Abstract
The tadpole larva of the basal chordate Ciona intestinalis has the most simplified, basic body-plan of chordates. Because it has a compact genome with a complete draft sequence, a large quantity of EST/cDNA information, and a short generation time, Ciona is a suitable model for future genetics. We establish here a transgenic technique in Ciona that uses the Tc1/mariner superfamily transposon Minos. Minos was integrated efficiently into the genome of germ cells and transmitted stably to subsequent generations. In addition, an enhancer-trap line was obtained. This is a demonstration of efficient, Minos-mediated transgenesis in marine invertebrates.
Keywords: ascidian, transgenic technique, enhancer trap
The ascidian Ciona intestinalis provides an appealingly simple experimental system for the investigation of the molecular mechanisms underlying cell-fate specification during development (1–3). The ascidian tadpole is composed of only ≈2,600 cells, and there is extensive information on the cell lineage of most major tissues and organs (4, 5). The blastomeres of early embryos are large and easy to manipulate, which permits the detailed visualization of differential gene expression during development (2). In addition, embryogenesis is rapid (≈18 h from fertilization to a free-swimming tadpole at 18°C); the entire life cycle takes <3 months, thereby facilitating genetic analyses (6). Furthermore, transgenic DNAs can be introduced into developing Ciona embryos by using simple electroporation methods to characterize cis-regulatory DNAs (7). Electroporation and microinjection methods have been used to produce mutant phenotypes by misexpressing or over-expressing a variety of regulatory genes that encode transcription factors or signaling molecules (8). In addition, morpholino oligo-nucleotides are useful in Ciona for disrupting gene function (9).
In addition, the determination of the draft genome sequence of C. intestinalis has revealed that it possesses the smallest, nonredundant set of genes for the chordate-specific features (10). Furthermore, a large cDNA/EST database that shows dynamic spatiotemporal expression of Ciona developmental genes has been compiled (11, 12). This system, therefore, is amenable to the application of genetical approaches to examining gene function. Mutagenesis using the chemical mutagen N-ethyl-N-nitrosourea has already been applied to Ciona savignyi (6, 13) and C. intestinalis (14). Identifying specific genes that are mutated by N-ethyl-N-nitrosourea, however, is very difficult. Another mutagenesis technique, such as insertional mutagenesis by germ-line transgenesis, may be more effective for studying gene function in this organism. We showed recently that Minos, a transposable element of the Tc1/mariner superfamily (15), retains high activity in Ciona (16); this finding suggested the possibility of using Minos as a tool for germ-line transgenesis. In this study, we determined that germ-line transgenesis of C. intestinalis by Minos is possible.
Materials and Methods
Construction of Vector. Genome DNA containing the CiTPO (thyroid peroxidase) promoter was isolated from the genomic DNA library of C. intestinalis. The promoter was ligated to gfp, which encodes enhanced GFP (BD Biosciences Clontech), and the construct was subcloned into the PstI–SmaI site of pMiLRneo (17) to create pMiLRCiTPO-gfp.
Introduction of Minos into Ciona. Minos transposase mRNA was synthesized in vitro by using a MEGAscript T3 kit (Ambion, Austin, TX). A mixture of 50 ng/μl pMiLRCiTPO-gfp and 50 ng/μl Minos transposase mRNA was microinjected into dechorionated, unfertilized eggs of Ciona. The method of microinjection was described previously (18). The injection volume was ≈5 pl. Inseminated and fertilized eggs were cultured on agar-coated Petri dishes filled with Millipore-filtered seawater until the swimming-larva stage and then transferred to new dishes to promote metamorphosis. After founders had developed mature sperm, they were mated with wild type to obtain F1 progeny. GFP-transmitting founders were named Mi[CiTPOgfp]1, Mi[CiTPOgfp]2, and so on. Electroporation was done as described (19).
Culturing Condition of C. intestinalis. Metamorphosed C. intestinalis individuals were cultured in 9-cm Petri dishes (three to five individuals per dish) for ≈10 days with 3% (vol/vol) full-grown diatom Chaetoceros gracilis in seawater (3% GSW). Seawater was changed every day. After 10 days, the Ciona were cultured with 0.2% Microfeast–Spirulina (MS) solution in 3% GSW. The mixture of 400 mg of Microfeast PZ-20 (Saltcreek, Salt Lake City) and 200 mg of Jade Spirulina (Saltcreek) in 50 ml of seawater was left for 5–10 min. The supernatant was used as MS. After juveniles had grown to ≈5 mm in length, the number was reduced (one to three individuals per dish) and the juveniles were cultured in 500 ml of 0.2% MS in 3% GSW. Seawater was changed every day. The concentration of MS and the amount of seawater were increased up to 0.6% and 1 liter, respectively, according to the size of individuals. Culturing was done at ≈18°C.
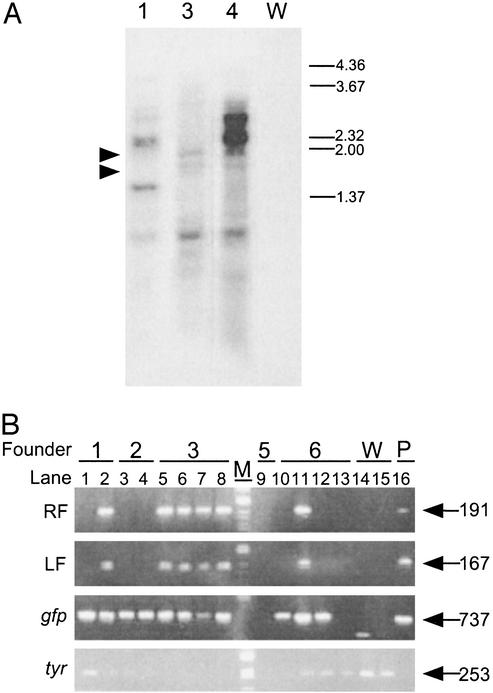
Southern Blot Analysis. The molecular experiments were carried out according to a previously described method (20). Genomic DNA was isolated from whole F1 animals grown to 3–5 cm. The total amount of DNA was cut by EcoRI, electrophoresed, and blotted on to Hybond N+ nylon membrane (Amersham Biosciences). The probe used is described in Fig. 1A. Membrane hybridized with 32P-labeled probes was washed under high stringency conditions.
Fig. 1.
Inheritance of GFP expression in Ciona F1 and F2 progeny. (A) Structure of a Minos construct, pMiLRCiTPO-gfp. The dotted line indicates the probe region used for Southern blot analysis shown in Fig. 2A. Arrows indicate the primer positions used for PCR analysis shown in Fig. 2B. LIR and RIR, left and right inverted repeats (IR) of Minos; pA, polyadenylation signal sequence; EI, EcoRI restriction site. (B–E) F1 progeny and the expression of GFP from Mi[CiTPOgfp]3 (B and C) and Mi[CiTPOgfp]2 (D and E). In C, GFP was seen at the anterior and posterior ends of the endostyle (red arrows). In E, endostyle (EN), peripharyngeal band (PB), dorsal tubercle (DT), and posterior margin of pharyngeal sac (PM) were marked by GFP. (F and G) GFP expression in F2 progeny derived from Mi[CiTPOgfp]3 (F) and Mi[CiTPOgfp]2 (G).
PCR Analysis. All PCR experiments were performed with rTaq DNA polymerase (Toyobo, Kyoto). Thermal asymmetric interlaced PCR (TAIL-PCR) was done as described (21). When only one side of the flanking sequences was determined by TAIL-PCR, the other side was amplified by specific primers. Primer sequences and detailed PCR conditions are available from Y.S. on request.
Results
Inheritance of GFP Expression to Next Generations. To effect germ-line transgenesis in C. intestinalis, a mixture of Minos transposon DNA and Minos transposase mRNA was introduced into Ciona eggs. The transposon includes a gfp gene with upstream sequence of CiTPO (Fig. 1 A). CiTPO encodes thyroid peroxidase (22) and its upstream sequence drives GFP expression at the edges of the endostyles of juveniles. The manipulated eggs were allowed to develop into founder animals, which grew up to produce mature sperm. Sperm was collected from these founders and used to fertilize wild-type eggs to obtain F1 progeny. The transmission of gfp to F1 progeny was observed first by assaying GFP expression. F1 progeny inherited GFP expression from 7 of 21 founders (Fig. 1 B–E). The ratio of F1 progeny with GFP signal to those without differed depending on the founders; the ratio, ranging from 1% to 20% (data not shown), suggests a mosaic mode of transgenesis of germ cells. Progeny derived from the founder, Mi[CiTPOgfp]2, showed an irregular pattern of GFP signal (Fig. 1 D and E). Instead of the edges of the endostyle, the whole ventral half of the endostyle, posterior margin of the branchial sac, peripharyngeal band, and dorsal tubercle showed strong GFP signals. It is possible that gfp is under the control of a specific enhancer for these tissues, as discussed below.
Next, the frequency of transgene transmission to F2 progeny was analyzed. We obtained F2 progeny by mating GFP-positive F1 to wild-type eggs. As shown in Fig. 1 F and G, the expression of GFP was inherited correctly by the F2 progeny. In most cases, ≈50% of the F2 progeny expressed GFP (Table 1); this result is consistent with Mendel's law of segregation for a single heterozygous locus. Therefore, GFP expressed in progeny might be derived from a single gfp insertion. Similarly, the inheritance ratio of GFP expression in F3 progeny was also ≈50% (Table 1).
Table 1. Inheritance ratio of GFP expression to F2 and F3 progeny.
| No. of progeny
|
||||
|---|---|---|---|---|
| Derived founder | Progeny | GFP positive | Total | GFP inheritance frequency |
| Mi[CiTPOgfp]1 | F2 | 44 | 93 | 0.47 |
| Mi[CiTPOgfp]2 | F2 | 95 | 158 | 0.60 |
| Mi[CiTPOgfp]2 | F2 | 102 | 136 | 0.75 |
| Mi[CiTPOgfp]2 | F2 | 79 | 144 | 0.54 |
| Mi[CiTPOgfp]3 | F2 | 46 | 94 | 0.49 |
| Mi[CiTPOgfp]3 | F2 | 34 | 87 | 0.39 |
| Mi[CiTPOgfp]4 | F2 | 16 | 37 | 0.43 |
| Mi[CiTPOgfp]4 | F2 | 4 | 31 | 0.12 |
| Mi[CiTPOgfp]5 | F2 | 34 | 60 | 0.56 |
| Mi[CiTPOgfp]1 | F3 | 35 | 67 | 0.52 |
| Mi[CiTPOgfp]2 | F3 | 58 | 106 | 0.54 |
| Mi[CiTPOgfp]2 | F3 | 38 | 78 | 0.48 |
| Mi[CiTPOgfp]2 | F3 | 41 | 88 | 0.46 |
| Mi[CiTPOgfp]3 | F3 | 50 | 109 | 0.45 |
| Mi[CiTPOgfp]3 | F3 | 19 | 49 | 0.38 |
Genomic Integration of Minos by Transposase. To reveal the nature of transposon insertion into the genome, Southern blot analysis was performed by using genomic DNA from F1 progeny. As shown in Fig. 2A, multiple bands of various sizes were observed in each lane. Such variance in size can be caused new restriction sites that result from random insertion of Minos into the genome. F1 progeny, derived from Mi[CiTPOgf]4, showed two strong bands of ≈2.2 and 3.0 kbp (Fig. 2 A, lane 4). The bands may derive from multiple concatamers of Minos and vector sequences at a site; this possibility was suggested also by PCR analysis (data not shown). Bands of ≈1.6 and 2.0 kbp were commonly found in lanes 3 and 4. These bands are likely to be caused by insertion in similar regions of the genome, such as repetitive sequences, or by the formation of a Minos concatamer. Thus, Southern blot analysis revealed the integration of Minos into several regions of the genome and the correct transmission to F1 progeny. The multiple bands in each lane suggest that multiple insertion events potentially occurred in the founder germ line. The multiplicity of Minos insertions was contradictory to the transmission rate of GFP expression. A possible explanation is that the expression of the gfp gene was subject to the locus effect, and only a part of inserted gfp genes was allowed to express GFP sufficient for observation.
Fig. 2.
Genomic integration of Minos revealed by Southern blot and PCR analyses. (A) Genomic DNA isolated from F1 progeny of Mi[CiTPOgfp]1, 3, and 4 (lanes 1, 3, and 4) and wild type (lane W) were digested with EcoRI and electrophoresed. Right numbers indicate the sizes of markers (in kbp). There are several bands characteristic to each F1 progeny, in addition to similarly sized bands of ≈1.6 and 2.0 kbp (arrowheads). (B) Several F1 progeny lacked plasmid sequences of pMiLRCiTPO-gfp flanking Minos IR. Genomic DNA isolated from F1 progeny from Mi[CiTPOgfp]1–3, -5, -6 (1–6), and wild type (W) were subjected to PCR analysis using primers described in Fig. 1 A. pMiLRCiTPO-gfp plasmid (P) was used as a positive control. Lane M indicates the marker. Right numbers show the size of bands (in bp). Note that several F1 (lanes 1, 3, 4, 10, and 12) lacked left flanking (LF) and right flanking (RF) sequences of IR of pMiLRCiTPO-gfp, although gfp was transmitted. PCR against tyrosinase gene (tyr) was done as an internal control.
Although Southern blot analysis showed integration of Minos DNA into the genome, it remained to be determined whether Minos integration was caused by transposase, plasmid insertion, or both. To determine the cause of integration, we performed PCR analysis with genomic DNA from F1 progeny. As shown in Fig. 2B, some progeny from Mi[CiTPOgfp]1, Mi[CiTPOgfp]2, and Mi[CiTPOgfp]6 did not contain plasmid sequences flanking the Minos IR, although gfp was transmitted to these progeny correctly. Therefore, the segregation pattern of gfp is independent of the plasmid sequence; this independence indicates that insertions were caused by transposase.
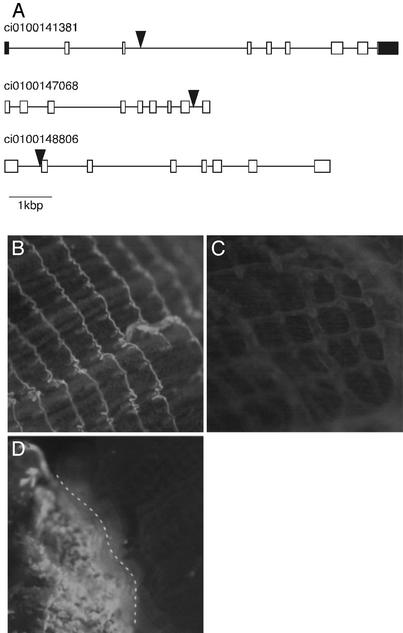
To address directly whether Minos insertions were caused by transposase, DNA sequences flanking Minos IR in the genome were determined with TAIL-PCR (21). TAIL-PCR is easier than inverse PCR, because pre-PCR treatment of genomic DNA is not a prerequisite. By using this method, we determined several sequences adjacent to IR to compare with the whole C. intestinalis genomic sequence (10). As a result, nine pairs of flanking sequences were mapped directly on the Ciona genome (Table 2). In all cases, TA dinucleotides were targeted and plasmid sequences adjacent to IR were absent, whereas the IR remained intact. The duplication of TA dinucleotides was observed in all cases in which both left and right flanking sequences were determined. Because the manner of integration of Minos was identical to that described (23, 24), we concluded that Minos was inserted correctly into genomic DNA by transposase. The nine integration sites were located on different scaffolds of the assembled genome sequence (http://genome.jgi-psf.org/ciona4), which suggests that Minos integration occurs in variable regions of the genome. One common feature of the integration sites was the enrichment of TA dinucleotides (two or more TAs adjacent to the targeted TA were observed); this common feature suggests that Minos targeted TA-rich sites preferentially. Among the nine integration sites, six insertion sites hit genes (Table 2 and Fig. 3A). This finding suggests that Minos can very efficiently disrupt Ciona genes.
Table 2. Genome sequences flanking the Minos IRs.
| Derived founder | Left flanking sequence | Minos | Right flanking sequence | Genome location* | Position of insertion† | Homology of targeted genes |
|---|---|---|---|---|---|---|
| Mi[CiTPOgfp]1 | CCAAGCTACTACTACTACTA | CGAG.. .CTCG | TACACACTATGTACATTGCT | 646 | UTR | Inhibitor of apoptosis protein 2 |
| Mi[CiTPOgfp]2 | CGAG.. .CTCG | TAGAGGTACAAAGATTCTGG | 7 | Intron | Musashi-2 | |
| Mi[CiTPOgfp]2 | ATTATTGGCTATGATATTTA | CGAG.. .CTCG | TAGTAACGTAATGTATTTCT | 55 | Intron | Prostaglandin-transporting peptide |
| Mi[CiTPOgfp]3 | TTGAGTAACTTTACAGAATA | CGAG.. .CTCG | TAGTGTCCTCTCCTACTGCT | ND‡ | UTR | No homology |
| Mi[CiTPOgfp]3 | AATGCTTATATATATATATA | CGAG.. .CTCG | TAGGACCACAGGAATTTATG | 35 | No | |
| Mi[CiTPOgfp]3 | CATCTCCCCCCACCCTACTA | CGAG.. .CTCG | TATAATAATATGATAAGCGG | 26 | No | |
| Mi[CiTPOgfp]3 | GTCAGTTAAAGAACTATATA | CGAG.. .CTCG | TATGCATGATAACGCAATAT | 671 | No | |
| Mi[CiTPOgfp]5 | CACGCCAACACACGTTTATA | CGAG.. .CTCG | 1056 | Intron | No homology | |
| Mi[CiTPOgfp]6 | ACTGTCGATGCATGTAAGTA | CGAG.. .CTCG | TATATATACTTAGAAAAACT | 528 | Intron | Putative transmembrane transporter FLIPT1 |
The number indicates the Scaffold numbers of the assembly of the C. intestinalis genome sequences
UTR and Intron indicate that Minos was inserted into the 3′ UTRs and introns of genes, respectively
ND, not detected. The insertion site is not included in the Ciona genome database, but hit to Ciona ESTs
Fig. 3.
Examples of genes targeted by Minos and of enhancer-trap events. (A) Genes disrupted by Minos. Exons and introns are shown by boxes and lines, respectively. Exons corresponding to UTRs are labeled in black. Large triangles indicate the insertion sites of Minos. Grail numbers indicate the gene model in the C. intestinalis genome database (http://genome.jgi-psf.org/ciona4). (B–D) Two examples of enhancer-trap events. GFP was expressed in the pharyngeal gill of F2 progeny derived from Mi[CiTPOgfp]2 (B), but not in Mi[CiTPOgfp]3 (C). (D) A founder animal expressing GFP in the pharyngeal gill. Because the Minos insertion probably occurred in half of the precursor cells of the pharyngeal gill, only half of the gill expressed GFP (left side of the broken line).
Enhancer Traps by Minos. Transgenesis by transposon can be applied to other genetic techniques, such as the enhancer trap. This technique is useful for generation of cell-type markers and for identification of novel genes by expression patterns (25). We obtained two lines of indirect evidence suggesting that Minos can be used as an enhancer trap in Ciona. One is the Mi[CiTPOgfp]2 line described above. The GFP-expression pattern in this line of juveniles was distinct from the other lines (Fig. 1B). In addition, GFP signal at the pharyngeal gill was observed in 1-month-old F2 progeny of Mi[CiTPOgfp]2, but never observed in F2 progeny of Mi[CiTPOgfp]3–5 (Fig. 3 B and C). These different patterns of expression suggest an enhancer-trap event in the Mi[CiTPOgfp]2 line. Two Minos insertions were identified in progeny from Mi[CiTPOgfp]2 (Table 2). The insertions occurred within genes (Fig. 3A). One targeted gene (ci0100141381) encodes a homologue of vertebrate Musashi-2 (26); the other (ci0100148806) encodes a homologue of prostaglandin transporter (27). The gfp inserted into the genome may be entrapped by an enhancer, which controls the expression of either of these two genes. C. intestinalis Musashi-2 homologue showed the expression in the endostyle and the peripharyngeal band of the adult body (data not shown). This evidence suggests that the insertion into this gene is a strong candidate of the enhancer-trap insertion. Another example of an enhancer trap is the expression of GFP at the pharyngeal gill in one founder (Fig. 3D). We obtained this founder during a trial of introduction of Minos DNA and transposase RNA by electroporation (19). Because no other founders obtained by microinjection or electroporation showed similar GFP expression at the pharyngeal gill, it is reasonable to conclude that expression at the gill was caused by the entrapment of gfp by a gill-specific enhancer. The enhancer-trap event occurred in somatic cells but not in germ cells because F1 progeny of this founder did not express GFP at the gill. Although two enhancer traps expressed GFP in the pharyngeal gill, the expression patterns were different; this difference suggests that different enhancers were involved in each case.
Small-Scale Culturing System of C. intestinalis. One important point for insertional mutagenesis is the short generation time. The C. intestinalis individuals used in this study were kept in 1 liter of seawater per one to three individuals and the seawater was changed every day. We gave them Microfeast and Spirulina, in addition to a living diatom, Chaetoceros gracilis, according to previous reports (28, 29). Under these conditions, some individuals mature to produce sperm within 1 month and eggs within ≈2 months. The generation time is much faster than that of zebrafish (≈3 months), and comparable with that of Arabidopsis thaliana (1–2 months). Although Ciona requires a considerable quantity of seawater for culturing, our system provides the possibility of fast and mass culturing of Ciona in a laboratory.
Discussion
Our current data show the integration of Minos transposon into the genomic DNA of Ciona germ cells and transmission to subsequent generations. We emphasize that the easy identification of the insertion sites was achieved by the TAIL-PCR method. Considering the high polymorphism rate of the C. intestinalis genome (10), we find this ease of identification to be an advantage over N-ethyl-N-nitrosourea-based mutagenesis. In addition, the causal genes of mutants provided by Minos are identified easily, which is a key process for mutagenesis studies. In the present study, we obtained up to four insertions derived from one founder, and one enhancer-trap line of 21 founders. For this high efficiency of Minos insertions in Ciona, the combination of Ciona and Minos provides an excellent system for large-scale mutagenesis. The high efficiency suggests also the possibility of applying Minos to other genetic techniques in Ciona. The cis element of CiTPO gene was used in this study for the driver of GFP. All of the individuals containing Minos insertions were not screened, however, because of the locus effect of insertion sites. The use of a stronger cis element or insulator is necessary for more efficient screening (identification of all of Minos-positive individuals by GFP signal). In this study, we presented evidence of germ-line transgenesis by using a transposon in deuterostomes other than vertebrates. Using Minos, a highly efficient transgenic technique can be applied to many other animals, such as echinoderms, cephalochordates, and other chordates, including vertebrates (17). Finally, large-scale mutagenesis in C. intestinalis may provide many interesting cues based on the biological and phylogenetic position of the ascidian, not only to gene function, but also to developmental and evolutionary biology.
Acknowledgments
We thank Dr. Apostolos G. Klinakis, Prof. Charalambos Savakis, and Minos BioSystems, Ltd., for providing us with Minos plasmids and useful suggestions; Dr. Shungo Kano for helpful discussions; Dr. Kentaro Shimizu for the instruction of TAIL-PCR technique; all members of the Otsuchi Marine Research Center (Ocean Research Institute, University of Tokyo), the Education and Research Center of Marine Bio-resources (Tohoku University); Dr. Shigeki Fujiwara for collection of Ciona adults; and Drs. Hiroki Takahashi and Michio Ogasawara for the use of the Ciona genomic DNA library and for assistance with isolation of the CiTPO promoter. This work was supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology Japan to Y.S. (0200967), S.C. (0060102) and N.S. (12358012).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TPO, thyroid peroxidase; IR, inverted repeat; MS, Microfeast–Spirulina; TAIL-PCR, thermal asymmetric interlaced PCR.
References
- 1.Satoh, N. (1994) Developmental Biology of Ascidians (Cambridge Univ. Press, New York).
- 2.Satoh, N. (2001) Differentiation (Berlin) 68, 1-12. [DOI] [PubMed] [Google Scholar]
- 3.Corbo, J. C., Di Gregorio, A. & Levine, M. (2001) Cell 106, 535-538. [DOI] [PubMed] [Google Scholar]
- 4.Conklin, E. G. (1905) J. Acad. Nat. Sci. 13, 1-119. [Google Scholar]
- 5.Nishida, H. (1987) Dev. Biol. 121, 526-541. [DOI] [PubMed] [Google Scholar]
- 6.Nakatani, Y., Moody, R. & Smith, W. C. (1999) Development (Cambridge, U.K.) 126, 3293-3301. [DOI] [PubMed] [Google Scholar]
- 7.Di Gregorio, A. & Levine, M. (2002) Differentiation (Berlin) 70, 132-139. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi, H., Hotta, K., Erives, A., Di Gregorio, A., Zeller, R. W., Levine, M. & Satoh, N. (1999) Genes Dev. 13, 1519-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Satou, Y., Imai, K. S. & Satoh, N. (2001) Genesis 30, 103-106. [DOI] [PubMed] [Google Scholar]
- 10.Dehal, P., Satou, Y., Campbell, R. K., Chapman, J., Degnan, B., De Tomaso, A., Davidson, B., Di Gregorio, A., Gelpke, M., Goodstein, D. M., et al. (2002) Science 298, 2157-2167. [DOI] [PubMed] [Google Scholar]
- 11.Satou, Y., Takatori, N., Yamada, L., Mochizuki, Y., Hamaguchi, M., Ishikawa, H., Chiba, S., Imai, K., Kano, S., Murakami, S. D., et al. (2001) Development (Cambridge, U.K.) 128, 2893-2904. [DOI] [PubMed] [Google Scholar]
- 12.Satou, Y., Yamada, L, Mochizuki, Y., Takatori, N., Kawashima, T., Sasaki, A., Hamaguchi, M., Awazu, S., Yagi, K., Sasakura, Y., et al. (2002) Genesis 33, 153-154. [DOI] [PubMed] [Google Scholar]
- 13.Moody, R., Davis, S. W., Cubas, F. & Smith, W. C. (1999) Mol. Gen. Genet. 262, 199-206. [DOI] [PubMed] [Google Scholar]
- 14.Sordino, P., Heisenberg, C., Cirino, P., Toscano, A., Giuliano, P., Marino, R., Pinto, M. R. & De Santis, R. (2000) Sarsia 85, 173-176. [Google Scholar]
- 15.Franz, G., Loukeris, T. G., Dialektaki, G., Thompson, C. R. L. & Savakis, C. (1994) Proc. Natl. Acad. Sci. USA 91, 4746-4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasakura, Y., Awazu, S., Chiba, S., Kano, S. & Satoh, N. (2003) Gene 308, 11-20. [DOI] [PubMed] [Google Scholar]
- 17.Klinakis, A. G., Zagoraiou, L., Vassilatis, D. K. & Savakis, C. (2000) EMBO Rep. 1, 416-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshida, S., Marikawa, Y. & Satoh, N. (1996) Development (Cambridge, U.K.) 122, 2005-2012. [DOI] [PubMed] [Google Scholar]
- 19.Corbo, J. C., Levine, M. & Zeller, R. W. (1997) Development (Cambridge, U.K.) 124, 589-602. [DOI] [PubMed] [Google Scholar]
- 20.Ausubel, M. F., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (1996) Current Protocols in Molecular Biology (Wiley, New York).
- 21.Liu, Y., Mitsukawa, N., Oosumi, T. & Whittier, R. F. (1995) Plant J. 8, 457-463. [DOI] [PubMed] [Google Scholar]
- 22.Ogasawara, M., Di Lauro, R. & Satoh, N. (1999) J. Exp. Zool. 285, 158-169. [DOI] [PubMed] [Google Scholar]
- 23.van Luenen, H. G. A., Colloms, S. D. & Plasterk, R. H. A. (1994) Cell 79, 293-301. [DOI] [PubMed] [Google Scholar]
- 24.Arca, B., Zabalou, S., Loukeris, T. G. & Savakis, C. (1997) Genetics 145, 267-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Kane, C. J. (1998) in Drosophila, ed. Roberts, D. B. (Oxford Univ. Press, New York), pp. 131-178.
- 26.Sakakibara, S., Nakamura, Y., Satoh, H. & Okano, H. (2001) J. Neurosci. 21, 8091-8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanai, N., Lu, R., Satriano, J. A., Bao, Y., Wolkoff, A. W. & Schuster, V. L. (1995) Science 268, 866-869. [DOI] [PubMed] [Google Scholar]
- 28.Kano, S., Chiba, S. & Satoh, N. (2001) Mar. Biotechnol. 3, 58-67. [DOI] [PubMed] [Google Scholar]
- 29.Cirino, P., Toscano, A., Caramiello, D., Macina, A., Miraglia, V. & Monte, A. (27November2002) Mar. Mod. Elec. Rec. [serial online]. Available at www.mbl.edu/BiologicalBulletin/MMER/cirino/CirTit.html.