Abstract
P element-mediated mutagenesis has been used to disrupt an estimated 25% of genes essential for Drosophila adult viability. Mutation of all genes in the fly genome, however, poses a problem, because P elements show significant hotspots of integration. In addition, advanced screening scenarios often require the use of P element-based tools like the generation of germ-line mosaics using FLP recombinase-mediated recombination or gene misexpression using the UAS/Gal4 system. These techniques are P element-based and can therefore not be combined with the use of P elements as mutagenic agents. To circumvent these limitations, we have developed an insertional mutagenesis system using non-P element transposons. An enhanced yellow fluorescent protein-marked piggyBac-based mutator element was mobilized by a piggyBac specific transposase source expressed from a Hermes-based jump-starter transposon marked with enhanced cyan fluorescent protein. In a pilot screen, we have generated 798 piggyBac insertions on FRT bearing third chromosomes of which 9% have sustained a putatively piggyBac-related lethal hit. The FRTs present on the target chromosome remained stably integrated during the screen and could subsequently be used to generate germ-line clones associated with maternal and zygotic phenotypes. PCR-based analysis of insertion loci shows that 57% of the insertions are in genes for which no P element insertions have been reported. Our data demonstrate the potential of this technique to facilitate the quest for saturation mutagenesis of the Drosophila genome. The system is Drosophila nonspecific and potentially applicable in a broad spectrum of nonmodel organisms.
At a time when whole genome sequences for the most important developmental biology model organisms have become available, the focus of developmental biologists is turning toward the functional analysis of whole genomes. The most straightforward strategy for the identification of gene functions relies on the inactivation of individual transcription units, by either random or site-directed mutagenesis. In Drosophila, an estimated 25% of all vital genes have been mutated to date (1, 2). Mutagenesis efforts have been based largely on chemically induced random mutagenesis by using ethane methyl sulfonate (3) or on transposon-mediated insertional mutagenesis by using P elements (4, 5). Unfortunately, both of these methods have limitations, which make the mutation of all genes in the Drosophila genome difficult to attain.
Mapping of chemically induced mutations at the DNA level is labor intensive and time consuming and therefore not practical on a genome-wide basis. In contrast, P element-induced mutations can easily and quickly be identified on a large scale. However, the tendency of P elements to integrate preferentially into certain hotspots (1) makes saturation mutagenesis difficult, if not impossible. Moreover, genetic screens based on the isolation of zygotic lethal mutations allow only the detection of the earliest function of a given gene. The analysis of later gene functions requires the generation of mosaic clones of homozygous mutant tissue in an otherwise phenotypically wild-type (heterozygous) background. Such mosaics are generated by using the site-specific recombination system based on the yeast FLP recombinase (FLP) and its recombination target sequences (FRTs) (6). Because genome-integrated FRTs are contained in P elements, they would be mobilized and lost during P element-mediated mutagenesis. Chromosomes carrying FRTs can therefore not be used in P element-based screens. Instead, preexisting P element insertions must be recombined with FRT containing chromosomes by meiotic recombination to make them accessible to clonal analysis, which is time consuming and labor intensive.
To overcome these limitations and combine the advantages of transposon-based mutagenesis with direct access to the FLP/FRT system [and other P element-based genetic tools like the UAS/Gal4 system (7)], we have adapted a non-P element-based germ-line transformation system (8, 9) to allow insertional mutagenesis in Drosophila. The system is based on the use of Hermes and piggyBac transposable elements (10, 11) that carry different spectral variants of the GFP as dominant transformation markers (8, 9, 12). In contrast to P elements, GFP-marked Hermes and piggyBac transposons are Drosophila nonspecific and are therefore expected to be applicable in a broad spectrum of nonmodel organisms (13).
Here, we show that a piggyBac-based system can be used to generate transposon insertions in the Drosophila genome with efficiency similar to P elements. P introduced FRTs present on the target chromosome remain stably integrated and can be used subsequently to induce FLP-mediated recombination. New piggyBac insertions are stable in the Drosophila genome but can be reverted when transposase is reintroduced. A Gal4 reporter gene in the mutator transposon can be used for misexpression studies and enhancer trapping (14–17). Analysis of insertion loci shows that piggyBac is less susceptible to hotspots and has an insertion preference significantly different from P elements. This piggyBac-based insertional mutagenesis system is therefore a valuable tool for a broad spectrum of genetic applications.
Materials and Methods
Genetics. Fly strains were reared under standard laboratory conditions. Drosophila germ-line transformation with piggyBac and Hermes vectors was performed as described (9). Filter sets used for the identification of the different fluorescent transformation markers have been described (13). Embryonic enhancer traps were visualized by crossing mutator-carrying males to UAS-lacZ-carrying virgins and performing a 5-bromo-4-chloro-3-indolyl β-D-galactoside staining (14). Germ-line clones were generated by crossing mutator-carrying virgins to P{hsFLP}22, y[1] w[*]; P{ovoD1–18}3L P{FRT(w[hs])}2A/TM3 males for 3L or P{hsFLP}22, y[1] w[*]; P{neoFRT}82B P{ovoD1–18}3R/ TM3 males for 3R (6). Heat shocks were performed at the third larval instar stage for 2 h at 37°C on 2 consecutive days.
Molecular Biology. Construction of pBac{3xP3-EYFP, p-GAL4Δ-K10}, and pHer{3xP3-ECFP, αtub-piggyBacK10} is described in detail in ref. 18. DNA sequences flanking recessive lethal piggyBac transposon insertions were amplified by inverse PCR as described (19). In brief, five fly equivalents of genomic DNA were cleaved with HaeIII and ligated. Flanking sequences were amplified by PCR [5 min, 95°C; 35× (30 s, 95°C; 1 min, 65°C; 2 min, 72°C); 7 min, 72°C] by using primers PLF (5′-CTTGACCTTGCCACAGAGGACTATTAGAGG-3′) and PLR (5′-CAGTGACACTTACCGCATTGACAAGCACGC-3′) for the 5′ junction and PRF (5′-CCTCGATATACAGACCGATAAAACACATGC-3′) and PRR (5′-AGTCAGTCAGAAACAACTTTGGCACATATC-3′) for the 3′ junction. Amplified DNA fragments were directly sequenced by using primers PLR and PRF, respectively. Sequences were analyzed by using blast searches of the Drosophila Genome Database at www.ncbi.nlm.nih.gov/BLAST.
Insertion sites of viable piggyBac insertions were mapped to the genomic sequence by the Berkeley Drosophila Genome Project, funded by National Institutes of Health Grant P50 HG00750 to G. M. Rubin and by the Howard Hughes Medical Institute through its support of work in the laboratories of G. M. Rubin and A. C. Spradling. Sequences flanking insertion sites were amplified by inverse PCR by G. Tsang, M. Evans, and G. Davis in the laboratory of G. M. Rubin at the University of California, Berkeley. PCR products were sequenced by S. Park, K. Wan, and R. A. Hoskins at Lawrence Berkeley National Laboratory under Department of Energy Contract DEAC0376SF00098, University of California. Mapping information was determined by using software written by G. Liao in the laboratory of G. M. Rubin, followed by manual curation by B. Levis and A. C. Spradling at the Carnegie Institution.
Results
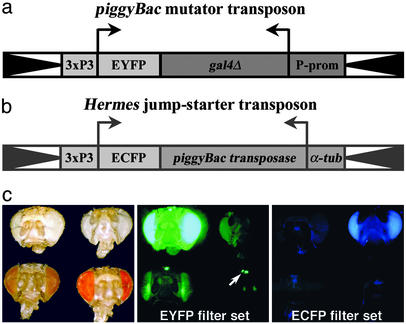
Mutator and Jump-Starter Transposons. To establish a transposon insertion system in Drosophila that can function independently of P element-based systems, we made use of the Hermes and piggyBac transposons, which had previously been shown to allow germ-line integration and transposition in Drosophila (20, 21). To be able to distinguish the Hermes and piggyBac transposons from each other and from P elements simultaneously present in the genome, we used the spectral variants of GFP, enhanced yellow fluorescent protein (EYFP) and enhanced cyan fluorescent protein (ECFP), as dominant markers (12). The mutator transposon (Fig. 1a) was marked with 3xP3-EYFP, which is expressed under control of the endogenous Pax6 gene of the fly in the larval light sensory organ, the adult compound eye and ocelli (Fig. 1c) (8), and parts of the nervous system (9). In addition, we included the heterologous transcriptional activator Gal4 to allow enhancer trapping (14–17).
Fig. 1.
(a) piggyBac-based mutator transposon marked with 3xP3-EYFP. (b) Hermes-based jump-starter transposon marked with 3xP3-ECFP.(c) Expression of 3xP3-EYFP and 3xP3-ECFP in unpigmented and pigmented eyes. (Left) Bright-field view of eyes expressing 3xP3-EYFP (upper left) or 3xP3-ECFP (upper right) in a white- background, and eyes expressing 3xP3-EYFP in the presence of one (lower left) or two copies of a white transgene (lower right). (Center) Same as Left viewed through the EYFP filter set; 3xP3-EYFP is detectable in white- eyes (upper left) or in eyes expressing one copy of a white transgene (lower left); in eyes expressing two copies of a white transgene (lower right), 3xP3-EYFP is detectable in the ocelli (arrow) and the pseudopupil of the compound eye (not visible in frontal view); 3xP3-ECFP is not detectable (upper right). (Right) Same as Left viewed through the ECFP filter set. 3xP3-EYFP is not detected (left and lower right), but 3xP3-ECFP is detected (upper right).
Mobilization of the mutator transposon requires the expression of a stable piggyBac transposase source in the germ line. To avoid mobilization of the piggyBac transposase source itself, we integrated the piggyBac transposase coding region into a Hermes transposon marked with 3xP3-ECFP (Fig. 1b). ECFP can be readily distinguished from the EYFP of the mutator transposon and allows selection against the jump-starter transposon in progeny flies carrying new transposon insertions (Fig. 1c). The piggyBac transposase was placed under control of the constitutively active α1-tubulin promoter (Fig. 1b) (22). The generation of Drosophila lines carrying these mutator and jump-starter constructs was described previously (18).
piggyBac-Mediated Mutagenesis in Drosophila. The piggyBac transposon insertion system was tested in a pilot mutagenesis screen. A piggyBac mutator transposon located on the X chromosome was mobilized from a piggyBac transposase source integrated on the second chromosome. Because one feature of the system was to function in the presence of P elements without interfering with their stability, we targeted third chromosomes carrying insertions of FRT-bearing P elements at the base of the left and right arms (6). A list of available stocks generated for this purpose is shown in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org, and the respective crossing scheme is given in Fig. 5, which is published as supporting information on the PNAS web site.
We established a total of 2,500 single dysgenic male crossings (SMCs), of which 1,741, corresponding to a jumping rate of 70%, yielded new autosomal piggyBac insertions. This rate depended on the number of progeny obtained from each SMC and climbed to >90% among vials, which produced 40 males or more. Therefore, the rate of recovery of piggyBac insertions is at least equal to that observed for P elements (23). A segregation analysis established that 798 (46%) new insertions were located on the third chromosome, suggesting that piggyBac insertions are distributed evenly between the autosomes. The data of the pilot screen are summarized in Table 1.
Table 1. Statistical overview over piggyBac insertions on the third chromosome.
| Category | n | % |
|---|---|---|
| Single male crosses | 2,500 | |
| Independent insertions recovered | 1,741 | 70 |
| Insertions on third chromosome | 798 | 46 |
| Insertions sequenced | 604 | |
| Repetitive | 46 | |
| Unique | 558 | |
| Intergenic insertions | 254 | |
| Insertions in transcripts | 304 | |
| Previously hit by P elements | 132 | 43 |
| No P element hit reported | 172 | 57 |
| Lethal insertions | 131 (72) | 16 (9) |
| Excised | 39 | |
| Reverted | 22 | 56 |
Numbers in parentheses represent values corrected for insertions likely to be due to piggyBac-associated lethal mutations based on excision data
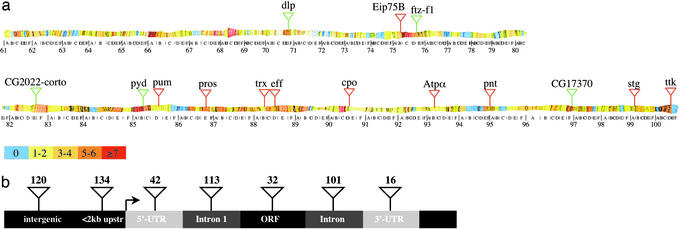
piggyBac and P Elements Have Distinct Insertion Preferences. An inherent problem in P element-based mutagenesis screens is the existence of hotspots of integration (1). To investigate whether the piggyBac system can be used to mutagenize genes that are not good targets for P elements, we determined the genomic flanking sequences of 604 piggyBac insertions. Mapping of these insertions to the annotated Drosophila genome sequence showed that the insertions are distributed evenly throughout the third chromosome (Fig. 2a; Table 3, which is published as supporting information on the PNAS web site). Three hundred and four of the 604 insertions are located within putative transcripts. Of those transcripts, 83 were hit two times or more, accounting for 27% of the insertions. In similar P element-mediated screens, genes hit six times or more have been shown to account for 39% of insertions integrated into vital genes (1). Conversely, 72% of the piggyBac insertions were single hits compared with 29% for P elements. The maximum number of piggyBac insertions found in the same transcription unit or intergenic region was four, equaling ≈1% of insertions. By comparison, P element hotspots account for up to 7% of insertions in P element-mediated mutagenesis (1). These data strongly suggest that piggyBac is significantly less susceptible to hotspots of integration.
Fig. 2.
(a) Distribution of lethal piggyBac insertions on chromosome 3. Color code represents number of piggyBac insertions isolated for each letter division of the third chromosome. Transposon insertions in red show the 10 genes most frequently hit by P elements (1). Transposon insertions in green show the genes most frequently hit by piggyBac in this screen. dlp, ftz-f1, and CG17370 were hit three times each, and pyd and the intergenic region between CG2022 and corto were hit four times. (b) Insertion preference of piggyBac within a schematic transcription unit.
Of the 10 transcription units most frequently hit by P elements (1), seven were not hit at all in our piggyBac screen, two were hit twice, and one was hit once (Fig. 2a). Using the data available in F LYBASE (http://flybase.bio.indiana.edu), we also determined that no P element insertions have been reported to date for 172 (57%) of the 304 piggyBac insertions located in putative genes. These data suggest that, in addition to the lack of obvious hotspots, the insertion preference of piggyBac is significantly different from P elements.
Forty-one (6.8%) of the 604 analyzed piggyBac transposons were found integrated into repetitive sequences and five insertions mapped to “Arm U,” which is mainly heterochromatic and could therefore not be assigned to unique chromosomal locations. Of the 558 piggyBac insertions, which mapped to unique loci, 254 (45%) are in intergenic regions (Fig. 2b). Among the latter, 134 (24% of all insertions) are integrated within 2 kb of putative transcripts. Together with the insertions located inside putative genes (54%), 78% of all mapped insertions were in the vicinity of genes, indicating that piggyBac has a strong preference to integrate into or near transcribed sequences.
We also investigated the insertion preference of piggyBac within putative transcription units. Among the 304 piggyBac transposons located inside predicted transcripts, 42 (8%) were in the 5′-UTR, 32 (6%) were in putative ORFs, 214 (38%) were in introns, and 16 (3%) were in the 3′-UTR (Fig. 2b). One hundred thirteen insertions, corresponding to 53% of intronic insertions and 19% of all insertions, were found in the first intron of a given gene. These data show that piggyBac has a strong preference to insert into introns near the 5′-end of a transcription unit.
Generation of Recessive Lethal Alleles. The value of piggyBac as a mutagenesis tool depends significantly on its ability to introduce mutations efficiently. In 131 (16%) of the 798 third chromosomal insertions, the target chromosome had sustained a recessive lethal hit (see Table 1). We selected 39 of the lethal lines and reintroduced the jump-starter transposon to determine whether the transposons could be remobilized and whether the lethality was associated with the piggyBac insertion. Excision of the piggyBac transposon was observed in all assays, indicating that piggyBac excision is highly efficient. Among 10 parallel assays set up for each line, the result was always identical, suggesting that imprecise excisions of piggyBac are rare. However, excision of the piggyBac transposon reversed the lethality of the target chromosome only in 22 (56%) of the 39 tested lines. Consequently, the rate of lethal hits caused by piggyBac must be corrected to 9%, which is comparable to the rate obtained with P elements (2).
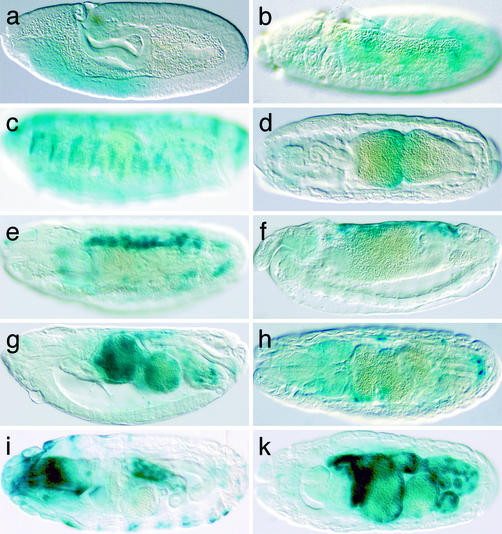
Enhancer Trapping. The piggyBac mutator transposon used contains the coding sequence of the heterologous transcriptional activator Gal4 to allow enhancer trapping (18). To confirm the Gal4 reporter gene function of the mutator transposon, we randomly selected 20 insertion lines from the lethal collection, crossed them to a P{UAS-lacZ} line, and performed 5-bromo-4-chloro-3-indolyl β-D-galactoside stainings on embryos. Tissue-specific lacZ expression was detected in 10 of the tested lines (Fig. 3) and was found associated with transposons inserted in introns, 5-UTRs, and upstream of genes. Several lines reproduced expression patterns of previously described genes. The earliest lacZ expression was detectable at stage 9 in embryos produced by line l(3)PL00759. This insertion lies in the first intron of hunchback (hb) (24) (Fig. 3a) and detects the early anterior expression domain of hb. Line l(3)PL00790 is inserted in the second intron of branchless (bnl) (25) and reproduces the epidermal expression of the Drosophila FGF homolog starting at stage 10 (Fig. 3b). Due to the stability of the Gal4 protein, the pattern remains visible until the end of embryogenesis (Fig. 3c). In l(3)PL00757, a piggyBac is found in the second intron of labial (lab) (26) and recapitulates the expression of this homeodomain protein in the second midgut constriction (Fig. 3d). In l(3)PL00779, the mutator transposon is inserted in the first intron of the GATA transcription factor gene serpent (srp) (27) and shows expression in the fat body and the amnioserosa (Fig. 3e). A common feature of all these expression patterns is a significant delay in the appearance of the lacZ expression compared with expression of the endogenous genes.
Fig. 3.
Enhancer trap expression patterns associated with piggyBac insertions. Shown are 5-bromo-4-chloro-3-indolylβ-d-galactoside stainings of embryos from piggyBac insertion lines crossed to P{UAS-LacZ}. (a) Line l(3)PL00759 inserted in hb.(b and c) Line l(3)PL00790 inserted in bnl.(d) Line l(3)PL00757 inserted in lab. (e) Line l(3)PL00779 inserted in srp. (f and g) Line l(3)PL00799 inserted in Mpk2. (h) Line l(3)PL00795 inserted in CG10823.(i) Line l(3)PL00724 inserted in CG10522. (k) Line l(3)PL00749 inserted in CG12546. Anterior is to the left, dorsal side up.
We also discovered tissue-specific enhancers of several previously uncharacterized genes. l(3)PL00799 is inserted into the 5′-UTR of the Mpk2 gene, which has been implicated in regulation of the Drosophila immune response by in vitro studies (28). The insertion in l(3)PL00799 detects an enhancer driving expression in the amnioserosa (Fig. 3f) and later in the embryonic midgut (Fig. 3g). These patterns are strikingly reminiscent of srp, which is expressed in a similar pattern in the amnioserosa and has been shown to be essential for the development of the embryonic midgut (27). In addition, srp is also involved in mediating the Drosophila immune response (29), suggesting a possible link between Mpk2 and srp during this process.
l(3)PL00795 is inserted in the first intron of CG10823 (30), a putative G protein-coupled receptor encoding gene and shows enhancer trap expression in a small ring of cells surrounding the hindgut and in small groups of cells throughout the embryo (Fig. 3h). l(3)PL00724 is inserted near the 5′-end of CG10522 (31), encoding a putative serine–threonine kinase and shows lacZ expression in epidermal stripes and in the hindgut epithelium (Fig. 3i). l(3)PL00749 is inserted just upstream of CG12546 and shows strong lacZ expression throughout the midgut and hindgut at stage 16 of embryogenesis (Fig. 3k). These data confirm the function of Bac{3xP3-EYFP, p-Gal4Δ-K10} as a tool for enhancer trapping studies in Drosophila.
Stability of P Elements in the Presence of piggyBac Transposase. An important question is whether the FRTs present on the target chromosomes remain stably integrated during piggyBac mobilization. During the course of the screen, several strains carrying FRT bearing P elements (see Table 2) and Bac{3xP3-EYFP, p-Gal4Δ-K10} or Her{3xP3-ECFP, αtub-piggyBac-K10} were kept for >50 generations. Spontaneous loss or mobilization of P elements or piggyBac was never observed. Furthermore, mobilization or loss of P{FRT} was not observed during the screen when P elements were combined with both Bac{3xP3-EYFP, p-Gal4Δ-K10} and Her{3xP3-ECFP, αtub-piggyBac-K10} at the same time. P elements therefore appear to be insensitive to the presence of piggyBac transposase. In contrast, spontaneous loss of Her{3xP3-ECFP,αtub-piggyBac-K10} was observed on several occasions, suggesting that Hermes transposons may not be stable in Drosophila. Hermes transposons have been reported to be unstable in the presence of hobo elements (32), which could explain this observation. This, however, poses no problem during piggyBac mutagenesis, because the transposase source is introduced only transiently during the screen.
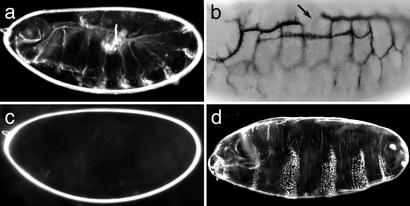
piggyBac-Induced Zygotic Lethal Mutations with Maternal Effect Phenotypes. To determine whether piggyBac insertions are associated with embryonic phenotypes, we performed cuticle preparations on 77 lethal insertion lines. Surprisingly, we found consistent phenotypes in only two lines. Homozygotes of l(3)PL00779, which is inserted in the first intron of srp, die with the u-shaped cuticle phenotype previously reported for srp mutants (Fig. 4a) (33). Immunohistochemical stainings using the trachea-specific antibody 2A12 confirmed that insertion l(3)PL00790 frequently shows defects in tracheal development similar to bnl alleles (Fig. 4b). These data indicate that insertions of Bac{3xP3-EYFP, p-Gal4Δ-K10} do create recessive lethal mutations associated with zygotic phenotypes even when inserted into introns. It may be noteworthy, however, that many piggyBac insertions in embryonic lethal genes appeared to create hypomorphic alleles. The insertions in bnl and 14-3-3ε (34) are semilethal, and homozygotes of the insertions in the embryonic lethal genes crumbs (crb) (35), lab, and hb die only at the larval or pupal stages, respectively.
Fig. 4.
Embryonic phenotypes associated with piggyBac insertions. (a) Cuticle phenotype of a homozygote of line l(3)PL00779 inserted in srp. (b) Embryo homozygous for line l(3)PL00790 inserted in bnl stained with 2A12 antibodies. (c) Cuticle phenotype of an embryo derived from a female carrying germ-line clones of line l(3)PL00784 inserted in 14-3-3 crossed to a wild-type male. (d) Cuticle phenotype of embryo maternally and zygotically mutant for line l(3)PL00739 inserted in ftz-f1.
We then asked whether the FRTs can be used to remove a potential maternal contribution of the mutated genes by inducing germ-line clones for the left or the right arm of the third chromosome by using the dominant female sterile-flipase (DFS-FLP) technique (6). In this technique, all females are heterozygous for the piggyBac insertion and for the dominant sterile mutation ovoD, which aborts oogenesis at an early stage. Therefore, females can not lay eggs unless a FLP-mediated recombination event occurs at the FRTs and creates germ-line cells, which have lost the ovoD mutation. We tested a total of 30 lines and found that in each case, germ-line clone females that laid eggs could be produced for at least one arm of the third chromosome, showing that the FRTs on the target chromosome remain functional after piggyBac mutagenesis.
In several cases, germ-line clones of the chromosome arm bearing a zygotic lethal piggyBac insertion were associated with maternal effect phenotypes. Line l(3)PL00784 has a piggyBac insertion in the first intron of the 14–3-3 gene. Germ-line clone females of this line produce embryos that die before they produce a larval cuticle and appear as “empty eggs” (Fig. 4c), as previously reported (36). Line l(3)PL00739 has a piggyBac insertion in the third intron of the gene ftz transcription factor 1 (ftz-f1) (37). ftz-f1 encodes a steroid hormone receptor like protein, which is required as a cofactor for the pair-rule transcription factor fushi tarazu (ftz) (38, 39). Embryos homozygous for l(3)PL00739 die at the end of embryogenesis without obvious cuticle defects (not shown). However, embryos maternally and zygotically mutant for l(3)PL00739 develop with a pair-rule type segmentation defect (Fig. 4d). Several other insertions causing maternal effect phenotypes were obtained in noncharacterized genes whose detailed analysis will be given elsewhere. These examples confirm that the FRTs remained functional after piggyBac mutagenesis and that piggyBac insertions can cause mutations associated with maternal effect phenotypes.
Discussion
In this study, we present a transposon mutagenesis system alternative to the existing P element-based system. We show that this piggyBac-based system functions efficiently to create new mutations in the presence of P elements and can therefore be applied with P element-based tools in the background. This feature allows the combination of insertional mutagenesis screens with UAS/Gal4 misexpression and FLP/FRT systems. Mutagenesis screens designed for mosaic tissue analysis or for the identification of genetic modifiers of misexpression phenotypes (40) are therefore no longer limited to chemical mutagens, which hamper the identification and characterization of the mutated genes. In addition, piggyBac is less susceptible to hotspots and has a significantly different insertion preference, which makes genes accessible to mutation that have not been hit by P elements.
The Mutator and Jump-Starter Transposons. piggyBac belongs to the TTAA-specific transposon family and was originally found in cell lines of the cabbage looper moth Trichoplusia ni (11). We chose piggyBac as a mutator transposon because no endogenous transposase sources are present in the Drosophila genome, and piggyBac insertions are stable (21). This was confirmed by our results, because we did not observe spontaneous mobility of piggyBac in the absence of a piggyBac-specific transposase source. Moreover, P element insertions were stable in the presence of piggyBac transposase. piggyBac seems therefore well suited as a tool for transposon tagging in Drosophila. To facilitate detection of the mutator transposon, we used the highly sensitive transformation marker 3xP3-EYFP (12). This marker allows easy distinction from the 3xP3-ECFP marker used in the jump-starter transposon and also from the white+ marker used in many P elements, which can be present in the genome at the time of piggyBac mobilization. One limitation of this marker is that it is difficult to detect in pigmented eyes. This problem can be remedied, however, by introducing a cn bw chromosome, which results in white eye color independent of the white locus, or the use of a UASw-RNAi transgene (41), which inactivates expression of Drosophila white genes.
The piggyBac mutator transposon is mobilized by a transposase source introduced into the genome via a Hermes transposon. The use of different transposons for mutator and transposase sources, which belong to different transposon superfamilies that cannot crossmobilize each other, makes it unnecessary to disable the transposon carrying the transposase source like in the case of the Δ2–3 P element transposase source (5). We marked the Hermes transposon with the less-sensitive 3xP3-ECFP, because it is not necessary to screen through large numbers of individuals for the transposase source. We also selected Hermes for the jump-starter transposon and not as a mutator, because Hermes has been reported to be unstable in the presence of hobo elements in the genome (32). We did, in fact, observe spontaneous loss of Hermes transposons from the jump-starter stock on several occasions. This, however, is not a significant problem during mutagenesis screens, because the transposase source is needed only transiently.
piggyBac-Mediated Mutagenesis on FRT Chromosomes. In this screen, insertions were recovered from 70–90% of all single dysgenic male crossings. This rate is equal or superior to the rate usually obtained in P element-mediated mutagenesis (23) and shows that piggyBac mutator elements are easily remobilized and that the piggyBac transposase source is highly effective.
Analysis of the flanking sequences of the piggyBac insertions showed that the insertions are distributed evenly over the third chromosome. We did not observe any obvious hotspots of integration, and 57% of the insertions are in genes that have not been targeted by P elements. This result suggests that the insertion preference of piggyBac is significantly different from P elements and promises that piggyBac will be a valuable tool to increase the coverage of genes tagged by transposon insertions in the Drosophila genome.
We also find that 70% of the insertions hitting putative genes are in introns, clearly distinguishing piggyBac from P elements, which preferentially insert in 5′-UTRs (42). This observation could suggest that the rate at which piggyBac induces lethal mutations might be low, because many transposons may be removed from the pre-mRNA during splicing, and a functional transcript may be generated. However, we find lethal insertions at a rate similar to that observed for P elements (1, 2). It is possible that the efficiency of piggyBac transposons to inactivate genes could even be increased by using vectors carrying additional splice donor and acceptor sites that lead to aberrant splicing or by including strong transcriptional terminators that abort a transcript before it can be spliced. The preference to insert into introns can also be an advantage in certain instances: piggyBac could serve as an excellent tool for the generation of gene trap (43) or protein trap (44) insertions and could make these approaches significantly more efficient than they are with P elements.
Another difference between piggyBac and P elements appears to be the precision of excision. Despite extensive efforts, we could not find evidence for imprecise excisions when we tried to revert piggyBac insertions. This result may preclude the possibility of converting nonlethal piggyBac insertions into lethal mutations unless special measures are taken, for example, by incorporating P element sequences into a piggyBac transposon. On the other hand, the precise excision mechanism may harbor advantages as well. piggyBac transposons leave no target site duplication (footprint) when they are excised (45) and, therefore, allow restoration of insertion loci to wild type even when the transposon is inserted in the ORF. In contrast to P elements, piggyBac is therefore less likely to cause secondary mutations by a hit-and-run mechanism, which can severely complicate the identification of mutations.
A major advantage is that piggyBac can be mobilized in the presence of P elements in the genome, which opens the possibility of combining existing P element-based tools with piggyBac-based mutagenesis screens. As we have shown here, piggyBac-mediated mutagenesis can easily be carried out to target FRT-bearing chromosomes, which makes the new insertions directly accessible to mosaic analyses using FLP/FRT-based recombination. This possibility avoids time- and labor-intensive recombination of preexisting P element insertions onto FRT chromosomes and facilitates easy identification of the mutated genes. Similarly, piggyBac-based mutagenesis could be combined with other P element-based tools like the UAS/Gal4 system. In a screen for new Gal4 insertions, like the one presented here, dysgenic males can be crossed directly to a UAS-GFP reporter, allowing the identification of enhancer traps at the larval or adult stages in the F1 generation (18). F1 screens for genetic modifiers of a UAS/Gal4-generated overexpression phenotype could be done by piggyBac mutagenesis, allowing quick identification of the mutated loci. Similarly, once the repertoire of available piggyBac transposons has been expanded, misexpression screens using an EP-type (46) piggyBac transposon could be conducted, crossing dysgenic flies directly to a Gal4 reporter, allowing the identification of UAS-tagged insertions without the prior establishment of individual lines. We expect that the availability of the piggyBac-based alternative insertional mutagenesis system will open the field to a variety of new applications, while at the same time decreasing the time and effort involved in the successful identification of novel gene functions.
Supplementary Material
Acknowledgments
We thank Monika Rosén and Darren Cleare for technical assistance and for critically reading the manuscript and Norbert Perrimon for providing the double FRT chromosomes. This work is supported by the Swedish Foundation for Strategic Research, Developmental Biology Program, which provided a junior research group to U.H.; the Swedish Research Council VR (U.H.); the Swedish Cancer Foundation (U.H.); the Swedish Medical Society (U.H.); the Craaford Foundation (U.H. and S.N.); the Åke Wiberg Foundation (S.N.); the Royal Physiographic Society in Lund (S.N.); the Robert Bosch Foundation, which provided a junior research group to E.A.W.; the Fonds der Chemischen Industrie; and the Bundesministerium für Bildung und Forschung Technologie (E.A.W.). E.A.W. is a European Molecular Biology Organization Young Investigator, and C.H. is a fellow of the Fonds der Chemischen Industrie.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: EYFP, enhanced yellow fluorescent protein; ECFP, enhanced cyan fluorescent protein.
References
- 1.Spradling, A. C., Stern, D., Beaton, A., Rhem, E. J., Laverty, T., Mozden, N., Misra, S. & Rubin, G. M. (1999) Genetics 153, 135-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peter, A., Schottler, P., Werner, M., Beinert, N., Dowe, G., Burkert, P., Mourkioti, F., Dentzer, L., He, Y., Deak, P., et al. (2002) EMBO Rep. 3, 34-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashburner, M. (1989) Drosophila: A Laboratory Handbook (Cold Spring Harbor Lab. Press, Plainview, NY).
- 4.Cooley, L., Kelley, R. & Spradling, A. (1988) Science 239, 1121-1128. [DOI] [PubMed] [Google Scholar]
- 5.Robertson, H. M., Preston, C. R., Phillis, R. W., Johnson-Schlitz, D. M., Benz, W. K. & Engels, W. R. (1988) Genetics 118, 461-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chou, T. B. & Perrimon, N. (1996) Genetics 144, 1673-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brand, A. H. & Perrimon, N. (1993) Development (Cambridge, U.K.) 118, 401-415. [DOI] [PubMed] [Google Scholar]
- 8.Berghammer, A. J., Klingler, M. & Wimmer, E. A. (1999) Nature 402, 370-371. [DOI] [PubMed] [Google Scholar]
- 9.Horn, C., Jaunich, B. & Wimmer, E. A. (2000) Dev. Genes Evol. 210, 623-629. [DOI] [PubMed] [Google Scholar]
- 10.Warren, W. D., Atkinson, P. W. & O'Brochta, D. A. (1994) Genet. Res. 64, 87-97. [DOI] [PubMed] [Google Scholar]
- 11.Cary, L. C., Goebel, M., Corsaro, B. G., Wang, H. G., Rosen, E. & Fraser, M. J. (1989) Virology 172, 156-169. [DOI] [PubMed] [Google Scholar]
- 12.Horn, C. & Wimmer, E. A. (2000) Dev. Genes Evol. 210, 630-637. [DOI] [PubMed] [Google Scholar]
- 13.Horn, C., Schmid, B. G., Pogoda, F. S. & Wimmer, E. A. (2002) Insect Biochem. Mol. Biol. 32, 1221-1235. [DOI] [PubMed] [Google Scholar]
- 14.O'Kane, C. J. (1998) in Drosophila: A Practical Approach, ed. Roberts, D. B. (IRL Press, Oxford), pp. 131-178.
- 15.Bier, E., Vaessin, H., Shepherd, S., Lee, K., McCall, K., Barbel, S., Ackerman, L., Carretto, R., Uemura, T., Grell, E., et al. (1989) Genes Dev. 3, 1273-1287. [DOI] [PubMed] [Google Scholar]
- 16.Bellen, H. J., O'Kane, C. J., Wilson, C., Grossniklaus, U., Pearson, R. K. & Gehring, W. J. (1989) Genes Dev. 3, 1288-300. [DOI] [PubMed] [Google Scholar]
- 17.Wilson, C., Pearson, R. K., Bellen, H. J., O'Kane, C. J., Grossniklaus, U. & Gehring, W. J. (1989) Genes Dev. 3, 1301-1313. [DOI] [PubMed] [Google Scholar]
- 18.Horn, C., Offen, N., Nystedt, S., Häcker, U. & Wimmer, E. A. (2003) Genetics, 647-661. [DOI] [PMC free article] [PubMed]
- 19.Huang, M. A., Rehm, E. J. & Rubin, G. M. (2000) in Drosophila Protocols, eds. Sullivan, W., Ashburner, M. & Hawley, S. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 429-437.
- 20.O'Brochta, D. A., Warren, W. D., Saville, K. J. & Atkinson, P. W. (1996) Genetics 142, 907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Handler, A. M. & Harrell, R. A., 2nd (1999) Insect Mol. Biol. 8, 449-457. [DOI] [PubMed] [Google Scholar]
- 22.Theurkauf, W. E., Baum, H., Bo, J. & Wensink, P. C. (1986) Proc. Natl. Acad. Sci. USA 83, 8477-8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berg, C. A. & Spradling, A. C. (1991) Genetics 127, 515-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tautz, D., Lehmann, R., Schnürch, H., Schuh, R., Seifert, E., Kienlin, A., Jones, K. & Jäckle, H. (1987) Nature 327, 383-389. [Google Scholar]
- 25.Sutherland, D., Samakovlis, C. & Krasnow, M. A. (1996) Cell 87, 1091-101. [DOI] [PubMed] [Google Scholar]
- 26.Diederich, R. J., Merrill, V. K., Pultz, M. A. & Kaufman, T. C. (1989) Genes Dev. 3, 399-414. [DOI] [PubMed] [Google Scholar]
- 27.Reuter, R. (1994) Development (Cambridge, U.K.) 120, 1123-1135. [DOI] [PubMed] [Google Scholar]
- 28.Han, Z. S., Enslen, H., Hu, X., Meng, X., Wu, I. H., Barrett, T., Davis, R. J. & Ip, Y. T. (1998) Mol. Cell. Biol. 18, 3527-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen, U. M., Kadalayil, L., Rehorn, K. P., Hoshizaki, D. K., Reuter, R. & Engstrom, Y. (1999) EMBO J. 18, 4013-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brody, T. & Cravchik, A. (2000) J. Cell Biol. 150, F83-F88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrison, D. K., Murakami, M. S. & Cleghon, V. (2000) J. Cell Biol. 150, F57-F62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sundararajan, P., Atkinson, P. W. & O'Brochta, D. A. (1999) Insect Mol. Biol. 8, 359-368. [DOI] [PubMed] [Google Scholar]
- 33.Jurgens, G., Wieschaus, E., Nusslein-Volhard, C. & Kluding, H. (1984) Roux Arch. Dev. Biol. 193, 283-295. [DOI] [PubMed] [Google Scholar]
- 34.Chang, H. C. & Rubin, G. M. (1997) Genes Dev. 11, 1132-1139. [DOI] [PubMed] [Google Scholar]
- 35.Tepass, U., Theres, C. & Knust, E. (1990) Cell 61, 787-799. [DOI] [PubMed] [Google Scholar]
- 36.Perrimon, N., Lanjuin, A., Arnold, C. & Noll, E. (1996) Genetics 144, 1681-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lavorgna, G., Ueda, H., Clos, J. & Wu, C. (1991) Science 252, 848-851. [DOI] [PubMed] [Google Scholar]
- 38.Weiner, A. J., Scott, M. P. & Kaufman, T. C. (1984) Cell 37, 843-851. [DOI] [PubMed] [Google Scholar]
- 39.Kuroiwa, A., Hafen, E. & Gehring, W. J. (1984) Cell 37, 825-831. [DOI] [PubMed] [Google Scholar]
- 40.Karim, F. D., Chang, H. C., Therrien, M., Wassarman, D. A., Laverty, T. & Rubin, G. M. (1996) Genetics 143, 315-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalidas, S. & Smith, D. P. (2002) Neuron 33, 177-184. [DOI] [PubMed] [Google Scholar]
- 42.Spradling, A. C., Stern, D. M., Kiss, I., Roote, J., Laverty, T. & Rubin, G. M. (1995) Proc. Natl. Acad. Sci. USA 92, 10824-10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zambrowicz, B. P., Friedrich, G. A., Buxton, E. C., Lilleberg, S. L., Person, C. & Sands, A. T. (1998) Nature 392, 608-611. [DOI] [PubMed] [Google Scholar]
- 44.Morin, X., Daneman, R., Zavortink, M. & Chia, W. (2001) Proc. Natl. Acad. Sci. USA 98, 15050-15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elick, T. A., Bauser, C. A. & Fraser, M. J. (1996) Genetica 98, 33-41. [DOI] [PubMed] [Google Scholar]
- 46.Rorth, P. (1996) Proc. Natl. Acad. Sci. USA 93, 12418-12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.