Abstract
Prestin, the fifth member of the anion transporter family SLC26, is the outer hair cell molecular motor thought to be responsible for active mechanical amplification in the mammalian cochlea. Active amplification is present in a variety of other auditory systems, yet the prevailing view is that prestin is a motor molecule unique to mammalian ears. Here we identify prestin-related SLC26 proteins that are expressed in the auditory organs of nonmammalian vertebrates and insects. Sequence comparisons revealed the presence of SLC26 proteins in fish (Danio, GenBank accession no. AY278118, and Anguilla, GenBank accession no. BAC16761), mosquitoes (Anopheles, GenBank accession nos. EAA07232 and EAA07052), and flies (Drosophila, GenBank accession no. AAF49285). The fly and zebrafish homologues were cloned and, by using in situ hybridization, shown to be expressed in the auditory organs. In mosquitoes, in turn, the expression of prestin homologues was demonstrated for the auditory organ by using highly specific riboprobes against rat prestin. We conclude that prestin-related SLC26 proteins are widespread, possibly ancestral, constituents of auditory organs and are likely to serve salient roles in mammals and across taxa.
Keywords: mechanosensation, Anopheles, Drosophila, Danio
The mammalian cochlea achieves its sensitivity and frequency selectivity by a process of active mechanical amplification that is based on voltage-dependent contractions of outer hair cells (for reviews see refs. 1–5). A molecular motor driving this cellular electromotility has been identified recently to be prestin, a membrane-based molecule that seems to change its conformation in a voltage-dependent way (6–9).
Apart from mammals, active mechanical amplification also improves hearing in other animals, i.e., lower tetrapods (for reviews see refs. 10 and 11) and insects (12, 13). In these animals, mechanical amplification is deemed to involve motor mechanisms other than prestin; proposed candidate motors include myosin-based adaptation motors and Ca2+-dependent reclosure motions of mechanotransduction channels in lower tetrapods (11), whereas microtubule-dependent, ciliary motors have been indicated to bring about active mechanical amplification in the auditory systems of insects (13). Hence, different taxa may have solved the need for sensitive hearing in analogous ways, with the prestin motor being an evolutionary innovation unique to the mammalian cochlea.
Prestin, encoded by 18 exons, shows highest homology to other members of the newly discovered solute carrier (SLC) anion transport family SLC26 within the sulfate transport region between residue 98–135 (6). Amino acid sequence and gene-structure analysis identified prestin to be the fifth member of this family (prestin, SLC26A5) (14). Apart from prestin, this family includes, among others, pendrin (PDS, SLC26A4) (15), down-regulated in adenoma (DRA, SLC26A3) (16), diastrophic dysplasia sulfate transporter (DTDST, SLC26A2) (17), and SLC26A6 (18). Functionally, SLC26 proteins have been proposed to serve as chloride-iodide transporters, Cl-/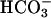 exchangers, or sulfate transporters and, in the case of prestin, motors (9, 14).
exchangers, or sulfate transporters and, in the case of prestin, motors (9, 14).
Using a 723-bp clone derived from the rat prestin cDNA sequence covering the deduced amino acids between exons 11 and 18, we have generated riboprobes for in situ hybridization. In the cochlea of rats, these probes selectively detect prestin (19) but not pendrin and members of another SLC family, the SLC4 family, of which AE1 (SLC4A1), AE2 (SLC4A2), and AE3 (SLC4A3) are expressed within the cochlea of rats (20–23). Interestingly, this highly specific riboprobe displayed positive hybridization in the auditory organ of mosquitoes, the Johnston's organ, which consists of thousands of radially arranged multicellular mechanoreceptor units (12). Therefore, we started to search genome databases for homologous sequences in mosquitoes and other species. Sequence alignment and gene cloning revealed prestin-homologous transcripts in nonmammalian vertebrates and insects. The transcripts were identified to be members of the SLC26 family and to be expressed in the auditory organs. These findings highlight the ubiquity of SCL26 proteins in auditory systems and raise the question about their functional roles in audition.
Materials and Methods
Animals. Wistar rats and C57BL/6 mice were purchased from Charles River Breeding Laboratories. Adult zebrafishes (Danio rerio) were obtained from a local supplier. Animals of the mosquito species Toxorhynchites brevipalpis and fruit flies Drosophila melanogaster (Oregon R) were taken from laboratory cultures. Animal experiments followed approved institutional protocols at the University of Tübingen (Tübingen, Germany).
Tissue Preparation. For in situ hybridization of postnatal cochleae of rats and mice, cochleae were isolated, fixed, decalcified, and cryosectioned as described (24, 25). For in situ hybridization of fly and mosquito antennae, heads were dissected and embedded in Tissue-Tek OCT compound (Sakura, Zoeterwoude, The Netherlands). The macular organs of the lagena and sacculus of zebrafish were dissected, fixed in 2% paraformaldehyde for 10 min, washed in PBS, and embedded. Cryosections were performed as described (24, 25).
Cloning and Sequencing of the Drosophila Prestin Homologue. For mRNA isolation, heads of adult D. melanogaster were dissected and immediately frozen in liquid nitrogen. The heads of up to 30 adult flies were pooled and stored at -70°C before use. mRNA was isolated and reverse-transcribed as described (19). By using the oligonucleotide primer DroF2 (5′-CAAAATGCCTAGTGATAAAGAAAGTCCAG-3′), spanning the region from nucleotides 179,547–179,575 deduced from the original D. melanogaster CG5485 sequence (FlyBase FBgn0036770), in combination with either the downstream primer DroR2 (5′-CATATTCCACACAATCGTGCAGGGTG-3′, nucleotides 181,987–182,012) or DroEcoRVR (5′-AGAGGAAGGTAGATATCCATGTGAAC-3′, nucleotides 181,308–181,333), a 2.21-kb and a 1.60-kb fragment of the CG5485 gene were amplified by PCR (annealing temperature, 55°C; number of cycles, 40). Both fragments were cloned into the pCRII TOPO Vector (Invitrogen) following manufacturer instructions.
Cloning and Sequencing of the Zebrafish Prestin Homologue. For mRNA isolation, the lagena and sacculus of adult D. rerio were dissected by using a stereomicroscope. Tissues were frozen immediately in liquid nitrogen. Tissues of six animals were pooled and mRNA was isolated and reverse-transcribed as described (19). By using the oligonucleotide primer DanF1 (5′-TGCAGCCATGGAGCACGTAACTGTTAG-3′) deduced from nucleotides 30,585–30,559 of the D. rerio genomic sequence z06s031971 (Ensembl Zebrafish Genome Browser, D. rerio, Assembly 06) together with the primer DanR4 (5′-CATCCCGTATGAGATACCAGTGGACAC-3′, nucleotides 22,474–22,500) and the primer combination DanF3 (5′-GTGTCCACTGGTATCTCATACGGGATG-3′, nucleotides 22,474–22,500) and DanR1 (5′-ATGAGGTCAGGGGTCACAGGCTCACTG-3′, nucleotides 13,975–14,001), two overlapping fragments spanning 2,151 bp of coding sequence altogether were amplified by PCR (annealing temperature, 60°C; number of cycles, 40) and cloned as described for the Drosophila gene product CG5485.
Riboprobe Synthesis and in Situ Hybridization. In situ hybridization with prestin-specific riboprobes in vertebrates was performed as described in ref. 19. For Drosophila-specific riboprobes, the 1.6-kb CG5485-PA cDNA fragment was used for in vitro transcription as described for the prestin-specific riboprobe. Sections of D. melanogaster heads were postfixed with 2% paraformaldehyde for 15 min and washed in PBS. In situ hybridization was performed as described (24, 25).
Sequence Analysis. Sequence analysis was performed by using programs in the Sequence Manipulation Suite (http://bioinformatics.org/sms/index.html), the advanced genebee clustalw 1.75 (http://drone1.genebee.msu.su/clustal/advanced.html), the ExPASy translate tool (www.expasy.org/tools/dna.html), the blast services (www.ncbi.nlm.nih.gov/blast/), the Ensembl Zebrafish and Mosquito Genome Browser (www.ensembl.org) as well as the FlyBase BDGB blast server (http://flybase.bio.indiana.edu). Pairwise alignments were obtained with the CLUSTALW program, and the identity scores were represented graphically in a dendrogram.
Results
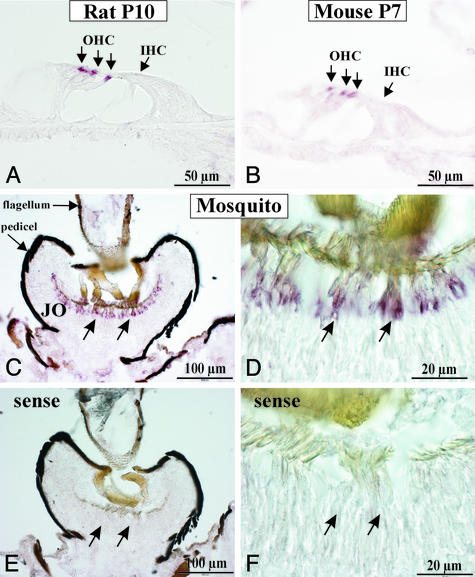
Prestin-Specific Riboprobes Detect Mechanosensory Cells in the Hearing Organs of Rodents and Mosquitoes. Using the 723-bp cDNA clone of rat prestin (19) for synthesis of digoxigenin-labeled riboprobes covering residues 465–704 of the deduced amino acids of rat prestin, we localized prestin mRNA in cochleae of rats and mice (Fig. 1 A and B). Hybridization confirmed the exclusive expression in outer hair cells previously described for rats (19). No hybridization signals were detected with corresponding sense probes (data not shown).
Fig. 1.
Expression of mRNA in cross sections of rat cochlea at postnatal day 10 (A), mouse cochlea at postnatal day 7 (B), and the mosquito T. brevipalpis (C and D) using in situ hybridization with rat prestin-specific riboprobes. Note the restricted localization of prestin mRNA to the outer hair cells (OHC) in rat and mouse cochlea (three arrows in row). IHC, inner hair cell. (C) In cross sections of T. brevipalpis antennae, hybridization signals occur in the second antennal segment (pedicel). (D) Higher magnification of positive signals localizes hybridization to chordotonal sensilla, the auditory mechanosensory units of Johnston's organ (JO). (E and F) No signals were observed with the sense probe.
To assess whether prestin is unique to the mammalian cochlea, we used rat prestin riboprobes to test hybridization in a mosquito, i.e., T. brevipalpis, a species that reportedly uses mechanical amplification to improve the sensitivity of its antennal ears (12). Positive hybridization signals were detected in the chordotonal mechanosensory units of Johnston's organ, the auditory sense organ in the antenna (Fig. 1 C and D), whereas sense probes failed to show positive signals (Fig. 1 E and F).
Apparently, these insects express a prestin homologue in their hearing organs, with sequence similarities within the region between exons 11 and 18 (amino acids 465–704) of mammalian prestin. We therefore searched for prestin-homologous genes in insects and nonmammalian vertebrates choosing two different strategies. First, we screened available genome databases for sequences homologous to the deduced amino acids spanning the region of the rat prestin riboprobe. Second, we cloned the presumptive prestin-homologous genes identified in Danio and Drosophila to investigate their expression in auditory organs.
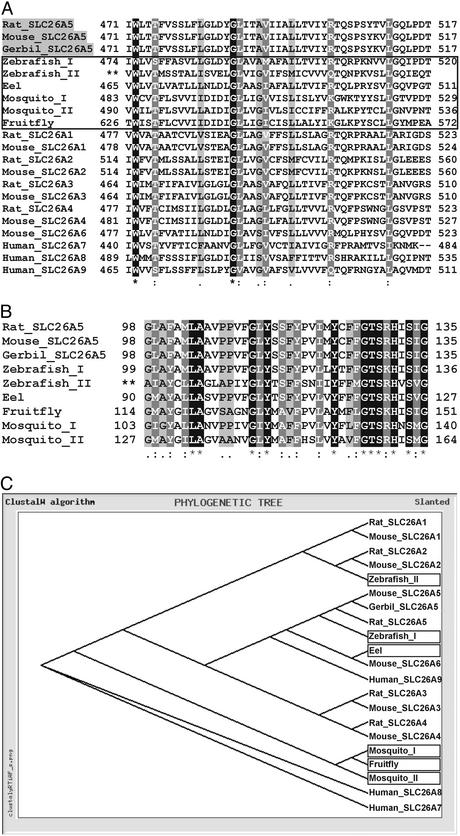
Sequence Alignments Among Various Species. By using the deduced amino acid sequence 465–704 of the rat prestin gene [Rattus norvegicus, GenBank accession no. NP_110467 (Rat_SLC26A5)], blast algorithms of the NCBI, Ensembl, and FlyBase genome browsers identified related sequences in D. rerio [genomic sequences z06s031971 (Zebrafish_I) and z06s015490 (Zebrafish_II)], Anguilla japonica [GenBank accession no. BAC16761 (Eel)], Anopheles gambiae [GenBank accession no. EAA07232, predicted gene product agCP10636 (Mosquito_I), and EA A07052, predicted gene product agCP7199 (Mosquito_II)] and D. melanogaster [GenBank accession no. AAF49285, predicted gene product CG5485-PA (Fruit fly)]. Alignment between these sequences and the best-matching amino acids stretches between 471 and 517 of rodent prestin SLC26A5 [R. norvegicus NP_110467 (Rat_SLC26A5); Mus musculus, GenBank accession no. AAG59999 (Mouse_SLC26A5); Meriones unguiculatus, GenBank accession no. AAF71715 (Gerbil_SLC26A5)] are shown in Fig. 2A (highlighted in gray). To further specify the identified gene sequences, the alignment was extended to the other members of the SLC26 family (A1–A9) (Fig. 2 A and C). Homology searches within the Ensembl Zebrafish Genome Database identified a best-matching region spanning translated nucleotides 18,884–18,783 and 18,481–18,410 of the Zebrafish_I sequence. The corresponding deduced amino acid residues are illustrated (Fig. 2 A), with numbers referring to the transcripts cloned in this study. A sulfate anion transport motif with high homology to the rat prestin amino acid sequence 99–136 was detected between the translated nucleotides 27541–27428, indicated as 99–136 (Fig. 2B, Zebrafish_I). This alignment identified the Zebrafish_I gene product to be closely related to prestin (SLC26A5) of rat, mice, and gerbil (Fig. 2C, Zebrafish_I), whereas another related genomic zebrafish sequence derived from Zebrafish_II (Fig. 2 A and B) best matches rodent SLC26A2 (Fig. 2C).
Fig. 2.
Sequence comparison and phylogenetic relationship of prestin-homologous genes. (A) Predicted amino acids 471–517 of rodent prestin (rat, mouse, and gerbil), which are part of our rat-specific prestin riboprobe, are aligned with deduced amino acids of DNA from different species (zebrafish, eel, mosquito, and fruit fly; boxed) as well as with representative members of the SLC26 family. (B) Alignment of rodent prestin (SLC26A5) with the same species within the highly conserved sulfate anion transport motif (amino acids 98–135). Identical amino acids (*) are shown in white type on a black background, strongly similar amino acids (:) are shown in white type on a gray background, and weakly similar amino acids (.) are shown in black type on a gray background. No identity or similarity is not highlighted. (C) The relationship between prestin-homologous genes from different species and from a selection of representative members of the SLC26 family. For the generation of the phylogenetic tree, the alignment shown in A was used.
Homology screening was extended to other species including freshwater eel A. japonica, the mosquito A. gambiae, and the fruit fly D. melanogaster. Again database analysis revealed best-matching sequences, which in all cases belonged to genes thus far uncharacterized. In the eel A. japonica a blast search identified the gene product Eel (Fig. 2 A and C), which is closest to Mouse_SLC26A6 (GenBank accession no. AAL13129; Fig. 2 A and C, Mouse_SLC26A6). In mosquitoes, the Ensembl Mosquito Genome Browser highlighted two related gene products, Mosquito_I and Mosquito_II (Fig. 2 A). Both products exhibit the sulfate anion transport motif (Fig. 2B, Mosquito_I, _II). In Drosophila, the FlyBase BDGB blast server identified the gene product Fruitfly (Fig. 2 A), again a gene endowed with the sulfate anion transport motif (Fig. 2B).
As expected for SLC26 members, all the gene products identified in fish and insects displayed the sulfate transporter and anti-sigma-factor antagonist domain (STAS) characteristic for this family (26). When sequences taken from the sulfate transporter motif (Fig. 2B) or full-length amino acid sequences of SLC26 members in all species shown in Fig. 2C were used for the generation of a dendrogram, a similar topography was observed (data not shown).
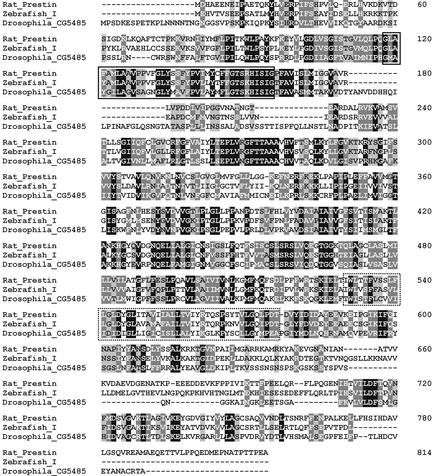
Cloning and Analysis of a Prestin Homologue in Drosophila. To analyze the expression of the SLC26 member of D. melanogaster, we cloned the predicted gene product CG5485-PA as described in Materials and Methods. Sequence analysis with the FlyBase BDGB blast server predicted an ORF of 2,484 bp encoding a 742-aa protein. A fragment of 2.21 kb and a fragment of 1.6 kb, both displaying complete identity with the corresponding nucleotides from the predicted gene product CG5485-PA, were cloned. The 2.21-kb fragment encodes amino acids 1–735 of the 742 predicted amino acids. Within the ORF, homology to SLC26A5 was found (Fig. 3), especially in regions 98–135 (Fig. 2B) and 471–517 (Fig. 2 A) of the rat prestin amino acid sequence. A sulfate transporter and anti-sigma-factor antagonist domain (STAS) between amino acids 587–709 further confirmed the gene to be an SLC26 member.
Fig. 3.
Sequence alignment of the predicted amino acid sequence of the Drosophila CG5485 and the D. rerio Zebrafish_I gene product with rat prestin. Identical amino acids are shown in white type on a black background, strongly similar amino acids are shown in white type on a gray background, and weakly similar amino acids are shown in black type on a gray background. Rat prestin and Drosophila_CG5485 share an identity of 26.5%, a strong similarity of 49.4%, and a weak similarity of 61.4%. Rat prestin and Zebrafish_I share an identity of 49.8%, a strong similarity of 68.4%, and a weak similarity of 76.6%. The conserved sulfate anion transport motif is boxed (compare with Fig. 2B); the region with highest homology within the rat prestin riboprobe is dot-boxed (compare with Fig. 2 A).
During the cloning of the CG5485-PA, an additional 400-bp band was recurrently obtained from RT-PCR amplification, indicating the presence of an alternatively spliced form of the gene. The transcript was cloned and sequenced. The transcript encodes a CG5485 subtype with a deletion of 1,855 nucleotides, affecting the deduced amino acids 8–627 and, possibly, leading to a truncated form of the protein.
Cloning and Analysis of a Prestin Homologue in D. rerio. To examine the expression of the putative zebrafish prestin-homologous gene referred to as Zebrafish_I, we derived primers from the genomic sequence z06s031971 (Ensembl Zebrafish Genome Browser) and isolated mRNA from the hearing organ (lagena and sacculus) for RT-PCR. Two overlapping fragments of 2,151 bp were cloned that encode an ORF for a protein of 714 aa. Sequence analysis with the Ensembl Genome Server revealed these transcripts to be identical with the respective gene encoded in the genomic sequence z06s031971. An alignment between the 2,151-bp fragment and the genomic sequence indicated the presence of at least 15 exons (data not shown). The nucleotide sequence of the 2,151-bp transcript was submitted to GenBank (accession no. AY278118). The cloning of the uttermost 3′ end containing a stop codon has not been completed yet. The homologies between the 2,142-bp sequence transcribed, starting with a putative ATG codon (M) at base pair 8, rat prestin and the Drosophila gene product CG5485 are shown in Fig. 3. Again, the assignment of this gene to the SLC26 family was additionally supported by the presence of a sulfate transporter and anti-sigma-factor antagonist domain (STAS) (Fig. 3, amino acids 650–711). Between rat prestin and Zebrafish_I, we observed an identity of 49.8%, a strong similarity of 68.4%, and a weak similarity of 76.6%.
Prestin-Homologous mRNA in Sensory Systems of Zebrafish and D. melanogaster. To evaluate whether either rat prestin or its Drosophila homologues display hybridization in the auditory organs of any of the other animals examined, in situ hybridization studies were performed by using riboprobes of in vitro-transcribed 1.6-kb cDNA fragments of Drosophila CG5485-PA along with rat prestin cDNA riboprobes.
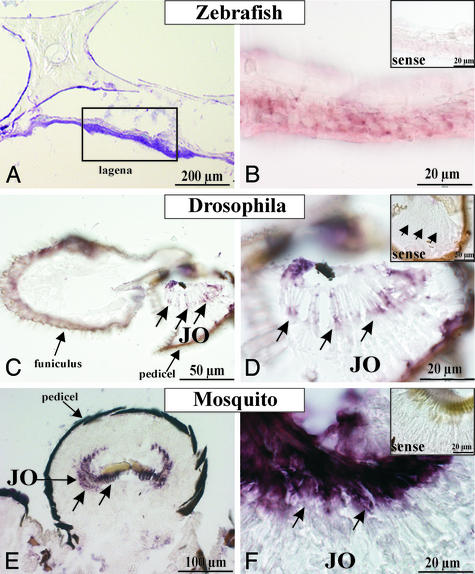
Rat-derived prestin riboprobes, but not Drosophila-derived riboprobes (data not shown), detected reactive cells distributed within the sensory epithelium of the zebrafish lagena (Fig. 4B), whereas appropriate sense probes were negative (Fig. 4B Inset). In turn in Drosophila, Drosophila CG5485-specific riboprobes but not rat prestin-specific riboprobes showed hybridization with the chordotonal sensilla of Johnston's organ (Fig. 4 C and D), localized in the second antennal segment, the pedicel (Fig. 4C), neighboring the third antennal segment, the funiculus (Fig. 4C). Furthermore, Drosophila CG5485-specific riboprobes strongly crosshybridized with mRNA within the pedicel of the mosquito T. brevipalpis (Fig. 4E). Hybridization signals appear within the Johnston's organ (Fig. 4 E and F). Radially arranged hybridization signals (Fig. 4F) suggest the localization within the radially arranged mechanoreceptor units. Sense probes gave no hybridization signals (Fig. 4 D and F Insets).
Fig. 4.
(A) Toluidin blue-stained section showing the sensory epithelium of the zebrafish lagena. (B) In situ hybridization with rodent-specific prestin riboprobes detects the mRNA encoding an SLC26A-homologous transcript in the sensory epithelium of the lagena of the zebrafish. (Inset) No signal was visible with the appropriate sense riboprobe. In situ hybridization with Drosophila CG5485-specific riboprobes detects reactive cells within the Johnston's organ (JO) in the second segment of antennae (pedicel) of D. melanogaster (C and D) and of the mosquito T. brevipalpis (E and F). (D and F Insets) Sense probes gave no signals.
Discussion
Prestin (SLC26A5) (14) is a motor protein specific to cochlear outer hair cells and thought to be responsible for active mechanical amplification in mammalian ears (6–9). Beside prestin, pendrin (SLC26A4) (15) is a member of the SLC26 family, displaying a defined expression pattern in the developing cochlea different from that of prestin. Thus far no further members of the SLC26 family have been identified in mechanosensitive organs, neither in mammals nor in nonmammalian vertebrates or insects. This study establishes the presence of SLC26 members for the auditory sense organs of nonmammalian vertebrates, i.e., teleost fish and insects, documenting that SLC26 proteins occur in a vast variety of ears.
Sequence alignment and comparative analysis (Fig. 2 A and C) show that the members of the SLC26 family identified in insects constitute a separate branch of the family tree, whereas those of fish cluster with pendrin (Eel), diastrophic dysplasia sulfate transporter (Zebrafish_II), and prestin (Zebrafish_I). This division seems to be reflected in the number of exons, which is small in insects and mammalian SLCA1/A2 (≥3 exons) but considerably higher for Zebrafish_I (≥15 exons) and mammalian pendrin and prestin (>18 exons) (14, 27). The insect branch is evolutionarily interesting. First, although the ears of insects and vertebrates have evolved independently, their auditory mechanosensors (chordotonal sensilla and hair cells, respectively) may share a common evolutionary origin (28). Such a scenario is supported by the fact that homologous genes control the developments of these sensors (29) and may explain the presence of closely related SLC26 members in insect and vertebrate ears. Second, because insects reportedly use active mechanical amplification to improve the sensitivity of their ears (12, 13), the identification of SLC26 members in insect auditory systems raises the possibility that these ears are endowed with prestin-like motors.
The closest relative of prestin was identified in fish. Evolutionarily, the auditory organs of teleost fish (sacculus and lagena) and mammals (cochlea) can be traced back to a single evolutionary origin (30). With an amino acid sequence identity of 49.8%, Zebrafish_I is closer to rat prestin than any other known member of the mammalian SLC26 family (rat pendrin, 36.4% identity). Notably, unlike tetrapods and insects, fish are not known to improve audition by active mechanical amplification. Physiological evidence, however, indicates that their hair cells can be motile. The hair bundles of fish hair cells have been observed to move both spontaneously and in response to electrical stimulation (31), similar to the mechanically active hair bundles of lower tetrapods (11). Such a form of cellular motility is notably different from the prestin-mediated contractions of mammalian outer hair cells, indicating that motors other than prestin are involved. Conversely, the identification of a close prestin homologue in the auditory system of fish suggests that prestin-like motors may be at work. Clearly, future molecular and functional studies are required to resolve this apparent conundrum. The presence of prestin-related SLC26 members in genetically accessible animals such as zebrafish and fruit flies may help resolving which motors are at work in ears and what their respective contributions are to the process of hearing.
Acknowledgments
We thank Frank Schnorrer (Max Planck Institute, Tübingen, Germany) and Hans Briegel (University of Zurich) for providing flies and mosquitoes, respectively, and Martina Knirsch for introducing us to dissecting fish ears. This work was supported by a Royal Society Research fellowship (to M.C.G.), the Swiss National Science Foundation (D.R., H.K., and A.M.), the University of Bristol (D.R.), the Interdisciplinary Center of Clinical Research (Tübingen, Germany), and Deutsche Forschungsgemeinschaft SFB 430/Kni-B3.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: SLC, solute carrier anion transport family.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY278118).
References
- 1.Dallos, P. J. (1992) J. Neurosci. 12, 4575-4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruggero, M. A. (1992) Curr. Opin. Neurobiol. 2, 449-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yates, G. K., Johnstone, B. M., Patuzzi, R. B. & Robertson, D. (1992) Trends Neurosci. 15, 57-61. [DOI] [PubMed] [Google Scholar]
- 4.Nobili, R., Mammano, F. & Ashmore, J. (1998) Trends Neurosci. 21, 159-167. [DOI] [PubMed] [Google Scholar]
- 5.Robles, L. & Ruggero, M. A. (2001) Physiol. Rev. 81, 1305-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng, J., Shen, W., He, D. Z., Long, K. B., Madison, L. D. & Dallos, P. (2000) Nature 405, 149-155. [DOI] [PubMed] [Google Scholar]
- 7.Ludwig, J., Oliver, D., Frank, G., Klöcker, N., Gummer, A. W. & Fakler, B. (2001) Proc. Natl. Acad. Sci. USA 98, 4178-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dallos, P. & Fakler, B. (2002) Nat. Rev. Mol. Cell Biol. 3, 104-111. [DOI] [PubMed] [Google Scholar]
- 9.Liberman, M. C., Gao, J., He, D. Z., Wu, X., Jia, S. & Zuo, J. (2002) Nature 419, 300-304. [DOI] [PubMed] [Google Scholar]
- 10.Manley, G. A. (2001) J. Neurophysiol. 86, 541-549. [DOI] [PubMed] [Google Scholar]
- 11.Hudspeth, A. J. (1997) Curr. Opin. Neurobiol. 7, 480-486. [DOI] [PubMed] [Google Scholar]
- 12.Göpfert, M. C. & Robert, D. (2001) Proc. R. Soc. London Ser. B 268, 333-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Göpfert, M. C. & Robert, D. (2003) Proc. Natl. Acad. Sci. USA 100, 5514-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng, J., Madison, L. D., Oliver, D., Fakler, B. & Dallos, P. (2002) Audiol. Neurootol. 7, 9-12. [DOI] [PubMed] [Google Scholar]
- 15.Everett, L. A., Glaser, B., Beck, J. C., Idol, J. R., Buchs, A., Heyman, M., Adawi, F., Hazani, E., Nassir, E., Baxevanis, A. D., et al. (1997) Nat. Genet. 17, 411-422. [DOI] [PubMed] [Google Scholar]
- 16.Kere, J., Lohi, H. & Hoglund, P. (1999) Am. J. Physiol. 276, G7-G13. [DOI] [PubMed] [Google Scholar]
- 17.Hastbacka, J., de al Chapelle, A., Mahtani, M. M., Clines, G., Reeve-Daly, M. P., Daly, M., Hamilton, B. A., Kusumi, K., Trivedi, B., Weaver, A., et al. (1994) Cell 78, 1073-1087. [DOI] [PubMed] [Google Scholar]
- 18.Lohi, H., Kujala, M., Kerkela, E., Saarialho-Kere, U., Kestila, M. & Kere, J. (2000) Genomics 70, 102-112. [DOI] [PubMed] [Google Scholar]
- 19.Weber, T., Zimmermann, U., Winter, H., Mack, A., Köpschall, I., Rohbock, K., Zenner, H. P. & Knipper, M. (2002) Proc. Natl. Acad. Sci. USA 99, 2901-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalinec, F., Kalinec, G., Negrini, C. & Kachar, B. (1997) Hear. Res. 110, 141-146. [DOI] [PubMed] [Google Scholar]
- 21.Mhatre, A. N., Charachon, G., Alper, S. L. & Lalwani, A. K. (1998) Biochim. Biophys. Acta 1414, 1-15. [DOI] [PubMed] [Google Scholar]
- 22.Zimmermann, U., Köpschall, I., Rohbock, K., Bosmann, G. J., Zenner, H. P. & Knipper, M. (2000) Mol. Cell. Biochem. 205, 25-37. [DOI] [PubMed] [Google Scholar]
- 23.Everett, L. A., Belyantseva, I. A., Noben-Trauth, K., Cantos, R., Chen, A., Thakkar, S. I., Hoogstraten-Miller, S. L., Kachar, B., Wu, D. K. & Green, E. D. (2001) Hum. Mol. Genet. 15, 153-161. [DOI] [PubMed] [Google Scholar]
- 24.Knipper, M., Gestwa, L., Ten Cate, W. J., Lautermann, J., Brugger, H., Maier, H., Zimmermann, U., Rohbock, K., Köpschall, I., Wiechers, B. & Zenner, H. P. (1999) J. Neurobiol. 38, 338-356. [PubMed] [Google Scholar]
- 25.Knipper, M., Zinn, C., Maier, H., Praetorius, M., Rohbock, K., Köpschall, I. & Zimmermann, U. (2000) J. Neurophysiol. 83, 3101-3112. [DOI] [PubMed] [Google Scholar]
- 26.Aravind, L. & Koonin, E. V. (2000) Curr. Biol. 10, R53-R55. [DOI] [PubMed] [Google Scholar]
- 27.Lohi, H., Kujala, M., Mäkelä, S., Lehtonen, E., Kestilä, M., Saarialho-Kere, U., Markovich, D. & Kere, J. (2002) J. Biol. Chem. 277, 14246-14254. [DOI] [PubMed] [Google Scholar]
- 28.Gillespie, P. G. & Walker, R. G. (2001) Science 413, 194-202. [DOI] [PubMed] [Google Scholar]
- 29.Hassan, B. A. & Bellen, H. J. (2000) Genes Dev. 14, 1852-1865. [PubMed] [Google Scholar]
- 30.Fritzsch, B., Beisel, K. W., Jones, K., Farinas, I., Maklad, A., Lee, J. & Reichardt, L. F. (2002) J. Neurobiol. 53, 143-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rüsch, A. & Thurm, U. (1990) Hear. Res. 48, 247-264. [DOI] [PubMed] [Google Scholar]