) and ML-distance methods. ML trees were constructed by using the program PROML in PHYLIP 3.6 (http://evolution.genetics.washington.edu/phylip.html) using the Dayhoff (PAM 001) amino acid substitution matrix, the global rearrangements option, one randomized sequence-input order, and an among-site rate variation model using a six rate category discrete approximation to the Γ distribution plus an invariable rate category. The relative rates for each category were estimated using TREE-PUZZLE 5.0 (25). ML-distance trees were inferred from Γ-corrected distance matrices (constructed with TREE-PUZZLE 5.0 with eight rate categories plus an invariable rate category) using BIONJ (26), weighted neighbor-joining (27), and Fitch–Margoliash (in PHYLIP 3.6). For Fitch analyses, the global rearrangements option was used, and the sequence-input order was randomized (with 5 or 10 jumbles). Support for ProML trees was obtained by bootstrapping 100 datasets. Due to the computational intensity of ML analyses, a uniform-rates model was used for bootstrapping ML trees, with the global rearrangements option and one randomized sequence input order. Support for ML-distance trees was obtained by bootstrapping (500 replicates) with PUZZLEBOOT (A. Roger and M. Holder; www.tree-puzzle.de) as described above. ML-distance trees were constructed (and bootstrapped) for all 78 datasets, and representative datasets were chosen for more comprehensive ML and ML-distance analysis.
Results
Plastid-Targeted Proteins in B. natans. From a survey of 3,995 B. natans EST sequences, 83 distinct genes encoding 78 putative plastid-targeted proteins were identified. Eighty-six percent were full-length or nearly full-length (possessing obvious N-terminal extensions), and 84% of these were predicted to function as signal peptides by IPSORT (22) and/or SIGNALP (21). Fourteen percent of the genes were 5′ truncated; some were truncated within N-terminal leaders with characteristics of transit peptides. Others were truncated within the mature protein, and their N-terminal extensions were thus not available for analysis. Nevertheless, they are assumed to encode proteins targeted to the endosymbiont compartment and plastid by virtue of their significant similarity to cyanobacterial homologs and/or proteins known to function in the plastids of plants and algae.
The 78 B. natans plastid-targeted proteins determined in this study (Fig. 1) encompass a wide range of functions associated with plastid biochemistry and molecular biology. Included among these proteins are a large number with basic housekeeping functions, such as mRNA processing (mRNA binding protein), translation (22 ribosomal proteins, threonyl-tRNA synthetase), posttranslational protein modification (peptide deformylase), and protein folding and import (cpn60, hsp70). As expected, a large number of the proteins play a role in photosynthesis. Components of both photosystem I (e.g., PsaD, PsaE, PsaF) and II (e.g., PsbO, PsbP, PsbW) were identified, as were three subunits of plastid ATP synthase (β′, γ, and δ), ferredoxin, ferredoxin-NADP oxidoreductase, the iron/sulfur subunit of the cytochrome b6f complex, and the small subunit of ribulose 1,5-bisphoshate carboxylase/oxygenase (RuBisCO). A variety of enzymes associated with other metabolic processes were also identified, such as enoyl-ACP reductase (fatty acid biosynthesis), phytoene synthase, and phytoene dehydrogenase (carotenoid biosynthesis). No ESTs were found encoding enzymes involved in the shikimate (aromatic amino acid biosynthesis) or the non-mevalonate isopentyl diphosphate (isoprenoid biosynthesis) pathways.
Fig. 1.
Plastid-targeted proteins in the chlorarachniophyte alga B. natans grouped according to their inferred evolutionary origin. The left column indicates the protein, the center column describes its phylogenetic placement, and the right column indicates the phylogenetic support for its position (percentage bootstrap support out of 500 replicates) or gives the relevant figure number. Proteins with N-terminal extensions predicted to contain plastid-targeting information (see text) are highlighted by filled boxes, those with N-terminal extensions lacking obvious plastid-targeting information are indicated by half-filled boxes, and proteins encoded by 5′-truncated cDNAs are indicated by open boxes. Sequences containing insertions/deletions consistent with the placement of the B. natans protein in phylogenetic analysis are highlighted with asterisks. Proteins were designated “green algal” or of ambiguous origin according to criteria described in the text. The B. natans ClpP protease seems to be nucleomorph-encoded (NM). Phylogenies of all genes putatively derived by lateral transfer are available in Figs. 2, 3, 5, and 6.
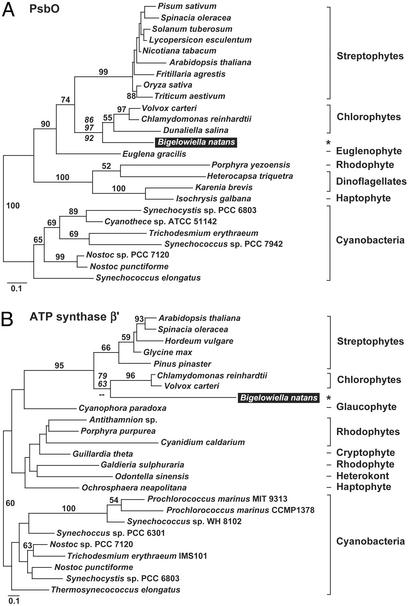
B. natans Plastid-Targeted Proteins Have Diverse Evolutionary Origins. To infer the evolutionary histories of the 78 B. natans plastid-targeted proteins, amino acid sequences were aligned with homologs from other organisms, and phylogenetic analyses were performed. ML-distance trees were inferred, and bootstrap analyses (500 replicates) were performed for all 78 datasets, and the results tabulated in Fig. 1. Twenty percent of the proteins yielded phylogenies that were ambiguous with respect to the origin of the B. natans sequence, due to the small size of the alignment, poor conservation of sequences, and/or insufficient taxonomic sampling. These proteins were not considered further. For the remaining 62 proteins, the expectation is that each would share a close relationship with chlorophyte homologs, because the B. natans endosymbiont was likely a chlorophyte. Accordingly, we conservatively considered plastid-targeted proteins to be chlorophyte-derived if they met any of several criteria. Most obviously, if a protein branched with a chlorophyte sequence in phylogenetic analyses and if the dataset contained at least one sequence from streptophytes, red algae, and cyanobacteria for appropriate comparison, it was considered chlorophyte in origin. ML phylogenies of two such examples are presented in Fig. 2. The phylogeny of PsbO (Fig. 2 A) shows the B. natans homolog branching robustly with the chlorophytes C. reinhardtii, Volvox carteri, and D. salina, with this clade forming a sister group to the streptophytes. The phylogeny of the β′ subunit of ATP synthase shows the same pattern (Fig. 2B), consistent with previous analyses of plastid- and nucleomorph-encoded genes (12, 13, 15). Other B. natans proteins (32%) produced phylogenies that were consistent with a green algal origin, but for reasons of taxonomic sampling (e.g., no chlorophyte, red algal, or cyanobacterial sequence available) or phylogenetic ambiguity (e.g., the sequence branched basal to chlorophytes and streptophytes), this finding could not be confirmed unambiguously. Without evidence to the contrary, these proteins are most likely derived from the endosymbiont, and are labeled “green algal” in Fig. 1. Altogether, these results indicate that the majority of plastid-targeted proteins in B. natans are probably of chlorophyte ancestry, and that the genes encoding them were presumably inherited from the chlorophyte endosymbiont, and transferred to the nucleus of the secondary host, as expected.
Fig. 2.
Protein ML phylogenies showing examples of B. natans plastid-targeted proteins with the expected relationship to chlorophyte green algae. (A) Oxygen-evolving enhancer 1 (PsbO) phylogeny (-lnL = 5399.95518). (B) ATP synthase β′ subunit phylogeny (-lnL = 5134.21803). B. natans plastid-targeted sequences are shaded black and highlighted with asterisks. ML bootstrap values are provided for all nodes >50%. Additional support values (italicized) are provided for the node specifically showing the placement of the B. natans sequences and are, from top to bottom, ML, weighted neighbor-joining, and Fitch–Margoliash. Scale bars indicate the expected number of amino acid substitutions (corrected) per site.
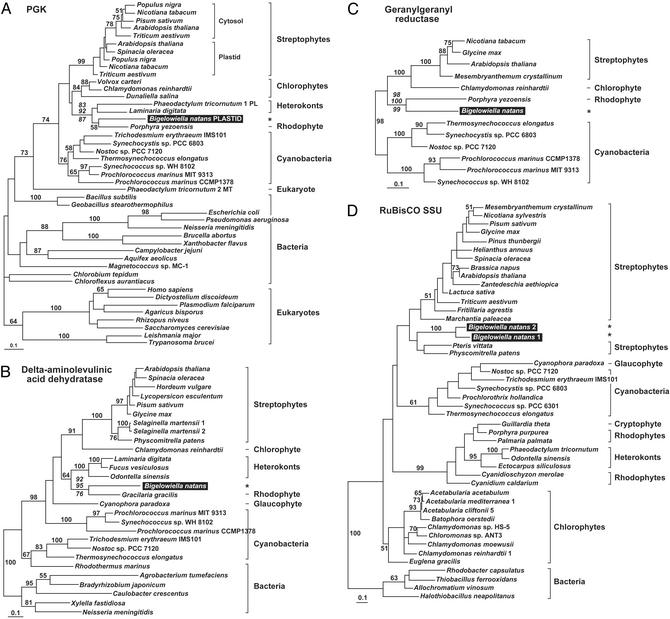
Surprisingly, however, 21% of the 62 informative proteins show phylogenetic affinities for lineages other than chlorophytes or green algae in general (Fig. 1), suggesting that they come from a source other than the endosymbiont. Interestingly, the majority of these are derived from some other algal plastid. Fig. 3 shows ML phylogenetic trees of four examples. In phosphoglycerate kinase (PGK) phylogenies, the B. natans homolog, which possesses a full-length leader, branches strongly within a clade containing the red alga P. yezoensis, as well as P. tricornutum and Laminaria digitata, two heterokont algae with red secondary plastids (Fig. 3A). The same pattern is seen in phylogenies of two enzymes involved in chlorophyll biosynthesis, delta-aminolevulinic acid dehydratase and geranylgeranyl reductase. In the delta-aminolevulinic acid dehydratase tree, the B. natans sequence forms a highly supported clade with the red alga Gracilaria gracilis, which, as expected, branches with the red secondary plastid-containing heterokonts (Fig. 3B). In this case, the red algal origin of the B. natans protein is further supported by a putatively homologous insertion shared between the B. natans and G. gracilis sequences (Fig. 4A). Interestingly, the plastid sequences in this phylogeny form a well-supported clade with a specific subset of cyanobacteria, unlike most phylogenies, which place plastids as a sister group to cyanobacteria as a whole. The taxonomic sampling available for geranylgeranyl reductase is relatively poor, but phylogenetic analysis reveals a very strongly supported relationship between B. natans and the red alga P. yezoensis, a clade well-separated from the streptophytes and the chlorophyte C. reinhardtii (Fig. 3C). We also identified two B. natans paralogs of the small subunit of RuBisCO that consistently branched near the base of the streptophyte algae (Fig. 3D), although this position was not well supported and the placement of the B. natans/streptophyte clade relative to other groups varied with different methods. The lack of resolution in these phylogenies is likely due to the fact that the RuBisCO small subunit is a very short protein and provides few sites for analysis (only 89 positions). However, independent evidence in support of the phylogeny comes from a variable region of the enzyme in which the B. natans sequences are distinctly similar to streptophyte homologs in length and sequence (Fig. 4B). In addition to these genes, seven others seem to be related to streptophytes, red algae, or algae with red algal secondary plastids (Fig. 1; and Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 3.
Protein ML phylogenies showing B. natans plastid-targeted proteins derived from other algae. (A) Phosphoglycerate kinase (PGK) (-lnL = 16275.24733). (B) Delta-aminolevulinic acid dehydratase (-lnL = 8636.18318). (C) Geranylgeranyl reductase (-lnL = 4490.47915). (D) Small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) (-lnL = 4103.65322). Bootstrap values and scale bars are as in Fig. 2. PL, plastid; MT, mitochondrial.
Fig. 4.
Representative alignments showing insertions/deletions supporting the phylogenetic position of B. natans plastid-targeted protein sequences. (A) Insertion shared between delta-aminolevulinic acid dehydratase sequences of B. natans and the red alga G. gracilis. (B) Variable region of RuBisCO small subunit proteins. The B. natans sequence shares length and sequence conservation with streptophyte homologs. Numbers for both correspond to coordinates within the Arabidopsis thaliana homolog.
More surprising still, two B. natans plastid-targeted proteins are not related to plastid sequences at all, but are instead related to bacterial proteins. The most striking example is the B. natans Calvin cycle enzyme ribulose-5-phosphate 3-epimerase, which branches very strongly within the Pseudomonadaceae and within the proteobacteria (Fig. 5), suggesting it was acquired recently by lateral gene transfer from a pseudomonad. The possibility that this gene represents a bacterial contaminant is extremely unlikely for two reasons. First, the B. natans gene encodes an N-terminal extension resembling other plastid-targeting peptides. Second, we used PCR to amplify the genomic copy of the gene and found it to possess several canonical spliceosomal introns, the hallmark of eukaryotic nuclear genes (data not shown). An equally compelling case is the B. natans GAPDH, a full-length gene that encodes a distinctive plastid-targeting leader sequence. This plastid-targeted GAPDH is not related to other plastid-targeted homologs, but instead branches with 100% support with genes from proteobacteria and Gram-positive bacteria (Fig. 6). In summary, many plastid-targeted proteins from B. natans are not derived directly from the endosymbiont, but instead come from the plastids of other algae or from bacteria.
Fig. 5.
Protein ML phylogeny (-lnL = 8618.55993) of ribulose 5-phosphate 3-epimerase. The B. natans plastid-targeted sequence is highlighted black. Bootstrap values and scale bars are as in Fig. 2. The B. natans protein is nested within the Pseudomonadaceae, indicative of lateral transfer from a pseudomonad.
The support for the position of the B. natans sequences in individual gene trees varies considerably, as does the diversity of taxa available for comparison. It is therefore possible that some of the unusual phylogenies are due to tree reconstruction artifacts. Nevertheless, even if some of the trees are misleading, there is still a large proportion of B. natans genes that were acquired by lateral transfer. This result is most apparent when the B. natans data are compared with the phylogenetic pattern observed for plastid-targeted proteins in the chlorophyte, C. reinhardtii. Ninety-one percent of our datasets contained a C. reinhardtii homolog, and these proteins typically branched as the sister to the streptophytes, as expected (e.g., Figs. 2, 3, and 5). In the few exceptions, it virtually always branched with other chlorophyte sequences (if present) and with the B. natans homolog, suggesting that the anomalous placement was a feature of the chlorophyte sequences as a whole. Therefore, in contrast to B. natans, there is no evidence for plastid-targeted proteins derived from exogenous sources in C. reinhardtii.
Discussion
Biochemical and molecular data suggest that the chlorarachniophyte endosymbiont is derived from a chlorophyte green alga (5, 11–15). Our analysis of 78 plastid-targeted proteins in B. natans shows that the evolutionary history of most such proteins is consistent with this conclusion. However, our analyses also suggest that a significant proportion (about 21%) of the nuclear genes encoding plastid-targeted proteins in B. natans are not endosymbiont-derived, but have been acquired by lateral gene transfer from a variety of sources. At face value, the evolutionary history of B. natans plastid-targeted proteins should be similar to homologs in the chlorophyte C. reinhardtii; however, we found no evidence for lateral transfer in this organism. This contrast is interesting in light of differences in the trophic behaviors of the two species. Unlike C. reinhardtii, chlorarachniophytes are capable of photosynthesis and phagocytosis. Chlorarachnion reptans has been shown to ingest a wide variety of algae (5), and partially digested and undigested bacteria have been observed inside food vacuoles in B. natans (28). The presence of a lateral transfer ratchet mechanism in phagotrophic eukaryotes has been suggested previously (29); this mechanism seems a likely explanation for the foreign plastid-targeted proteins in B. natans and the apparent lack of such proteins in C. reinhardtii.
The fact that most of the foreign genes identified in B. natans come from other algae may be explained by two factors. First, plastids use a restricted set of proteins, and other algae are the most likely source of genes encoding proteins that could conceivably function in the B. natans plastid. Second, plastid-targeted proteins must encode both a signal peptide and a transit peptide. Homologs from other algae would already possess transit peptides and perhaps also signal peptides, which may facilitate the integration of such a gene if the endogenous targeting machinery can recognize these leaders. That said, de novo acquisition of signal and transit peptides has occurred for at least some of the laterally transferred proteins identified in B. natans. This finding is most obvious in the case of transfers involving bacterial genes (ribulose-5-phosphate 3-epimerase and GAPDH), but also applies to genes transferred from plastid genomes. For example, the B. natans gene encoding ribosomal protein L1 seems to have been acquired by lateral gene transfer from a red alga or red algal plastid-containing organism (Figs. 1 and 6), and, in such organisms, rpL1 is encoded in the plastid genome, not the nucleus.
At present, lateral transfers involving plastid proteins are more easily recognized than those involving cytosolic ones, simply because of the increased taxonomic sampling of plant and algal plastid proteins. There is, however, no obvious reason to assume that the acquisition of foreign genes by chlorarachniophytes is limited to those involved in plastid function. Indeed, it has already been shown that the chlorarachniophyte cytosolic enolase is derived from a streptophyte (30). It is interesting to note that a recent analysis of the Arabidopsis genome showed that the plant cytosol contains a significant number of proteins derived from the ancestral plastid (31). Our work extends this observation to the plastid itself by showing that the B. natans organelle is a mosaic of proteins with different evolutionary histories.
Analyses of bacterial and archaeal genomes have made it abundantly clear that lateral gene transfer is an important force in prokaryotic evolution (32), but whether this extends to eukaryotes is less obvious. Although a handful of gene transfers involving eukaryotes have been documented (e.g., refs. 33 and 34), the full impact of lateral gene transfer in eukaryotic genomes is largely unexplored. Recently, however, a survey of diplomonad genes identified numerous instances of lateral gene transfer (35), and a comprehensive study of flowering plants has revealed rampant transfer between their mitochondrial genomes (36). Whether lateral transfer will prove to be as important in eukaryotes as in prokaryotes remains to be seen, but the wealth of eukaryotic genome sequence data on the horizon should soon provide an answer. At present, it seems that lateral gene transfer has been a factor in the evolution of eukaryotic genomes, but that its impact may vary from lineage to lineage.
Supplementary Material
Acknowledgments
We thank G. I. McFadden and P. R. Gilson for a B. natans cDNA library and N. M. Fast and B. S. Leander for helpful comments on the manuscript. This work was supported by Grant 227301-00 from the Natural Sciences and Engineering Research Council of Canada (to P.J.K.). J.M.A. is supported by postdoctoral fellowships from the Canadian Institutes of Health Research and the Killam Foundation. P.J.K. is a scholar of the Canadian Institute for Advanced Research, the Canadian Institutes of Health Research, and the Michael Smith Foundation for Health Research.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ML, maximum likelihood; RuBisCO, ribulose 1,5-bisphoshate carboxylase/oxygenase.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY267621–AY267704).
See commentary on page 7419.
References
- 1.Delwiche, C. F. & Palmer, J. D. (1997) in Origins of Algae and Their Plastids, ed. Bhattacharya, D. (Springer, New York), pp. 53-86.
- 2.McFadden, G. I. (2001) J. Phycol. 37, 951-959. [Google Scholar]
- 3.Moreira, D., Le Guyader, H. & Phillippe, H. (2000) Nature 405, 69-72. [DOI] [PubMed] [Google Scholar]
- 4.Archibald, J. M. & Keeling, P. J. (2002) Trends Genet. 18, 577-584. [DOI] [PubMed] [Google Scholar]
- 5.Hibberd, D. J. & Norris, R. E. (1984) J. Phycol. 20, 310-330. [Google Scholar]
- 6.McFadden, G. I., Gilson, P. R., Hofmann, C. J., Adcock, G. J. & Maier, U. G. (1994) Proc. Natl. Acad. Sci. USA 91, 3690-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Archibald, J. M., Longet, D., Pawlowski, J. & Keeling, P. J. (2003) Mol. Biol. Evol. 20, 62-66. [DOI] [PubMed] [Google Scholar]
- 8.Bhattacharya, D., Helmchen, T. & Melkonian, M. (1995) J. Eukaryot. Microbiol. 42, 64-68. [DOI] [PubMed] [Google Scholar]
- 9.Cavalier-Smith, T. (1998) Biol. Rev. Camb. Philos. Soc. 73, 203-266. [DOI] [PubMed] [Google Scholar]
- 10.Keeling, P. J. (2001) Mol. Biol. Evol. 18, 1551-1557. [DOI] [PubMed] [Google Scholar]
- 11.Durnford, D. G., Deane, J. A., Tan, S., McFadden, G. I., Gantt, E. & Green, B. R. (1999) J. Mol. Evol. 48, 59-68. [DOI] [PubMed] [Google Scholar]
- 12.Ishida, K., Cao, Y., Hasegawa, M., Okada, N. & Hara, Y. (1997) J. Mol. Evol. 45, 682-687. [DOI] [PubMed] [Google Scholar]
- 13.Ishida, K., Green, B. R. & Cavalier-Smith, T. (1999) Mol. Biol. Evol. 16, 321-331. [Google Scholar]
- 14.McFadden, G. I., Gilson, P. R. & Waller, R. F. (1995) Arch. Protistenkd. 145, 231-239. [Google Scholar]
- 15.Van de Peer, Y., Rensing, S. A. & Maier, U.-G. (1996) Proc. Natl. Acad. Sci. USA 93, 7732-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin, W. & Herrmann, R. G. (1998) Plant Physiol. 118, 9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruce, B. D. (2001) Biochim. Biophys. Acta 1541, 2-21. [DOI] [PubMed] [Google Scholar]
- 18.Ishida, K., Green, B. R. & Cavalier-Smith, T. (2000) J. Phycol. 36, 1135-1144. [Google Scholar]
- 19.McFadden, G. I. (1999) J. Eukaryot. Microbiol. 46, 339-346. [DOI] [PubMed] [Google Scholar]
- 20.van Dooren, G. G., Schwartzbach, S. D., Osafune, T. & McFadden, G. I. (2001) Biochim. Biophys. Acta 1541, 34-53. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen, H., Engelbrecht, J., Brunak, S. & von Heijne, G. (1997) Protein Eng. 10, 1-6. [DOI] [PubMed] [Google Scholar]
- 22.Bannai, H., Tamada, Y., Maruyama, O., Nakai, K. & Miyano, S. (2002) Bioinformatics 18, 298-305. [DOI] [PubMed] [Google Scholar]
- 23.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeanmougin, F., Thompson, J. D., Gouy, M., Higgins, D. G. & Gibson, T. J. (1998) Trends Biochem. Sci. 23, 403-405. [DOI] [PubMed] [Google Scholar]
- 25.Strimmer, K. & von Haeseler, A. (1996) Mol. Biol. Evol. 13, 964-969. [Google Scholar]
- 26.Gascuel, O. (1997) Mol. Biol. Evol. 14, 685-695. [DOI] [PubMed] [Google Scholar]
- 27.Bruno, W. J., Socci, N. D. & Halpern, A. L. (2000) Mol. Biol. Evol. 17, 189-197. [DOI] [PubMed] [Google Scholar]
- 28.Moestrup, Ø. & Sengco, M. (2001) J. Phycol. 37, 624-646. [Google Scholar]
- 29.Doolittle, W. F. (1998) Trends Genet. 14, 307-311. [DOI] [PubMed] [Google Scholar]
- 30.Keeling, P. J. & Palmer, J. D. (2001) Proc. Natl. Acad. Sci. USA 98, 10745-10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, W., Rujan, T., Richly, E., Hansen, A., Cornelsen, S., Lins, T., Leister, D., Stoebe, B., Hasegawa, M. & Penny, D. (2002) Proc. Natl. Acad. Sci. USA 99, 12246-12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doolittle, W. F., Boucher, Y., Nesbø, C. L., Douady, C. J., Andersson, J. O. & Roger, A. J. (2003) Philos. Trans. R. Soc. London B 358, 39-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andersson, J. O. & Roger, A. J. (2001) Eukaryot. Cell 1, 304-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qian, Q. & Keeling, P. J. (2001) Protist 152, 193-201. [DOI] [PubMed] [Google Scholar]
- 35.Andersson, J. O., Sjögren, Å. M., Davis, L. A. M., Embley, T. M. & Roger, A. J. (2003) Curr. Biol. 13, 94-104. [DOI] [PubMed] [Google Scholar]
- 36.Bergthorsson, U., Adams, K. L., Thomason, B. & Palmer, J. D. (2003) Nature, in press. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.