Abstract
The RUNX transcription factors are important regulators of lineage-specific gene expression. RUNX are bifunctional, acting both as activators and repressors of tissue-specific target genes. Recently, we have demonstrated that Runx3 is a neurogenic transcription factor, which regulates development and survival of proprioceptive neurons in dorsal root ganglia. Here we report that Runx3 and Runx1 are highly expressed in thymic medulla and cortex, respectively, and function in development of CD8 T cells during thymopoiesis. Runx3-deficient (Runx3 KO) mice display abnormalities in CD4 expression during lineage decisions and impairment of CD8 T cell maturation in the thymus. A large proportion of Runx3 KO peripheral CD8 T cells also expressed CD4, and in contrast to wild-type, their proliferation ability was largely reduced. In addition, the in vitro cytotoxic activity of alloimmunized peritoneal exudate lymphocytes was significantly lower in Runx3 KO compared with WT mice. In a compound mutant mouse, null for Runx3 and heterozygous for Runx1 (Runx3-/-;Runx1+/-), all peripheral CD8 T cells also expressed CD4, resulting in a complete lack of single-positive CD8+ T cells in the spleen. The results provide information on the role of Runx3 and Runx1 in thymopoiesis and suggest that both act as transcriptional repressors of CD4 expression during T cell lineage decisions.
The mammalian RUNX3/AML2 gene resides on human chromosome 1p36.1 and mouse chromosome 4, respectively (1–4). It belongs to the RUNX family of transcription factors, which contains three genes. The two other family members, RUNX1 and RUNX2, play fundamental roles in hematopoietic and osteogenic lineage-specific gene expression, and when mutated, are associated with human diseases (5, 6). The three RUNX genes are regulated at the transcriptional level by two promoters, and at the translational level by internal ribosome entry site (IRES)- and cap-dependent translation control (7–14). The gene products of RUNX bind to the same DNA motif and activate or repress transcription of target genes through recruitment of common transcriptional modulators (15–18). Despite this occurrence, each of the Runx genes has well defined biological functions reflected in a different expression pattern of the genes (19–23) and distinct phenotypes of the corresponding knockout mice (6, 24–27).
During mouse embryogenesis Runx3 is expressed in hematopoietic organs, epidermal appendages, developing bones, and sensory ganglia (20). Studies in knockout (KO) mice revealed that Runx3 is a neurogenic-specific transcription factor required for development and survival of TrkC neurons in the dorsal root ganglia. In the absence of Runx3 these neurons die, leading to disruption of the stretch reflex neuronal circuit, and consequently to severe ataxia (25, 26). Intriguingly, in one strain of Runx3 KO, the gastric mucosa of newborn mice exhibits hyperplasia due to stimulated proliferation and suppressed apoptosis of stomach epithelial cells (27).
It has previously been reported that in adults RUNX3 is highly expressed in the hematopoietic system as well as in various hematopoietic cell lines (1, 10, 28–31). Using adult Runx3 KO mice (25) we now report that Runx3 function is essential for the maturation of single-positive (SP) CD8+ T cells. In the absence of Runx3, peripheral CD8+ T cells are diminished and the cytolytic activity of T lymphocytes is impaired. Runx1 is essential for embryonal definitive hematopoiesis (24). We now show that haploinsufficiency of Runx1 in compound mutant Runx3-/-; Runx1+/- mice annulled production of peripheral CD8+ T cells, indicating that Runx1 is also involved in the development of CD8+ T cells. While this work was in progress Taniuchi et al. (32) described similar studies in immunodeficient Rag2 null mice reconstituted with fetal liver cells derived from Runx3 KO embryos. The differences between the two sets of data are discussed.
Methods
Mice. Runx3 KO mice were generated as described (25). For generation of Runx3-/-; Runx1+/- mice (compound Runx3/Runx1), Runx3+/- mice were mated with Runx1+/- mice (33). Analysis of lymphocyte development in compound Runx3/Runx1 was carried out with 9- to 12-day-old progenies because of high mortality of these mice.
Flow Cytometry. Cells were liberated from thymus and spleen, suspended in fluorescence-activated cell sorter (FACS) buffer (1% BSA/0.05% sodium azide in PBS) and incubated with mAbs for 30 min on ice, washed, and when necessary, incubated with secondary reagents for 30 min on ice. Flow cytometry was performed by using a FACSCalibur (Becton Dickinson) equipped with CELLQUEST software (Becton Dickinson). mAbs were as follows: CD4-biotinilated (GK1.5), CD8-Percp (53-6.7, PharMingen), and T cell receptor (TCR)αβ-FITC (H57-597), CD24-phycoerythrin (PE) (3c-F1), CD25-FITC(7D4), and CD44-PE (Pgp1,Ly-24, Southern Biotechnology Associates).
Immunohistochemistry. Thymocytes from 6-week-old mice were stained with anti-CD4 and anti-CD8 Abs. Four thousand cells from each subset [double-negative (DN) CD4-CD8-, double-positive (DP) CD4+CD8+,SPCD4+, and SP CD8+] were sorted onto poly-(L-lysine)-coated slides, air dried fixed for 20 min in 3% paraformaldehyde (PFA), 2% sucrose in PBS and treated for 15 min in 1% peroxide in 50% methanol. After 1-h blocking in 0.1% Triton and 20% goat serum, slides were incubated overnight with primary Ab (Affinity-purified RUNX1 1:100; purified RUNX3 1:1,000) in 0.1% Triton and 3% goat serum. Biotinylated secondary antibodies and ABC complex from the Vectastain kit (Vector Laboratories) were used for detection. Immunohistochemistry on paraffin-embedded thymuses was previously described (20). For detecting CD4 in cytolytic T lymphocytes (CTLs) conjugated with specific target cell, conjugates of immune peritoneal exudates CTLs and Fas (CD95)-expressing, leukemia L1210 target cells (LF+) were formed (34, 35) and examined by immunofluorescence. Four 10-μl drops containing ≈1 × 103 cells of conjugate suspension were placed on poly-(L-lysine)-coated slides. Cells were fixed with 2% PFA/PBS for 20 min at room temperature, washed three times with PBS, stained with purified rat anti-mouse CD4 (L3T4) mAb (PharMingen) for 30 min at 4°C, and then with donkey anti-rat Cy3 (Jackson ImmunoResearch) for 15 min at room temperature. Cells were viewed by using Zeiss confocal laser scanning microscopes at ×20 magnification. Nine fields were scored for CD4+ and CD4-.
Cytotoxicity and Proliferation Assays. Alloreactive peritoneal exudate lymphocytes (PELs) were produced in 6- to 10-week-old mice, purified and tested for their cytotoxic activity by the 51Cr-release assay as described (36). For monitoring CD8+ T cell proliferation, cells were liberated from spleen and RBC removed by treatment with ACK buffer (0.15 M NH4Cl/1 mM KHCO3/0.1 mM Na2 EDTA). CD8+ T cells were purified by using a magnetic cell sorting (MACS) separation system according to the manufacturer's instructions (Miltenyi Biotec, Auburn, CA). Cells were cultured at a density of 1 × 105 cells per well in a 96-well plate. Plate-bound anti-CD3 mAb (2 μg/ml) was used to activate T lymphocytes. Either soluble anti-CD28 mAb (2 μg/ml) or recombinant mouse IL-2 (PeproTech, Rocky Hill, NJ; 20 ng/ml) was used for costimulation. [3H]Thymidine (1 μCi; 1 Ci = 37 GBq) was added for the last 6 h of the 72-h incubation period. Cells were collected on a filter plate (Unifilter Tm-96, Packard) and radioactivity was measured by using a microplate scintillation counter (TopCount, Packard).
Results
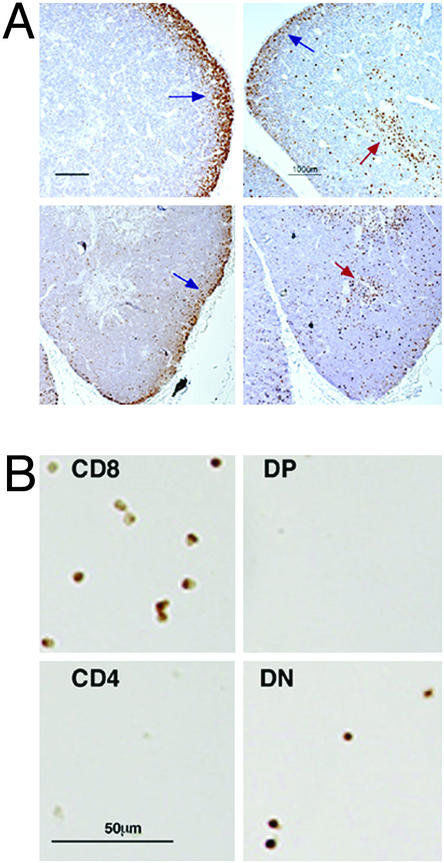
Runx1 and Runx3 Expression in Adult Thymus Is Confined Mainly to Cortex and Medulla, Respectively. In adults, Runx3 is highly expressed in spleen, thymus, and blood (1, 28, 29, 31), whereas in embryos, it is detected in liver hematopoietic precursors and in thymocytes (20). We have previously shown (25) that targeting of Runx3 exon 2 abolished expression in thymus. To define the expression pattern of Runx3 and Runx1 in the developing thymus, sections of fetal and adult thymuses and FACS-sorted thymocytes were examined by immunohistochemistry (Fig. 1). Late fetal thymic development [embryonic day (E)15.5 until birth] is characterized by a rapid accumulation of CD4+CD8+ DP thymocytes, onset of positive and negative selection, and regional demarcation of cortex versus medulla (37). At E18, we found that Runx3 is highly expressed in the newly formed medulla as well as in cortical thymocytes (Fig. 1 A). In adults, Runx3 expression is mainly detected in the medulla, where SP T cells mature. Much less Runx3 was detected in the cortex representing expression in immature (DN) T cells or in other cell types such as macrophages and B cells. On the other hand, Runx1 expression was mainly seen in the cortex both at E18 and at 6 weeks thymus (Fig. 1 A). To define the expression of Runx3 in isolated T cell subsets, CD4/CD8 FACS-sorted thymocytes were examined by immunohistochemistry. Consistent with its intrathymic localization, Runx3 was detected in SP CD8+ and in DN thymocytes, but was absent from SP CD4+ and DP CD4+CD8+ thymocytes (Fig. 1B).
Fig. 1.
Expression of Runx3 and Runx1 in the thymus. Runx3 is expressed mainly in medullary SP CD8+ and in cortical DN CD4-CD8- thymocytes, whereas Runx1 expression is found mainly in cortical thymocytes. (A) Paraffin sections of thymuses of E18 embryos (Upper) and 6-week-old mice (Lower) were stained with anti-Runx1 (Left) and anti-Runx3 (Right) antibodies. Cortex (blue arrow) and medulla (red arrow) are indicated. (B) Thymocytes from 6-week-old mice were sorted onto (poly-l)lysine-coated slides and were stained with anti-Runx3 antibodies.
In Runx3-KO Mice Maturation of CD8+ T Cells Is Impaired and Peripheral SP CD8+ Cells also Express CD4. Runx3 is highly expressed in embryo and adult thymus. Differentiation of DP CD+CD8+ thymocytes to SP CD8+ cells involves silencing of CD4, and two RUNX-binding sites were previously identified within the core sequence of the CD4 silencer (32, 38). Therefore, we investigated the involvement of Runx3 in thymopoiesis, particularly in the regulation of CD4 expression. Because, in embryos, Runx3 is expressed in cortical thymocytes and in isolated DN CD4-CD8- thymocytes, we first asked whether Runx3 was required for CD4 silencing in immature DN thymocytes. Thymocytes were stained with CD4, CD8, CD3, CD25, and CD44 mAbs and CD4 expression in different DN subsets was examined (see Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org). No consistent, quantifiable differences in CD4 expression between WT and KO were noted in any of the DN thymocytes examined (see Fig. 7, which is published as supporting information on the PNAS web site), indicating that although Runx3 is expressed in DN thymocytes, it is apparently not required for CD4 silencing at the immature DN stage.
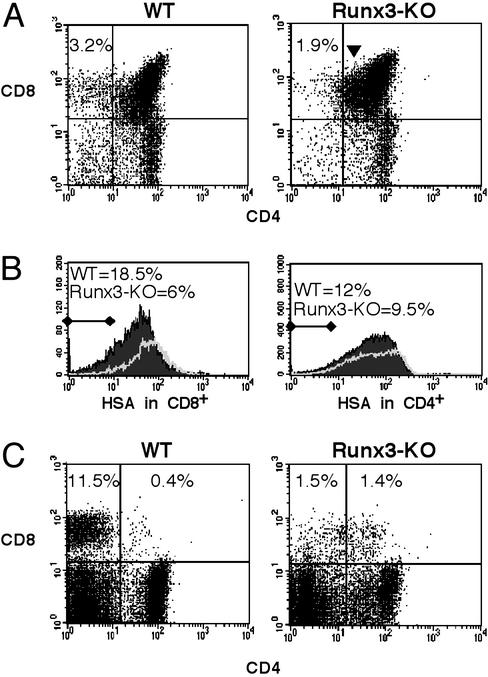
We next assessed the CD4/CD8 distribution in Runx3-KO thymus (Fig. 2A). The percentage of SP CD8+ cells in thymuses of KO mice (1.32 ± 0.59%, n = 9) was reduced compared with WT littermates (2.9 ± 0.85%, n = 9). Moreover, in KO thymus, a relatively large population of CD8+CD4lo cells was observed. These cells are intermediates in the development of SP CD8+ cells from CD8+CD4+ DP precursors, and their accumulation indicates a block in maturation of SP CD8+ thymocytes. A positive selection of DP CD4+CD8+ T cells into SP CD4+ or CD8+ functional T cells is accompanied by loss of expression of the heat-stable antigen (HSA) (39). We found that the percentage of SP CD8+HSA-/lo in the KO was 3-fold lower than in WT (Fig. 2B), indicating a delayed maturation of CD8+ cells in the KO. In contrast, SP CD4 thymocytes, exhibited comparable levels of HSA expression in KO and WT mice (Fig. 2B) suggesting that Runx3 was mainly involved in the maturation of CD8 thymocytes. The changes seen in T cell populations in Runx-KO thymus were even more prominent in peripheral T cells. The percentage of SP CD8 T cells in KO spleen was reduced ≈7-fold as compared with WT (Fig. 2C). Importantly, this reduction was associated with a significant increase in DP CD8+CD4+ cells. Of note, ≈60% (59 ± 11) of peripheral CD8+ T cells in all Runx3-KO mice analyzed (n = 10) also expressed CD4, whereas only few such cells were found in WT (Fig. 2). Most of CD4 expressing CD8+ cells in Runx3 KO had TCR and HSA levels comparable to SP CD8+ cells (see below), whereas the few residual DP cells found in WT spleen, were immature TCRloHSAhi thymic emigrants (not shown).
Fig. 2.
Impaired development of SP CD8+ T cells in Runx3-KO mice. (A) FACS analysis of CD4 and CD8 expression in thymus and spleen of WT and Runx3-KO mice. Numbers at the top of each dot plot refer to the percentage of SP CD8+ cells; KO 1.32 ± 0.59%, n = 9 versus WT 2.9 ± 0.85%, n = 9 (P = 0.00007). CD8+CD4lo cells are indicated by an arrowhead. (B) FACS analysis of HSA expression in CD8+ and CD4+ thymocytes. Thymocytes stained for CD4, CD8, CD24 (HSA), and SP CD8+ cells were gated for HSA expression profile. Numbers at the top indicate the percentage of HSA-/lo SP CD8+ cells (Left) or SP CD4+ cells (Right) in WT (filled curve) and Runx3-KO (solid line). (C) CD4/CD8 distribution among peripheral T cells. Splenocytes were stained with CD4 and CD8 mAbs and analyzed. Numbers at the top of the dot plots refer to the percentage of SP CD8+ and DP CD4+CD8+ splenocytes, respectively. The percentage of CD8+ cells in Runx3 KO mice that also expressed CD4 was similar (i.e., 59 ± 11%, n = 10).
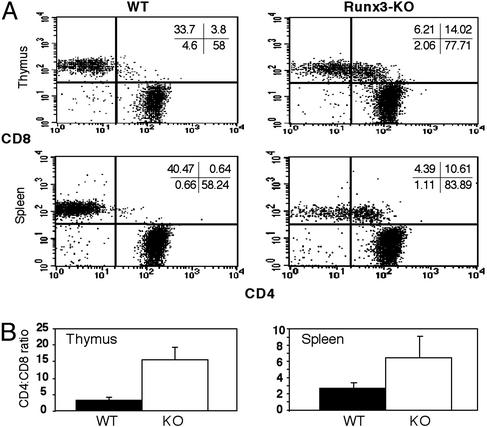
We next examined the expression of CD4 in mature CD8+ T cells. FACS analysis of TCRhighHSA-/low cells revealed that in thymus and spleen of Runx3-KO mice, most of the mature CD8+ cells also expressed CD4 (Fig. 3). In the KO thymus, this occurrence resulted in a 5-fold reduction as compared with WT in TCRhighHSA-/low SP CD8+ T cells (Fig. 3A), and the ratio of CD4/CD8 SP cells was ≈15, compared with the normal ratio of 3 seen in WT (Fig. 3B). Lack of CD4 silencing was also manifested in a pronounced (>3-fold) increase of mature DP CD4/CD8 cells in the KO thymus (Fig. 3A). Splenocytes from Runx3 KO mice also showed a marked decrease in TCRhighHSA-/low SP CD8 cells, a prominent increase in the DP compartment, and a higher CD4/CD8 ratio. Likewise, 74 ± 5.9% (n = 6) of TCRhighHSA-/low CD8 cells in KO spleen also expressed CD4 (Fig. 3B). On the other hand, the overall developmental program of the CD4 lineage in KO and WT was comparable with a small insignificant (P = 0.07) increase in SP CD4+ in KO spleen (Fig. 3).
Fig. 3.
Increased CD4/CD8 ratio in mature thymic and splenic T cells of Runx3-KO mice. Cells were stained for CD4, CD8, CD24 (HSA), and TCRαβ and analyzed. (A) CD4/CD8 dot plots of gated TCRhighHSA-/lo T cells. Runx3-KO cells exhibit a reduction in TCRhighHSA-/low SP CD8+ T cell both in thymus (approximately ×5) and spleen (approximately ×10). Numbers at the top right corner refer to percentage of cells in each of the four compartments. Note the prominent mature DP CD4+CD8+ T cell population in Runx3-KO thymus and spleen. In Runx3-KO periphery, 74 ± 5.9% of TCRhighHSA-/lo CD8+ cells also expressed CD4 (n = 6). (B) CD4/CD8 ratio in TCRhighHSA-/lo T cells is increased in thymus and spleen of Runx3-KO mice. (n = 4, P < 0.05).
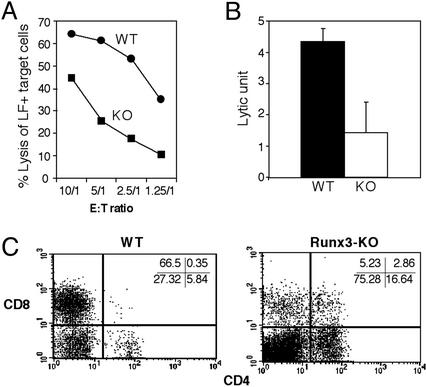
Runx3 KO PELs Display Reduced Cytolytic Activity in Vitro. The marked reduction in mature SP CD8 cells in the KO posed a question as to whether their cytolytic function (CTL activity) was affected. Alloreactive CTLs are generated in response to allogeneic normal or malignant tissues. A convenient method for generating CTLs in vivo utilizes the i.p. injection of allogeneic tumor cells. PELs collected shortly after rejection provide a good source of highly potent, specific CTLs. PELs effectively bind to and lyse target tumor cells in vitro, as determined by the conjugation and 51Cr-release assays, respectively (40). Runx3-KO mice and WT littermates were primed by a s.c. injection of allogeneic LF+ tumor cells, followed by an i.p. injection of ≈25 × 106 tumor cells 11 days later. Four to 5 days after last injection of tumor cells, PELs were collected and incubated in vitro with 51Cr-labeled LF+cells at various effector-to-target ratios and the amount of radioactivity released (CTL activity) was determined. Compared with WT littermates, Runx3-KO PELs exhibited a 3-fold reduction in the calculated lytic units at all four effector-to-target ratios (Fig. 4 A and B). CD4/CD8 FACS analysis of PELs from Runx3-KO and WT mice revealed a >10-fold reduction in the number of CD8+ CTLs in the peritoneal cavity of Runx3-KO mice (Fig. 4C), which was consistent with the reduced CTL activity detected. Nevertheless, this greatly reduced population of CD8+ CTL effector cells was still capable of rejecting both primary (s.c.) and secondary (i.p.) tumor injections in 73% of challenged Runx3-KO mice (n = 11).
Fig. 4.
In vitro cytotoxic activity of WT and Runx3-KO peritoneal CTLs. Primed mice were injected with 25 × 106 LF+ allogeneic tumor cells, and 4–5 days later PELs were retrieved and analyzed for CTL activity. (A) Percent of target LF+ cell lysis as a function of E to T ratio (E, effector PELs; T, target LF+ tumor cells). The data of one of three experiments with similar results are presented. (B) An ≈3-fold reduction in lytic units of Runx3-KO CTLs as compared with WT CTLs. A lytic unit is defined as the number of effector cells required to lyse 35% of target cells under specified assay conditions. The results are based on four WT and four Runx3-KO mice in three independent experiments. P = 0.005, based on double-sided Student's t test. (C) CD4/CD8 distribution in peritoneal lymphocytes. Peritoneal cells were retrieved 5 days after i.p. injection of 25 × 106 LF+ tumor cells, incubated for 1 h at 37°C to deplete most macrophages and B cells, and subjected to flow cytometry by using CD4 and CD8 mAbs.
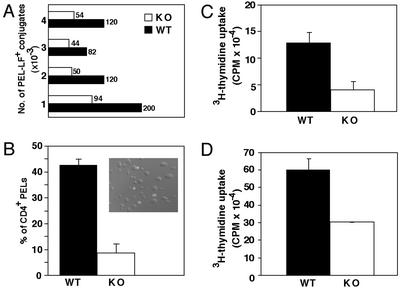
Expression of CD4 in Runx3-KO CTLs Does Not Preclude Conjugation with Target Cells. The ability of Runx3 KO mice to effectively reject injected tumors, despite marked reduction in SP CD8 cells, raised the possibility that in vivo CD4 expressing KO CTLs participate in tumor rejection. To examine the ability of DP CD4/CD8 CTLs to conjugate with target tumor cells, we stained an equal number of PELs and target tumor cells with anti-CD4 Ab and the number of CD4-positive conjugates monitored (Fig. 5A). Runx3 KO PELs formed ≈50% fewer conjugates with LF+ tumor cells compared with WT PELs (Fig. 5A). More than 40% of Runx3-KO PEL-LF+ conjugates stained positively for CD4 compared with only 8% of WT (Fig. 5B). The results indicated that coexpression of CD4 in the CD8+ alloreactive CTLs did not interfere with their conjugating ability, and can explain the retained cytolytic activity in Runx3 KO, despite marked reduction in SP CD8+ cells.
Fig. 5.
Conjugation to tumor cells and proliferation capabilities of WT and KO CD8+ T cells. (A and B) Fewer Runx3 KO PEL-LF+ conjugates are formed in vitro and many are CD4+.(A) Equal numbers of LF+ tumor cells and PELs were incubated for 5 min at 25°C and the number of PEL-LF+ conjugates counted per 1 × 106 cells was determined. Each combined (WT/Runx3-KO) histogram (experiments 1–4) represents a separate experiment. (In experiment 1, cells were incubated for 7 min.) (B) Percentage of CD4 expressing CTLs counted after immunofluorescent staining. Nine fields in each experiment were counted for CD4+ and CD4-PEL-LF+ conjugates (n = 3). (Inset) A confocal image depicting CD4 expressing CTLs conjugated to tumor cells. (C and D) Peripheral Runx3-KO CD8+ T cells are less proliferative than WT. CD8+ splenocytes were purified and subjected to an in vitro proliferation assay. Runx3-KO SP CD8+ splenocytes exhibit a 2- to 3-fold reduction in proliferation after stimulation with CD3/CD28 mAbs (C) or with CD3 mAb + IL-2 (D).
Proliferation Ability of Runx3 KO SP CD8 Cells Is Reduced Compared with WT. Naive CD8 T cells that encounter peptide:MHC class I complexes on the surface of antigen-presenting cells such as dendritic cells, are induced to produce IL-2, proliferate, and eventually differentiate into cytotoxic CD8 T cells. Because 10-fold fewer CD8+ effector cells were recruited to the peritoneal cavity of Runx3-KO, compared with WT mice after injection of allogeneic tumor (Fig. 4C), we asked whether the proliferative capacity of the KO CD8+ cells was compromised. Splenic SP CD8+ cells were isolated and their proliferation assayed by using either plate-bound anti-CD3 mAb and soluble anti-CD28 mAb (Fig. 5C), or anti-CD3 mAb and IL-2 (Fig. 5D) as stimulating reagents. The results show a 2- to 3-fold reduction in 3H-thymidine incorporation into KO SP CD8+ cells compared with WT (Fig. 5 C and D), indicating that peripheral KO CD8+ T cells have a reduced proliferative capacity. Proliferation of CD8+ cells was also assessed by 5- (and 6-)carboxyfluorescein diacetate, succinimidyl ester (CFSE) labeling. After 3 days in culture, ≈60% of KO CD8+ splenocytes have not yet divided, compared with only 17% of such cells in WT (see Fig. 8, which is published as supporting information on the PNAS web site). The reduced proliferative capacity of KO CD8+ cells may underlie the significant reduction in the number of SP CD8+ CTLs recruited to the peritoneal cavity during tumor rejection.
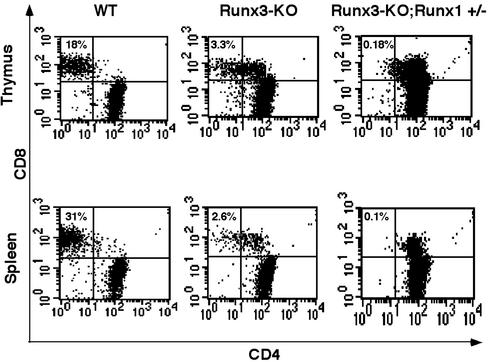
Haploinsufficiency of Runx1 in Compound Mutant Runx3-/-;Runx1+/- Mice Exacerbates the Developmental Defect of SP CD8 Cells. As indicated earlier, Runx1 is highly expressed in the thymus, mainly in the cortex, of both embryos and adults (Fig. 1). Runx1 KO mice lack definitive hematopoiesis and die at E12.5 (33), which precludes the study of Runx1 function in thymopoiesis. To investigate the combined activity of Runx1 and Runx3 in T cell development, we generated a compound mutant strain deficient in Runx3 and heterozygous for Runx1. Heterozygotes Runx3+/- and Runx1+/- mice were crossed and compound mutant Runx3-KO;Runx1+/- (compound Runx3/Runx1) mice were analyzed and compared with Runx3 KO mice. Compound Runx3/Runx1 mice lack Runx3 and have a markedly reduced Runx1 (not shown). Specifically, these mice are smaller, have a high mortality rate, and most die within 2 weeks. FACS analysis revealed that all CD8+ T cells in compound Runx3/Runx1 mice also expressed CD4, resulting in a complete loss of mature TCRhighHSA-/low SP CD8 cells in both thymus and spleen (Fig. 6). Additionally, most of the remaining SP CD8+ thymocytes of compound Runx3/Runx1 mice were TCR- (data not shown). No significant effect of Runx1 haploinsufficiency was evident on either maturation or proliferation of CD4 cells, including on expression of TCRαβ genes. The data indicate that in addition to Runx3, Runx1 as well is involved in development of mature CD8 cells through repression of CD4 expression.
Fig. 6.
Reduced expression of Runx1 in Runx3 KO further impairs differentiation of CD8+ T cells. Thymocytes and splenocytes of compound mutant Runx3/Runx1, Runx3-KO, and WT were subjected to flow cytometry by using CD4, CD8, CD24 (HSA), and TCRαβ markers. CD4/CD8 dot plots of gated TCRhighHSA-/lo T cells are shown. Numbers at the top of each dot plot refer to the percentage of SP CD8 T cells. Note the complete absence of TCRhighHSA-/lo SP CD8 cells in spleens of compound mutant Runx3/Runx1.
Discussion
Expression of Runx1 and Runx3 is spatially and/or temporally separated during mouse embryogenesis, even in tissues where both genes are expressed (20). Here we have shown that in embryonal and adult thymus Runx1 is mainly expressed in the cortex, whereas Runx3 is mainly found in the medulla and its expression is confined to SP CD8+ cells. An intronic region of 430 bp, known as the CD4 silencer, negatively regulates the expression of CD4 during T cell development (41–43). The silencer contains two oppositely oriented Runx-binding sites, as well as binding sites for several other transcription factors, including cMyb and Ets, which are known to cooperate with RUNX in transcription regulation (32, 38, 44). The in vivo functionality of these RUNX-binding sites was recently demonstrated through transfection experiments and gene targeting (44). It was previously reported that Runx1 and Runx3 are able to act as transcriptional repressors through recruitment of the corepressor Gro/TLE (17). Because Runx3 is specifically expressed in CD4-CD8+ T cells and absent in CD4 expressing cells, it was interesting to determine whether Runx3 participates in transcriptional repression of the CD4 gene during thymopoiesis.
Expression of Runx3 in medullary SP CD8+ T cells, but not in medullary SP CD4+ or cortically DP CD4+CD8+ T cells, suggested a role for Runx3 in maturation of CD8+ T cells. It also indicated that Runx3 does not play a role in positive selection or in SP CD4+ T cell maturation. Our data show a delayed maturation of CD8+ in Runx3 KO mice, reflected in reduced numbers of CD8+HSA-/low T cells in thymus and a marked decrease in mature TCRhighHSA-/low thymic and peripheral CD8+ cells. Significantly, the reduction in peripheral CD8+ was accompanied by an increase in CD4 expressing mature (TCRhighHSA-/low) CD8+ cells, so that in the KO, ≈60% of peripheral CD8+ were also CD4 positive. These data indicate that Runx3 functions as a negative regulator of CD4 expression and when absent, transcriptional repression of CD4 is abrogated resulting in CD4-expressing mature CD8 cells.
Using the Runx3 KO strain generated by Li et al. (27), Taniuchi et al. (32) have recently reported that Runx3 was required for lineage-specific CD4 silencing. To circumvent the neonatal lethality of this Runx3 KO strain, they used Rag2-/- mice, reconstituted with Runx3 KO fetal liver cells. Under these conditions, CD4 was derepressed in a variegated manner and the degree of derepression varied between animals. Notably, in 10% of the reconstituted mice, all peripheral CD8+ T lymphocytes expressed CD4. Conversely, there was only a small variation in CD4 derepression among 10 Runx3 KO mice examined in our study and ≈60% of their peripheral CD8+ T cells also expressed CD4. Additionally, in Rag-/- mice reconstituted with Runx3 KO fetal liver cells, only a small nonsignificant change in the CD4/CD8 ratio among mature thymocytes was observed (32), whereas in Runx3 KO, a marked increase in this ratio was observed (from 3 in WT to 15 in KO). These differences could be attributed to the Rag2-/- reconstituted system versus adult Runx3 KO mice, or to the different targeting strategy used for generating the two Runx3 KO strains (25, 27).
Li et al. (27) targeted Runx3 by introducing a LacZ-neo cassette in-frame at the carboxyl end of the runt domain (RD), creating a RD-LacZ fusion protein. Given the fact that the carboxyl terminus of the RD is important for DNA binding (45), the fused RD-LacZ protein is expected to bind DNA very poorly, if at all. However, because this fused product retains most of the RD, it could still bind the Runx partner protein CBFβ (46), and thereby exert a dominant-negative effect (47). A potential target for such a negative effect is the family member Runx1, which, as reported by Taniuchi et al. (32) and shown here, cooperates with Runx3 in regulating CD4/CD8 linage-specific decisions during thymopoiesis. Of note, as shown here, haploinsufficiency of Runx1 in Runx3 KO mice resulted in a complete loss of mature TCRhighHSA-/low CD8 cells in both thymus and spleen.
Thymic organogenesis involves signals derived from hematopoietic cells to induce proper differentiation of the epithelial compartment (48). Lymphoid organs of Rag-/- mice are small and hypoplastic (49), and blocking thymic differentiation at the CD3-CD44-CD25+ (DN3) stage, disrupts cortical structure and give rise to a small medulla (50, 51) Early developing triple-negative thymocytes provide signals to thymic cortical epithelial cells, which allow further progression in thymopoiesis to DP, TCRαβ-expressing thymocytes. These thymocytes in turn, crosstalk to medullary epithelial cells, leading to the generation of correct microenvironments in the medulla, thereby enabling final steps in the maturation of SP T cells (48, 51). It is possible that when transplanted into adult-irradiated Rag2-/- mice (32), Runx3-KO fetal liver cells are unable to provide sufficient cues to allow a completely normal development of thymic microenvironment.
As noted above, a substantial and relatively uniform decrease in peripheral SP CD8+ T cells was observed in Runx3-KO mice. Nevertheless, both SP CD8+ and DP CD4+CD8+ peripheral T cells in the KO expressed high levels of TCR, indicating they have undergone positive selection and matured. After in vivo tumor challenge, significantly fewer CD8+ CTLs were recruited to the peritoneal cavity of Runx3-KO mice. However, these CTLs could form conjugates with tumor cells in vitro, indicating that Runx3 null CTLs were capable of performing the first step in target cell lysis. Furthermore, most Runx3-KO mice challenged with tumor in vivo rejected the allogeneic tumor and survived. Together our data show that Runx3 is not required for CTL function per se, but rather for clonal expansion of CD8+ CTLs after tumor injection. These findings are consistent with previous reports demonstrating that CD4 expressing CD8+ CTLs retain their cytotoxic activity against allogeneic target cells (43), and that reconstituted Rag2-/- Runx3 KO CTLs exhibit normal perforin induction and are fully functional in a redirected CTL-mediated lysis assay (32).
An in vitro proliferation assay of peripheral CD8+ cells revealed that Runx3 KO cells respond poorly to mitogenic stimulation. Similar data were obtained by Taniuchi et al. (32) for CD8+ T cells derived from Runx3 KO reconstituted Rag2-/- mice. This reduced proliferative capacity may explain the significantly reduced numbers of CTLs recruited to the peritoneal cavity of our KO mice on tumor injection. This conclusion is supported by the CFSE proliferation assay (see Supporting Text).
Runx1 is highly expressed in immature cortical thymocytes and much less in the thymic medulla, the site of SP T cell maturation. Runx1 KO is embryonic lethal (24), therefore we studied its role in thymopoiesis, using the compound mutant Runx3-/-;Runx1+/- mice. Total number of thymocytes and splenocytes in compound Runx3/Runx1 was markedly reduced compared with Runx3-KO littermates. Diminished activity of Runx1, in addition to null Runx3, resulted in a complete loss of CD4 repression in mature CD8+ peripheral T cells so that all CD8+ cells also expressed CD4. In thymus of compound mutant mice, most SP CD8+ were TCR-, and in spleen a complete obliteration of mature SP CD8+ T cell occurred. The data indicate that Runx1 cooperates with Runx3 in regulating the development of CD8 T cells through transcriptional repression of CD4. It would be interesting to determine whether Runx1 activity is confined to the DN stage (32) or whether it also contributes to the repression of CD4 in mature CD8 cells.
The data provide information about the role of Runx3 and Runx1 in thymopoiesis and shed light on their activity as transcriptional repressors. It would be interesting to further evaluate the status of immune function in Runx3 KO mice, as defect in T cell lineages decisions during thymopoiesis might have far reaching consequences on the physiology of Runx3 mutant mice.
Supplementary Material
Acknowledgments
We thank Judith Chermesh, Rafi Saka, Shoshana Grossfeled, and Ethi Yael for expert assistance; Ayala Sharp and Eitan Ariel for help in FACS analysis; Nancy Speck for the Runx1 mutant mice; and Zelig Eshhar and Amiela Globerson for discussions. This work was supported by grants from the Commission of the European Union, the Israel Science Foundation, and the Shapell Family Biomedical Research Foundation at the Weizmann Institute.
Abbreviations: CTL, cytolytic T lymphocyte; HAS, heat-stable antigen; PEL, peritoneal exudate lymphocyte; KO, knockout; DP, double positive; DN, double negative; SP, single positive; LF, Fas (CD95)-expressing, leukemia L1210 target cells; TCR, T cell receptor; RD, runt domain.
References
- 1.Levanon, D., Negreanu, V., Bernstein, Y., Bar-Am, I., Avivi, L. & Groner, Y. (1994) Genomics 23, 425-432. [DOI] [PubMed] [Google Scholar]
- 2.Avraham, K. B., Levanon, D., Negreanu, V., Bernstein, Y., Groner, Y., Copeland, N. G. & Jenkins, N. A. (1995) Genomics 25, 603-605. [DOI] [PubMed] [Google Scholar]
- 3.Bae, S. C., Takahashi, E., Zhang, Y. W., Ogawa, E., Shigesada, K., Namba, Y., Satake, M. & Ito, Y. (1995) Gene 159, 245-248. [DOI] [PubMed] [Google Scholar]
- 4.Calabi, F., Rhodes, M., Williamson, P. & Boyd, Y. (1995) Genomics 26, 607-610. [DOI] [PubMed] [Google Scholar]
- 5.Tracey, W. D. & Speck, N. A. (2000) Semin. Cell. Dev. Biol. 11, 337-342. [DOI] [PubMed] [Google Scholar]
- 6.Karsenty, G. (2000) Semin. Cell. Dev. Biol. 11, 343-346. [DOI] [PubMed] [Google Scholar]
- 7.Ghozi, M. C., Bernstein, Y., Negreanu, V., Levanon, D. & Groner, Y. (1996) Proc. Natl. Acad. Sci. USA 93, 1935-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geoffroy, V., Corral, D. A., Zhou, L., Lee, B. & Karsenty, G. (1998) Mamm. Genome 9, 54-57. [DOI] [PubMed] [Google Scholar]
- 9.Pozner, A., Goldenberg, D., Negreanu, V., Le, S.-Y., Elroy-Stein, O., Levanon, D. & Groner, Y. (2000) Mol. Cell. Biol. 20, 2297-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bangsow, C., Rubins, N., Glusman, G., Bernstein, Y., Negreanu, V., Goldenberg, D., Lotem, J., Ben-Asher, E., Lancet, D., Levanon, D. & Groner, Y. (2001) Gene 279, 221-232. [DOI] [PubMed] [Google Scholar]
- 11.Rini, D. & Calabi, F. (2001) Gene 273, 13-22. [DOI] [PubMed] [Google Scholar]
- 12.Xiao, Z. S., Liu, S. G., Hinson, T. K. & Quarles, L. D. (2001) J. Cell Biochem. 82, 647-659. [DOI] [PubMed] [Google Scholar]
- 13.Stewart, M., MacKay, N., Cameron, E. R. & Neil, J. C. (2002) J. Virol. 76, 4364-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levanon, D., Glusman, G., Bangsow, T., Ben-Asher, E., Male, D. A., Avidan, N., Bangsow, C., Hattori, M., Taylor, T. D., Taudien, S., et al. (2001) Gene 262, 23-33. [DOI] [PubMed] [Google Scholar]
- 15.Ito, Y. (1999) Genes Cells 4, 685-696. [DOI] [PubMed] [Google Scholar]
- 16.Ito, Y. & Bae, S. C. (1997) in Oncogenes as Transcriptional Regulators, eds. Yaniv, M. & Ghysdael, J. (Birkhauser, Basel), Vol. 2, pp. 107-132. [Google Scholar]
- 17.Levanon, D., Goldstein, R. E., Bernstein, Y., Tang, H., Goldenberg, D., Stifani, S., Paroush, Z. & Groner, Y. (1998) Proc. Natl. Acad. Sci. USA 95, 11590-11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruhn, L., Munnerlyn, A. & Grosschedl, R. (1997) Genes Dev. 11, 640-653. [DOI] [PubMed] [Google Scholar]
- 19.North, T., Gu, T.-L., Stacy, T., Wang, Q., Howard, L., Binder, M., Marin-Padilla, M. & Speck, N. A. (1999) Development (Cambridge, U.K.) 126, 2563-2575. [DOI] [PubMed] [Google Scholar]
- 20.Levanon, D., Brenner, O., Negreanu, V., Bettoun, D., Woolf, E., Eilam, R., Lotem, J., Gat, U., Otto, F., Speck, N. & Groner, Y. (2001) Mech. Dev. 109, 413-417. [DOI] [PubMed] [Google Scholar]
- 21.Simeone, A., Daga, A. & Calabi, F. (1995) Dev. Dyn. 203, 61-70. [DOI] [PubMed] [Google Scholar]
- 22.Stricker, S., Fundele, R., Vortkamp, A. & Mundlos, S. (2002) Dev. Biol. 245, 95-108. [DOI] [PubMed] [Google Scholar]
- 23.Chen, S., Gu, T. T., Sreenath, T., Kulkarni, A. B., Karsenty, G. & MacDougall, M. (2002) Connect. Tissue Res. 43, 338-344. [DOI] [PubMed] [Google Scholar]
- 24.Speck, N. A. (2001) Curr. Opin. Hematol. 8, 192-196. [DOI] [PubMed] [Google Scholar]
- 25.Levanon, D., Bettoun, D., Harris-Cerruti, C., Woolf, E., Negreanu, V., Eilam, R., Bernstein, Y., Goldenberg, D., Xiao, C., Fliegauf, M., et al. (2002) EMBO J. 21, 3454-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inoue, K., Ozaki, S., Shiga, T., Ito, K., Masuda, T., Okado, N., Iseda, T., Kawaguchi, S., Ogawa, M., Bae, S. C., et al. (2002) Nat. Neurosci. 5, 946-954. [DOI] [PubMed] [Google Scholar]
- 27.Li, Q. L., Ito, K., Sakakura, C., Fukamachi, H., Inoue, K., Chi, X. Z., Lee, K. Y., Nomura, S., Lee, C. W., Han, S. B., et al. (2002) Cell 109, 113-124. [DOI] [PubMed] [Google Scholar]
- 28.Levanon, D., Bernstein, Y., Negreanu, V., Ghozi, M. C., Bar-Am, I., Aloya, R., Goldenberg, D., Lotem, J. & Groner, Y. (1996) DNA Cell Biol. 15, 175-185. [DOI] [PubMed] [Google Scholar]
- 29.Meyers, S., Lenny, N., Sun, W.-H. & Hiebert, S. W. (1996) Oncogene 13, 303-312. [PubMed] [Google Scholar]
- 30.Shi, M. J. & Stavnezer, J. (1998) J. Immunol. 161, 6751-6760. [PubMed] [Google Scholar]
- 31.Le, X., Groner, Y., Kornblau, S., Gu, Y., Hittelman, W., Levanon, D., Mehta, K., Arlinghaus, R. & Chang, K. (1999) J. Biol. Chem. 274, 21651-21658. [DOI] [PubMed] [Google Scholar]
- 32.Taniuchi, I., Osato, M., Egawa, T., Sunshine, M. J., Bae, S. C., Komori, T., Ito, Y. & Littman, D. R. (2002) Cell 111, 621-633. [DOI] [PubMed] [Google Scholar]
- 33.Wang, Q., Stacy, T., Binder, M., Marin-Padilla, M., Sharpe, A. H. & Speck, N. A. (1996) Proc. Natl. Acad. Sci. USA 93, 3444-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berke, G. & Fishelson, Z. V. (1975) J. Exp. Med. 142, 1011-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berke, G. & Gabison, D. (1975) Eur. J. Immunol. 5, 671-675. [DOI] [PubMed] [Google Scholar]
- 36.Rosen, D., Li, J. H., Keidar, S., Markon, I., Orda, R. & Berke, G. (2000) J. Immunol. 164, 3229-3235. [DOI] [PubMed] [Google Scholar]
- 37.Manley, N. R. (2000) Semin. Immunol. 12, 421-428. [DOI] [PubMed] [Google Scholar]
- 38.Duncan, D. D., Adlam, M. & Siu, G. (1996) Immunity 4, 301-311. [DOI] [PubMed] [Google Scholar]
- 39.Hough, M. R., Takei, F., Humphries, R. K. & Kay, R. (1994) J. Exp. Med. 179, 177-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berke, G., Rosen, D. & Ronen, D. (1993) Immunology 78, 105-112. [PMC free article] [PubMed] [Google Scholar]
- 41.Sawada, S., Scarborough, J. D., Killeen, N. & Littman, D. R. (1994) Cell 77, 917-929. [DOI] [PubMed] [Google Scholar]
- 42.Siu, G., Wurster, A. L., Duncan, D. D., Soliman, T. M. & Hedrick, S. M. (1994) EMBO J. 13, 3570-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zou, Y. R., Sunshine, M. J., Taniuchi, I., Hatam, F., Killeen, N. & Littman, D. R. (2001) Nat. Genet. 29, 332-336. [DOI] [PubMed] [Google Scholar]
- 44.Taniuchi, I., Sunshine, M. J., Festenstein, R. & Littman, D. R. (2002) Mol. Cell 10, 1083-1096. [DOI] [PubMed] [Google Scholar]
- 45.Bartfeld, D., Shimon, L., Couture, G. C., Rabinovich, D., Frolow, F., Levanon, D., Groner, Y. & Shakked, Z. (2002) Structure (London) 10, 1395-1407. [DOI] [PubMed] [Google Scholar]
- 46.Bravo, J., Li, Z., Speck, N. A. & Warren, A. J. (2001) Nat. Struct. Biol. 8, 371-378. [DOI] [PubMed] [Google Scholar]
- 47.Michaud, J., Wu, F., Osato, M., Cottles, G. M., Yanagida, M., Asou, N., Shigesada, K., Ito, Y., Benson, K. F., Raskind, W. H., et al. (2002) Blood 99, 1364-1372. [DOI] [PubMed] [Google Scholar]
- 48.Klug, D. B., Carter, C., Gimenez-Conti, I. B. & Richie, E. R. (2002) J. Immunol. 169, 2842-2845. [DOI] [PubMed] [Google Scholar]
- 49.Naquet, P., Naspetti, M. & Boyd, R. (1999) Semin. Immunol. 11, 47-55. [DOI] [PubMed] [Google Scholar]
- 50.Shinkai, Y., Rathbun, G., Lam, K. P., Oltz, E. M., Stewart, V., Mendelsohn, M., Charron, J., Datta, M., Young, F., Stall, A. M., et al. (1992) Cell 68, 855-867. [DOI] [PubMed] [Google Scholar]
- 51.van Ewijk, W., Hollander, G., Terhorst, C. & Wang, B. (2000) Development (Cambridge, U.K.) 127, 1583-1591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.