Abstract
Despite their high degree of genomic similarity, reminiscent of their relatively recent separation from each other (≈6 million years ago), the molecular basis of traits unique to humans vs. their closest relative, the chimpanzee, is largely unknown. This report describes a large-scale single-contig comparison between human and chimpanzee genomes via the sequence analysis of almost one-half of the immunologically critical MHC. This 1,750,601-bp stretch of DNA, which encompasses the entire class I along with the telomeric part of the MHC class III regions, corresponds to an orthologous 1,870,955 bp of the human HLA region. Sequence analysis confirms the existence of a high degree of sequence similarity between the two species. However, and importantly, this 98.6% sequence identity drops to only 86.7% taking into account the multiple insertions/deletions (indels) dispersed throughout the region. This is functionally exemplified by a large deletion of 95 kb between the virtual locations of human MICA and MICB genes, which results in a single hybrid chimpanzee MIC gene, in a segment of the MHC genetically linked to species-specific handling of several viral infections (HIV/SIV, hepatitis B and C) as well as susceptibility to various autoimmune diseases. Finally, if generalized, these data suggest that evolution may have used the mechanistically more drastic indels instead of the more subtle single-nucleotide substitutions for shaping the recently emerged primate species.
The draft sequence of the human genome is now at the final stages of being transformed into a definitive blueprint available for high-resolution comparative analysis (1). Perhaps the most biologically enticing comparative genomics experiment would be the one between our genome and that of our closest evolutionary relative, the chimpanzee (Pan troglodytes). The chimpanzee is believed to share ≈98.77% nucleotide and >99% amino acid identity with us (2, 3). However, there are important biomedical (as well as obvious morphological and cognitive) differences between the two species, which thus far have eluded any molecular explanation within this supposedly 1% diversity range. Among these are our differential handling of a number of infectious agents, e.g., HIV (progression to AIDS) (4), late complications of hepatitis B and C (5, 6), as well as susceptibility to Plasmodium falciparum (7), which are of utmost public health importance. The molecular basis of these distinctive traits is thought to be in large part encoded within the MHC, where MHC class I molecules sample pathogen-derived antigenic peptides for recognition by the CD8+ αβ T cell receptor expressing cytotoxic T cells (8).
We have already reported the complete sequence and gene map of the 3.7-Mb human chromosome 6p21.3-located MHC (alternatively called the human leukocyte antigen or HLA) gene complex (9, 10). This is a gene-rich (224 identified loci) highly polymorphic (with some MHC genes having >400 alleles) genomic segment that is associated with a myriad (>100) of mostly autoimmune but also infectious disorders for which our molecular knowledge, for the most part, remains rudimentary. It is precisely this extremely high level of MHC polymorphism and heterozygosity that is believed to confer a selective advantage to the host in encountering the extraordinarily diverse pathogen-derived antigenic repertoire (8). The human MHC is composed of three distinct regions, designated from the centromere to the telomere as the class II, III, and I regions. The telomeric 1.8-Mb class I region harbors two notable (but not only) multicopy gene families, HLA and MIC (10, 11), which are thought to have arisen from repeated gene duplications (10, 12) and which engage a host of critical immune receptors: the T cell receptor as well as Ig and lectin-like inhibitory and activatory receptors (13, 14).
Despite the facts that structural and/or functional orthologues for all human HLA genes have been found in chimpanzee (15–19) (HLA-A/B/C/E/F/G vs. Patr-A/B/C/E/F/G) and that there is no doubt that the MHC biology between these two close species is nearly interchangeable, the genomic architecture of chimp MHC is unknown, although it is assumed to be closely linear to that of human. Our aim was to capitalize on our detailed knowledge of the human HLA region to jump-start a large-scale comparative genomic analysis with regard to that of the chimpanzee (P. troglodytes) MHC (called Patr). Not only will the chimpanzee MHC sequence provide an in-depth analysis of this important genomic region between two such closely related species, but it also has the intrinsic power to unravel the molecular basis for some important biological differences between us and the chimpanzee. In this regard, we present 1.75 Mb of continuous genomic sequence linking the Lymphotoxin B (LTB) gene in the telomeric area of the class III region to Patr-F locus (chimpanzee HLA-F orthologue) at the telomeric end of the MHC class I region.
Materials and Methods
Bacterial Artificial Chromosome (BAC) Clones and Construction of a Contig Map. Two BAC libraries, RPCI-43 and CHORI-251, constructed from white blood cells of the same male chimpanzee, were obtained from the BACPAC Resource Center at the Children's Hospital Oakland Research Institute (Oakland, CA). Hybridization screenings were performed following the recommended protocols. Hybridization probes, ≈2 kb in length, were PCR-generated from the human HLA and MIC genes (exons 2–4) as well as several MHC-based sequence tagged sites by using cloned human genomic DNA as a template. The final contig map was constructed by comparison with the complete sequence of the human MHC (9, 10).
DNA Sequencing and Analysis. Fourteen chimpanzee BAC clones that covered ≈1.75 Mb from the LTB to Patr-F genes were completely and bidirectionally shotgun sequenced with an average redundancy of 7.0×, which was sufficient for assembly and analysis of the entire sequence using previously established procedures (10, 20). The chimpanzee sequence was compared with our previously published human sequence (GenBank accession nos. AP000502–000521) (10, 20). Sequence alignments were performed and homologies determined by using the programs contained within the GENETYX Ver. 11 (www.sdc.co.jp/genetyx) and DNASIS (Hitachi, Tokyo) software packages. Dot matrix analysis was performed by using HARRPLOT Ver. 2 as part of the GENETYX package. The nucleotide diversity (21) profile was constructed after determining the percent nucleotide difference between the human and chimpanzee sequences for a sliding window of 1 kb. The diversity profile was then drawn by using the graphics output of Microsoft EXCEL. All indels were removed from the alignments to standardize the number of nucleotides examined within each window. Finally, repetitive elements were identified within the contiguous sequences by using the REPEATMASKER webserver (A. F. A. Smit and P. Green, http://ftp.genome.washington.edu/cgi-bin/RepeatMasker).
Results and Discussion
Comparative MHC Genomics Reveals Indels as the Major Evolutionary Driving Force Between Human and Chimpanzee. The 1,750,601-bp MHC sequence was obtained from 14 overlapping chimpanzee BAC clones and encompasses the telomeric part of the class III as well as the entire Patr class I regions linking LTB, 70-kb centromeric to the class III/class I boundary, to Patr-F at the telomeric end of the class I region. The length of the entire Patr class I region proper, from Patr-MIC to Patr-F, is 1,671,882 bp, ≈95 kb shorter than the corresponding 1,796,912-bp HLA class I region. Fig. 1 depicts a detailed comparative genomic map between these two MHC regions. Analysis of the repeat content reveals an occupancy rate of 52.03% of the region by such sequences as compared with 51.11% for the human counterpart. These were respectively composed of 17.66% (chimpanzee)/16.79% (human) short interspersed elements (SINEs), 17.87%/18.10% long interspersed elements (LINEs), and 12.98%/12.88% LTR elements. A detailed breakdown of the repeat content of the entire region is provided in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). As expected, there is considerable similarity between the genomic organization of human and chimpanzee MHCs. The chimpanzee sequence contains 41 putative coding genes and 59 noncoding or pseudogenes, which are matched with orthologous loci in identical orientations within the human MHC (9, 10). Interestingly, a detailed sequence analysis revealed the existence of 64 indels, each >100 bp in length (Fig. 1). Most of these indels include repetitive elements, such as Alu, LINE, LTR, etc., as well as the frequent insertion of the repeated sequence, SVA, within the chimpanzee sequence. Importantly, the indels were directly responsible for the major differences observed between the two species. These include the loss of three human pseudogenes, DHFRP, HCGII-4, and MICF, and the presence of only a single chimpanzee MIC gene in the region corresponding to the two human functional MICA and MICB genes, at the centromeric end of the class I region. This single chimpanzee MIC gene, Patr-MIC, was therefore produced as result of a large 95-kb deletion between the corresponding human MICA and MICB genes following a scenario that we reconstitute below.
Fig. 1.
Comparative genomics of human and chimpanzee MHC class I region. The relative location of the 41 genes (orange box, MHC gene; red box, non-MHC gene) and 59 pseudogenes (purple box, MHC gene; gray box, non-MHC gene) for human (Bottom) and chimpanzee (Top) sequences are shown, left to right, in the direction from the centromere to the telomere, respectively. A total of 64 indels of >100 bp observed between the two sequences (chimpanzee serving as reference) are indicated by blue (deletions) and red (insertions) triangles or lines. One duplicated and one inverted region were indicated as a yellow triangle (duplication in chimpanzee) and a green box (inversion), respectively.
A 95-kb Deletion Between the Human MICA and MICB Genes Leads to the Generation of a Single Chimeric Patr-MIC Gene. The human MICA and MICB genes are believed to result from a genomic duplication that occurred ≈33–44 million years ago (Mya) (22, 23), hence well before the separation of the chimpanzee from the human lineage ≈6 Mya (24) (Fig. 2A). Therefore, we asked from which ancestral MIC gene (A or B) this single Patr-MIC gene was originated, i.e., which MIC gene was deleted from the chimpanzee genome? Because the human MICA and MICB genes have a relatively high sequence similarity, it was not possible to settle the issue by dot plot analysis (data not shown). Consequently, and to thoroughly address this question, we performed both structural as well as similarity analyses of the human and chimpanzee MIC genes. Structural analysis showed that the 5′ flanking sequences and the first intron of the Patr-MIC gene have retained all of the signature retroelements characteristic of the orthologous region of human MICA, whereas the 3′ flanking sequences of the Patr-MIC display all of the characteristic retroelements of the human MICB (Fig. 2B). In accordance with this retroelement profiling, similarity investigations unveiled that exon 1 to intron 2 of Patr-MIC show greater sequence similarity with corresponding regions of MICA rather than with MICB, whereas the opposite is seen for Patr-MIC exons 5 and 6 as well as the 3′ noncoding region (Table 2, which is published as supporting information on the PNAS web site). Finally, the polymorphic (GCT)n (n = 4, 5, 6, 9, 10) short-tandem repeat, which exists only within the fifth exon (transmembrane domain) of MICA but not MICB (19), is also absent from the same exon in Patr-MIC. However, because the sequences between exon 3 and intron 4 of the Patr-MIC are equally homologous to orthologous regions within MICA and MICB, one could not establish the exact position of the recombinational event. Nevertheless, on the basis of sequence differences, we have narrowed the recombination breakpoint down to a segment located between the ends of MICA's second and MICB's fourth introns.
Fig. 2.
Structural analysis of MIC genes. (A) Evolutionary time line for duplication of the ancestral MICA and MICB genes, insertion of AluY and AluS elements, and separation of the human and chimpanzee lineages. The time line (millions of years ago) is shown along the vertical axis. (B) Comparative genomic map of the exon, intron, and retroelement (Alu, L1, LTR, and MIR) organization of human MICA, MICB, and Patr-MIC genes.
The existence of a single functional MIC gene centromeric of the major classical class I locus, Patr-B here, is not exclusive to chimpanzee, because it has been recognized in other primates, including humans. The gorilla, indeed, appears to have only one MIC gene with a strong sequence similarity to the human MICA (25, 26). In humans, individuals carrying the HLA-B*4801 allele (rare in Caucasians but more common in Northeast Asians as well as Native Americans) have lost the MICA locus also due to a large 100-kb genomic deletion surrounding and including this locus (albeit the genomic breakpoints are distinct from those observed in chimpanzee) (27, 28). All in all, it is quite intriguing that an equal-sized deletion involving this very same region and genes (MICA/B) has happened at distinct points in time in several different primate species. This very phenomenon might also be the reason why rodents are devoid of MIC genes, because the putative location of functional mouse MICA and MICB genes, the segment linking H2-D (equivalent to HLA-B or Patr-B) and BAT1, is substantially shortened compared with the human MHC: 40 instead of 173 kb (11, 29, 30). The molecular basis of the existence of such an apparently “deletion-prone” segment between MICA and MICB remains to be established, but this could be due to the existence of a HERV-L sequence, which contains a 2.5-kb AT-rich insertion in its 5′ LTR, which might therefore serve as a recombination hot spot (23, 31).
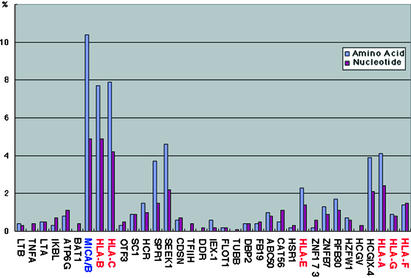
Nucleotide, Amino Acid, and Structural Similarities Between Human and Chimpanzee Orthologous Sequences. Fig. 3 compiles our similarity analysis with respect to nucleotide and amino acid diversity among 35 orthologous human/chimpanzee genes identified here, of a total of 41 putative coding sequences. The average nucleotide and amino acid identities were 98.9% and 98.3%, respectively (Table 3, which is published as supporting information on the PNAS web site). This relatively lower amino acid identity might be the result of positive selection aimed to maintain genetic polymorphism in the MHC (that is MHC class I) genes (32). Indeed, once genes were divided into MHC (hereafter designating MHC class I or MHC-I) and non-MHC loci, it was found that sequence identities were 99.3%/99.1% (nucleotide/amino acid) for the 28 non-MHC genes and “only” 97.1%/95.0% for the seven MHC-I genes, including the solo MIC gene. Furthermore, when MHC-I genes themselves were subdivided into classical/polymorphic (HLA-ABC-Patr-ABC), nonclassical/nonpolymorphic (HLA-EFG-Patr-EFG), as well as nonclassical but polymorphic (MICA/MICB-Patr-MIC), the nucleotide/amino acid identities were 96.2%/93.4%, 98.8%/98.5%, and 95.1%/89.6%, respectively. This analysis of nucleotide and amino acid similarities implies that, in addition to positive selection acting on MHC genes, a resolute degree of purifying selection acts primarily on the non-MHC genes to maintain their structural conservation (32). This makes sense, because most of these non-MHC genes are involved in basic (homeostatic) cellular functions that require interindividual as well as interspecies homogeneity. In contrast, MHC-I genes have to constantly adapt themselves to the microbiological habitat of every species (exceptions to this observed dichotomy are SPR1, SEEK1, and HCGIX-4; further functional characterization of these loci might answer this apparent discrepancy).
Fig. 3.
Differences in the coding genes between human and chimpanzee. Sequence differences for amino acid (blue) and nucleotide (purple) are indicated by percentages. MHC genes except MIC are shown in red, MIC (MICA/B fusion) in blue, and non-MHC genes in black.
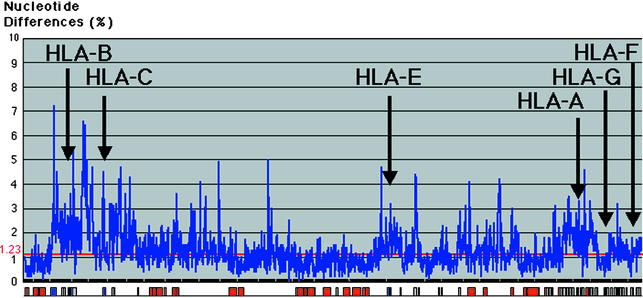
Comparative Nucleotide Diversity Profiling. A “nucleotide diversity (substitution: single-nucleotide polymorphism) profile” was generated across the entire 1.68-Mb gap-free (indels excluded) aligned genomic sequence by using a sliding window of 1,000 bp (Fig. 4). The average degree of nucleotide identity between the chimpanzee and the human for this region (again excluding indels) is 98.6%, which is similar to the earlier estimation of 98.77% (1.23% nucleotide difference) (7). However, this nucleotide difference is not constant across the entire MHC. For instance, within the two non-MHC gene-rich clusters (Fig. 4, left, LTB to BAT1 gene; center, IEX-1 to HSR1 gene), it is of ≈0.7%, which is five to nine times less than the average nucleotide difference of 6.7–3.5% around the classical MHC genes. This variation in nucleotide difference implies again that purifying selection is acting to maintain conservation much more strongly throughout these non-MHC gene-rich clusters (including their intergenic regions), whereas in contrast, the classical class I gene regions (including their intergenic sequences) show a lower degree of similarity, probably as a result of overdominant selection necessary to maintain polymorphism (33).
Fig. 4.
Diversity profile between human and chimpanzee MHC. The aligned sequence (excluding indels) is shown along the horizontal axis and the percent nucleotide differences calculated per 1 kb of nonoverlapping windows are shown along the vertical axis. The relative positions of the coding (red box), noncoding (gray box), and MHC (blue box) sequences are shown along the horizontal axis.
As expected, the genomic segments surrounding the MHC genes, except for the nonclassical HLA/Patr-G loci, reveal continuous high diversity profile, especially around the classical class I loci. This high degree of nucleotide variation may be the result of positive selection, the existence of multicopied sequences as well as hitchhiking effect due to the accumulative effect of balancing selection acting on the MHC loci in linkage disequilibrium (33). In contrast, the 35 kb surrounding HLA/Patr-G genes is highly conserved, displaying only a 0.9% nucleotide difference in contrast to the situation next to other MHC genes. This low level of nucleotide variation between HLA/Patr G genes might be in connection with the biology of the HLA/Patr-G molecule implicated at maternofetal immunity. Finally and interestingly, the diversity profile between the chimpanzee and human sequences closely resembles that previously obtained between different human MHC haplotypes (33–37).
Interestingly, once the indels are taken into account, the above-observed 98.6% sequence identity drops to only 86.7% (substitution, 1.4%; indels, 11.9%). This indel-included 86.7% identity may be a better representation of whole-genome sequence similarity between the human and the chimpanzee, as confirmed by a recently published study comparing a number of fragmented chimpanzee sequences with their human counterparts (38).
Further Analysis on Mismatched Sequence Between the Human and the Chimpanzee. More precise comparative analysis of indels and substitutions (transitions and transversions) was carried out by using mismatched sequences between the two species. When the 1,870,955-bp human and 1,750,601-bp chimpanzee sequences were aligned, the length of entire mismatched sequences was of 252,252 bp with substitutions representing 9.6% (24,221 bp) and indels 90.4% (228,031 bp). Thus, the major difference between the human and chimpanzee sequences is overwhelmingly attributable to indels. With regard to substitutions, they were further classified into transitions (16,680 bp, 6.6%) and transversions (7,541 bp, 3.0%). Among indels, single-nucleotide indels were represented only by 1,230 bp (0.5%), because most indels were longer than a single base pair. Fig. 5A shows a diversity profile of transitions, transversions, and single-nucleotide indels using a sliding window of 1,000 bp across the entire aligned sequence. The 1.91-Mb segment was divided into the MHC class I (multicopy) and non-MHC (single-copy) gene regions (Fig. 5A). Fig. 5B is the percentage of continuous indels that override 2-bp length. Diversity profiles give very similar patterns between transitions and transversions, although transitions, as expected, occurred more frequently than transversions (Fig. 5A). When focused on single-nucleotide indels (1,230 bp), one notices only a single peak between HLA-B/Patr-B and HLA-C/Patr-C (Fig. 5A), with the remainder of the region showing an even alteration of ≈0.06% with no significant peaks (even within the polymorphic MHC genes). However, when longer indels are included, indels were apparently accumulated in the MHC class I-harboring regions (1,013,321 bp in total), 191,512 bp (84.4% of total 226,801 bp indels) as compared with those in the non-MHC segments (897,959 bp), 35,289 bp (15.6%).
Fig. 5.
Structural differences between human and chimpanzee sequences. (A) Diversity profile of nucleotide differences between the human and chimpanzee genomic sequences using a sliding window of 1,000 bp across the entire 1.68-Mb aligned genomic sequence, including single-nucleotide indels. The aligned sequence is shown along the horizontal axis, and the percent nucleotide differences calculated per 1 kb of nonoverlapping windows are shown along the vertical axis. Diversity profiles of a single-nucleotide indel and substitutions by transition and transversion are indicated by blue, pink, and yellow lines, respectively. The relative positions of the MHC class I (multicopy) gene (brown) and non-MHC (single copy) gene (light blue) regions are shown along the horizontal axis. (B) Alteration of percentages of continuous indels that override 2-bp length.
The GC contents of the human and chimpanzee sequences were similar to each other, 45.9% and 46.1%, respectively. These contents were much higher than the average GC content of the entire human genome (41.0%) but lower than that expected from random nucleotide distribution (50%). By investigation of each of 24,221 substitutions, transition (T⇔C, A⇔G) and transversion (T, C⇔A, G) were found to contribute to 68.9% and 31.1% of the total substitutions, respectively (Fig. 6). This percentage of transition in the total substitutions is ≈10% higher than that reported in the previous studies using 16 pseudogenes for sequence comparison (59.3%) (36, 37). When considering individual transition and transversion pathways, T⇔C and A⇔G were found to have almost similar percentages of the total substitutions between them, but G⇔C (9.1%) and A⇔T (6.1%) gave higher and lower percentages as compared with G⇔T and A⇔C as well as those obtained in the previous studies, respectively (38, 39). Further, although the MHC gene regions tend to maintain a high degree of genetic polymorphism, the ratios within nucleotide substitutions from and to each base were almost the same between the MHC class I (multicopy) gene and non-MHC (single-copy) gene regions (Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 6.
Grouping of nucleotide substitutions.
In summary, this work reports the sequence of one-half of the chimpanzee MHC, which to date represents the longest continuous sequence within this species, our closest evolutionary relative. Comparative genomics with the orthologous human MHC class I region unveils a wealth of information, the most salient being the existence of a large number of indels that appear to be the main driving force behind the observed differences between the two species. Hence our perceived sequence divergence of only 1% between these two species appears to be erroneous, because this work, along with another recently published analysis, puts both species much further apart, >10% here and ≈5% in another recently published study (40), albeit the latter study compared shorter segments of both genomes. This relatively high and previously unexpected degree of sequence divergence might have functional implications not only within the coding sequences itself but also within regulatory elements (41, 42). Within the MHC per se, the most notable effect of indels appears to be the generation of a single chimeric Patr-MIC by fusion of MICA and MICB. This, along with other indels as well as nucleotide substitutions [which could be dubbed “transspecies single-nucleotide polymorphisms (SNPs)”], might therefore directly contribute to the patent difference between these two closely linked species with regard to susceptibility to a number of infectious as well as autoimmune disorders, most of which are primarily linked to the MHC. The study of these transspecies SNPs might further help to pinpoint the most ancient and perhaps functionally relevant human SNPs among the increasing numbers that are being continuously identified.
Supplementary Material
Acknowledgments
We thank Drs. N. Takahata, Y. Satta, N. Saito, and A. Fujiyama for discussions and advice. S.B. thanks the “Séquençage à Grande Echelle” [Institut National de la Santé et de la Recherche Médicale (INSERM)/Centre National de la Recherche Scientifique/Ministère de la Recherche] and Association pour la Recherche sur le Cancer for support. H.I. and S.B. acknowledge an INSERM/Japan Society for the Promotion of Science collaborative grant. This work was supported in part by 2001 Tokai University School of Medicine Research Aid.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: BAC, bacterial artificial chromosome.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AB100082–100087 and BA000041).
References
- 1.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 2.Fujiyama, A., Watanabe, H., Toyoda, A., Taylor, T. D., Itoh, T., Tsai, S. F., Park, H. S., Yaspo, M. L., Lehrach, H., Chen, Z., et al. (2002) Science 295, 131-134. [DOI] [PubMed] [Google Scholar]
- 3.Varki, A. (2000) Genome Res. 10, 1065-1070. [DOI] [PubMed] [Google Scholar]
- 4.Koopman, G., Haaksma, A. G., ten Velden, J., Hack, C. E. & Heeney, J. L. (1999) AIDS Res. Hum. Retroviruses 15, 365-373. [DOI] [PubMed] [Google Scholar]
- 5.Dienes, H. P., Purcell, R. H., Popper, H. & Ponzetto, A. (1990) J. Hepatol. 10, 77-84. [DOI] [PubMed] [Google Scholar]
- 6.Shouval, D., Chakraborty, P. R., Ruiz-Opazo, N., Baum, S., Spigland, I., Muchmore, E., Gerber, M. A., Thung, S. N., Popper, H. & Shafritz, D. A. (1980) Proc. Natl. Acad. Sci. USA 77, 6147-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ollomo, B., Karch, S., Bureau, P., Elissa, N., Georges, A. J. & Millet, P. (1997) Am. J. Trop. Med. Hyg. 56, 440-445. [DOI] [PubMed] [Google Scholar]
- 8.Hill, A. V. (1998) Annu. Rev. Immunol. 16, 593-617. [DOI] [PubMed] [Google Scholar]
- 9.The MHC Sequencing Consortium (1999) Nature 401, 921-923. [DOI] [PubMed] [Google Scholar]
- 10.Shiina, T., Tamiya, G., Oka, A., Takishima, N., Yamagata, T., Kikkawa, E., Iwata, K., Tomizawa, M., Okuaki, N., Kuwano, Y., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 13282-13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bahram, S. (2000) Adv. Immunol. 76, 1-60. [DOI] [PubMed] [Google Scholar]
- 12.Kulski, J. K., Gaudieri, S., Martin, A. & Dawkins, R. L. (1999) J. Mol. Evol. 49, 84-97. [DOI] [PubMed] [Google Scholar]
- 13.Garboczi, D. N. & Biddison, W. E. (1999) Immunity 10, 1-7. [DOI] [PubMed] [Google Scholar]
- 14.Held, W., Coudert, J. D. & Zimmer, J. (2003) Curr. Opin. Immunol. 15, 233-237. [DOI] [PubMed] [Google Scholar]
- 15.Balner, H., van Vreeswijk, W., Dersjant, H., d'Amaro, J., van Leeuwen, A. & van Rood, J. J. (1972) Transplant. Proc. 4, 42-48. [PubMed] [Google Scholar]
- 16.Adams, E. J. & Parham, P. (2001) Immunogenetics 53, 200-208. [DOI] [PubMed] [Google Scholar]
- 17.Adams, E. J., Cooper, S., Thomson, G. & Parham, P. (2000) Immunogenetics 51, 410-424. [DOI] [PubMed] [Google Scholar]
- 18.de Groot, N. G., Otting, N., Doxiadis, G. G., Balla-Jhagjhoorsingh, S. S., Heeney, J. L., van Rood, J. J., Gagneux, P. & Bontrop, R. E. (2002) Proc. Natl. Acad. Sci. USA 99, 11748-11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Groot, N. G., Otting, N., Arguello, R., Watkins, D. I., Doxiadis, G. G., Madrigal, J. A. & Bontrop, R. E. (2000) Immunogenetics 51, 398-409. [DOI] [PubMed] [Google Scholar]
- 20.Mizuki, N., Ando, H., Kimura, M., Ohno, S., Miyata, S., Yamazaki, M., Tashiro, H., Watanabe, K., Ono, A., Taguchi, S., et al. (1997) Genomics 42, 55-66. [DOI] [PubMed] [Google Scholar]
- 21.Nei, M. & Li, W. H. (1979) Proc. Natl. Acad. Sci. USA 76, 5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaudieri, S., Giles, K. M., Kulski, J. K. & Dawkins, R. L. (1997) Hereditas 127, 37-46. [DOI] [PubMed] [Google Scholar]
- 23.Yamazaki, M., Tateno, Y. & Inoko, H. (1999) J. Mol. Evol. 48, 317-327. [DOI] [PubMed] [Google Scholar]
- 24.Glazko, G. M. & Nei, M. (2003) Mol. Biol. Evol. 20, 424-434. [DOI] [PubMed] [Google Scholar]
- 25.Steinle, A., Groh, V. & Spies, T. (1998) Proc. Natl. Acad. Sci. USA 95, 12510-12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pellet, P., Vaneensberghe, C., Debre, P., Sumyuen, M. H. & Theodorou, I. (1999) Eur. J. Immunogenet. 26, 239-241. [DOI] [PubMed] [Google Scholar]
- 27.Komatsu-Wakui, M., Tokunaga, K., Ishikawa, Y., Kashiwase, K., Moriyama, S., Tsuchiya, N., Ando, H., Shiina, T., Geraghty, D. E., Inoko, H., et al. (1999) Immunogenetics 49, 620-628. [DOI] [PubMed] [Google Scholar]
- 28.Ota, M., Bahram, S., Katsuyama, Y., Saito, S., Nose, Y., Sada, M., Ando, H. & Inoko, H. (2000) Tissue Antigens 56, 268-271. [DOI] [PubMed] [Google Scholar]
- 29.Wroblewski, J. M., Kaminsky, S. G., Milisauskas, V. K., Pittman, A. M., Chaplin, D. D., Spies, T. & Nakamura, I. (1990) Immunogenetics 32, 200-204. [DOI] [PubMed] [Google Scholar]
- 30.Wroblewski, J. M., Kaminsky, S. G. & Nakamura, I. (1994) Immunogenetics 39, 276-280. [DOI] [PubMed] [Google Scholar]
- 31.Shiina, T., Tamiya, G., Oka, A., Yamagata, T., Yamagata, N., Kikkawa, E., Goto, K., Mizuki, N., Watanabe, K., Fukuzumi, Y., et al. (1998) Genomics 47, 372-382. [DOI] [PubMed] [Google Scholar]
- 32.Hughes, A. L. & Nei, M. (1988) Nature 335, 167-170. [DOI] [PubMed] [Google Scholar]
- 33.Satta, Y., Li, Y. J. & Takahata, N. (1998) Front. Biosci. 3, d459-d467. [DOI] [PubMed] [Google Scholar]
- 34.Guillaudeux, T., Janer, M., Wong, G. K., Spies, T. & Geraghty, D. E. (1998) Proc. Natl. Acad. Sci. USA 95, 9494-9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaudieri, S., Kulski, J. K., Dawkins, R. L. & Gojobori, T. (1999) Gene 238, 157-161. [DOI] [PubMed] [Google Scholar]
- 36.Gaudieri, S., Dawkins, R. L., Habara, K., Kulski, J. K. & Gojobori, T. (2000) Genome Res. 10, 1579-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Satta, Y. & Takahata, N. (2000) in The Major Histocompatibility Complex: Evolution, Structure, and Function, ed. Kasahara, M. (Springer, Tokyo), pp. 178-185.
- 38.Gojobori, T., Li, W. H. & Graur, D. (1982) J. Mol. Evol. 18, 360-369. [DOI] [PubMed] [Google Scholar]
- 39.Li, W. H., Gojobori, T. & Nei, M. (1981) Nature 292, 237-239. [DOI] [PubMed] [Google Scholar]
- 40.Britten, R. J. (2002) Proc. Natl. Acad. Sci. USA 99, 13633-13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.King, M. C. & Wilson, A. C. (1975) Science 188, 107-116. [DOI] [PubMed] [Google Scholar]
- 42.Enard, W., Khaitovich, P., Klose, J., Zollner, S., Heissig, F., Giavalisco, P., Nieselt-Struwe, K., Muchmore, E., Varki, A., Ravid, R., et al. (2002) Science 296, 340-343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.