Abstract
According to the present view, metastasis marks the end in a sequence of genomic changes underlying the progression of an epithelial cell to a lethal cancer. Here, we aimed to find out at what stage of tumor development transformed cells leave the primary tumor and whether a defined genotype corresponds to metastatic disease. To this end, we isolated single disseminated cancer cells from bone marrow of breast cancer patients and performed single-cell comparative genomic hybridization. We analyzed disseminated tumor cells from patients after curative resection of the primary tumor (stage M0), as presumptive progenitors of manifest metastasis, and from patients with manifest metastasis (stage M1). Their genomic data were compared with those from microdissected areas of matched primary tumors. Disseminated cells from M0-stage patients displayed significantly fewer chromosomal aberrations than primary tumors or cells from M1-stage patients (P < 0.008 and P < 0.0001, respectively), and their aberrations appeared to be randomly generated. In contrast, primary tumors and M1 cells harbored different and characteristic chromosomal imbalances. Moreover, applying machine-learning methods for the classification of the genotypes, we could correctly identify the presence or absence of metastatic disease in a patient on the basis of a single-cell genome. We suggest that in breast cancer, tumor cells may disseminate in a far less progressed genomic state than previously thought, and that they acquire genomic aberrations typical of metastatic cells thereafter. Thus, our data challenge the widely held view that the precursors of metastasis are derived from the most advanced clone within the primary tumor.
The prevailing paradigm of carcinogenesis suggests that epithelial cells sequentially accumulate multiple genetic and epigenetic changes underlying the disorganization of tissue morphology and uncontrolled growth (1). The model predicts that certain additional genomic events initiate invasiveness of the tumor and subsequently progression to metastasis. However, this hypothesis of an “additional event” initiating metastasis relatively late in tumorigenesis was recently challenged by another model based on global gene expression analysis of primary breast cancers in which groups of genes were identified that predicted the development of distant metastasis (2). Thus it was concluded that the proclivity to metastasize is acquired early during multistep tumorigenesis, although it manifests only much later after mutation of other genes (3).
A closer analysis of the natural course of breast cancer reveals several inconsistencies with both models. Why, after curative resection of a primary tumor and its regional lymph nodes, does it often take years or decades until clinical metastases appear? It is difficult to understand how a cell that has been selected at the primary site for acquisition of self-sufficiency in growth signals, unlimited proliferative potential, sustained angiogenesis, and unresponsiveness to growth inhibitory signals (4) can suppress these activities and remain dormant for years. Furthermore, within the spectrum of clinical courses, there is a very distinct scenario of cancer of unknown primary (CUP). This syndrome comprises up to 7% of hospitalized cancer patients who are first diagnosed with distant metastases, whereas their primary tumor remains unrecognized (5, 6). CUP therefore demonstrates that a large tumor extent apparently is not an absolute requirement for a systemic spreading to occur. The inconsistencies among the various models (7–9) and these few examples emphasize the need for studying the development of systemic cancer from its very beginning to a fully expressed manifestation.
It is self-evident that the progenitors of the later-arising metastases must be present among those cells that have disseminated to distant sites before the surgeon resected the primary tumor. In breast cancer, the probability of detecting such progenitors is highest in bone marrow, because the skeleton is the preferred site of metastasis. In this strictly mesenchymal organ, single tumor cells can be detected in 20–60% of carcinoma patients without manifest metastasis by epithelial-specific cytokeratin (CK) antibodies, while being virtually absent in large cohorts of donors without epithelial malignancy (10, 11). Although CK is not a marker of malignancy, a large body of follow-up studies demonstrates that the finding of CK-positive cells in bone marrow has a strong prognostic impact on relapse-free as well as overall survival in breast cancer and many other types of cancer (10, 11). Moreover, in breast cancer, their presence is a strong prognosticator specifically for the development of skeleton metastasis (10, 12). Their extreme rareness (10-5 to 10-6) has so far precluded their detailed characterization. Double-staining techniques, however, have provided unequivocal evidence for their epithelial origin, such as display of histogenetic markers like prostate-specific antigen in the case of prostate cancer (13), lack of expression of the common leukocyte antigen CD45, and down-regulation of MHC class I molecules (14), whereas their malignant nature was established by tumor-specific chromosomal abnormalities visualized by fluorescence in situ hybridization (15).
For a deeper understanding of the evolution of systemic cancer, we performed a comprehensive genomic analysis of bone marrow-derived CK-positive cells by single-cell comparative genomic hybridization (CGH) (16). The cells were isolated from breast cancer patients presenting with or without manifest metastasis and compared with their matched primary tumor. Therefore, as a link between the primary tumor and the later-arising metastases, they should provide interesting new information on a critical stage of cancer progression.
Methods
Tumor Cell Detection and Sample Preparation. Bone marrow was aspirated from 386 unselected breast cancer patients after informed consent was obtained. The status of the patients' disease was assessed postoperatively or at the time of bone marrow aspiration, and all patients were staged according to the standards of the International Union Against Cancer (UICC). Of the patients who had no evidence of metastasis, 44% of the samples were drawn before surgery, 39% within the next month, and 17% 1 mo to 10 yr after surgery of the primary tumors. The time points of sampling had no influence on the classification result by the applied algorithm. Preparation of bone marrow suspensions was performed as published (10). Staining, tumor cell isolation, and comparative genomic hybridization were performed by applying the alkaline phosphatase antialkaline phosphatase technique with the CK antibodies CK2 (Chemicon) and A45-B/B3 (Micromet, Munich), as published in ref. 17, and the CK antibody A45-B/B3 Fab fragment conjugated to alkaline-phosphatase (Micromet). Microdissection of primary tumors and tissue preparation were done as published (18), and PCR and CGH of the isolated DNA were identically performed as for the single cells.
Loss of Heterozygosity (LOH) Analysis for RB1CC1 and the Cadherin Locus on 16q22. LOH analysis on the primary PCR products from microdissected tumors and single cells was performed as described (16). Microsatellite markers for RB1CC1 were as published by Chano et al. (19). Primer sequences for the microsatellite markers on chromosome 16 were: D16S3095, 5′-TATAGTTTGTGTCCCCCGAC and 5′-TCAGTTGGAAGATGAGTTGG-3′; D16S485 5′-AGGCAATTTGTTACAGAGCC-3′ and 5′-AGTAATAATGTACCTGGTACC-3′; D16S511, 5′-CAGAGGCCTCATTTTCTAACC-3′ and 5′-TGCTACATAAAGAGGGAGGC-3′; and D16S505, 5′-GGCCCTAAATCCAGTGCTG-3′ and 5′-CTGCCTCCATACGTGGAGG-3′. Polyacrylamide gels were stained by SYBR-gold (Molecular Probes) and scanned by using a FluorImager (Molecular Dynamics).
Bioinformatic Evaluation. The ranking was achieved by a forward selection procedure based on mutual information between feature and target (20, 21). Briefly, we selected 46 genomic loci as features, mostly comprising chromosome arms, unless they were more clearly defined on the basis of the aberrations shown in Fig. 2B. Second, we defined the presence of clinically evident metastasis as target and ranked all 46 genomic loci according to the amount of information provided by their status (gain, loss, or unchanged) for the identification of a cell that was isolated from a M1 patient (for details, see Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org).
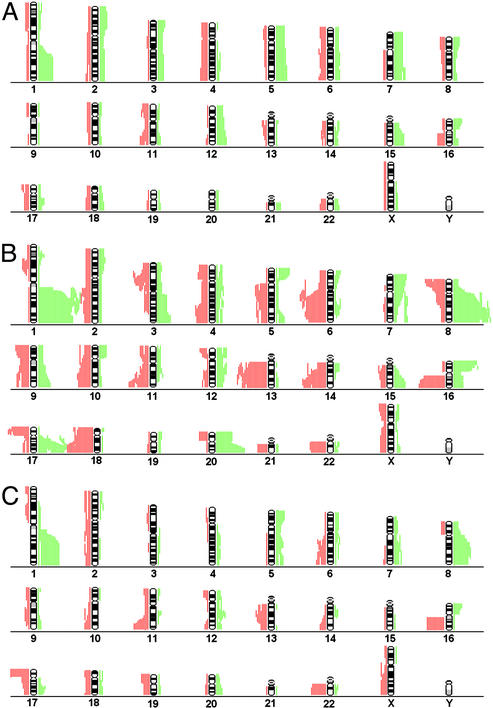
Fig. 2.
Chromosomal aberrations of disseminated tumor cells and primary tumors. Cumulative illustration of CGH aberrations for each chromosome of 40 disseminated tumor cells from 30 patients with minimal residual disease (A) and 47 single cells from 23 patients with clinical metastasis (B). The extension of each green or red bar depicts the chromosomal amplification or loss of an individual cell (or primary tumor), respectively. To depict the diversity of karyotypes retrieved from disseminated tumor cells and to avoid a bias in favor of patients with multiple cells, sister cells with identical CGH-profiles were excluded. (C) Chrormosomal aberrations of 27 matched primary tumors.
Several classifiers were trained to select the subset of loci that predict metastatic disease with highest accuracy. As classifiers, we used simple multivariate logistic functions. A final subset of features was selected that corresponds to the determined model with maximal model evidence. Using the selected set of features, we built a classifier that assigns each cell its probability of being isolated from an M1 patient. Several classifiers with different numbers of input features were trained. The weight vector w of the classifier is determined by using the Bayesian evidence approach (21, 22) to minimize the cross-entropy error function. To select the final subset of features, the model evidence P (D | M) is computed within the Bayesian framework for each classifier, where M consists of the parameters describing the model.
The classifier was 9-fold crossvalidated by splitting the data in nine equal parts and using each part once for testing and the remaining eight parts for training. Generalization performance was averaged over these nine crossvalidation sets. Furthermore, leave-one-out errors were determined by using one cell for testing and the remaining cells for training. The overall error was then averaged over all single experiments. Receiver operating characteristic curves (23) confirmed the model selection process (Fig. 7, which is published as supporting information on the PNAS web site). The Kolmogorov–Smirnov (24) test was used to test for significant differences between the predictive power of models.
A classifier based on clinical variables of the primary tumor (tumor size, lymph node status, tumor grade, or oestrogen/ progesteron/her2 receptor status) was built and tested as described for the genomic loci of the isolated tumor cells. In this comparison, a patient or cell was predicted as M1 when the predicted probability was >0.5.
Results
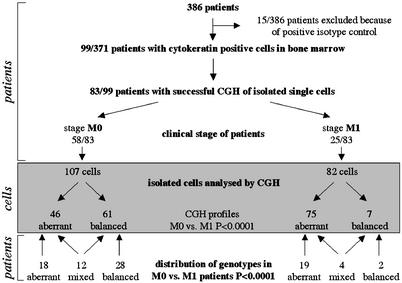
Detection and Isolation of Single Disseminated Tumor Cells. We screened the bone marrow aspirates of altogether 386 breast cancer patients, of whom 15 patients had to be excluded because positive cells were found that stained with an antibody directed against an irrelevant antigen, the so-called isotype control. Ninety-nine of the remaining patients had CK-positive cells in their bone marrow aspirates (26.7%). After isolation from adhesion slides, single-cell CGH (16, 17) was successfully performed in 83 of 99 patients (83.8%). Of the 83 patients, 58 presented without evidence of metastatic disease (UICC stage M0), whereas 25 suffered from manifest metastasis (UICC stage M1; Fig. 1).
Fig. 1.
Flow diagram of sample preparation and analysis. Bone marrow samples of 386 breast cancer patients were screened for CK-positive cells. Stained cells were isolated and individually analyzed by CGH. The genomic characterization of cells from patients with (M1) and without (M0) manifest metastasis revealed significant differences between the two groups.
Genotypes of Cells Isolated from M0 and M1 Patients. In total, 189 cells could be further analyzed by CGH, of which 107 were derived from patients in stage M0 and 82 from patients in stage M1 (Fig. 1). The number of cells with and without chromosomal aberrations is given in Fig. 1. We concentrated first on 121 cells with definite pathologic CGH abnormalities (Fig. 1). The comparison of such cells from the two clinical groups (M0, n = 46; M1, n = 75) revealed an increase of the mean number of chromosomal aberrations per cell from 5.9 (± 4.1 SD) to 12.8 (± 5.4 SD) in disease progression (P < 0.0001). Certain chromosomal aberrations were much more frequent in M1 cells, such as amplification of 1q, 8q, 16p, 17q, and 20q and loss of 6q, 13q, 14q, 16q, and 18q. In contrast, very few chromosomal regions were more often affected in M0 cells, such as amplifications of chromosome 5cen-5q23.3 or 18q (Fig. 2 A and B). The relative frequencies of all genomic aberrations are presented as Table 1, which is published as supporting information on the PNAS web site.
Surprisingly, 68 CK-positive cells from 46 patients were found to have no CGH-detectable chromosomal aberrations (i.e., balanced CGH profiles; Fig. 1). Sixteen patients showed a mixed population of CGH-balanced and -unbalanced cells (Fig. 1), whereas from 30 patients, we could isolate only CK-positive cells with balanced CGH-profiles. Twenty-eight of these 30 patients were in stage M0, whereas only two had clinical manifest metastasis (P < 0.0001). Because we could not positively prove the malignant nature of each individual CK-positive cell with a balanced profile for the moment, we proceeded solely with cells harboring CGH abnormalities. It is of interest, however, that CGH can detect chromosomal gains or losses only if they comprise chromosomal material >10–20 Mb (25).
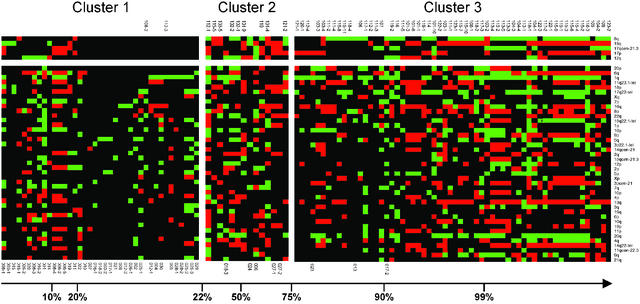
Stage-Defining Genomic Aberrations. We then tried to extract a subset of genomic changes that differentiate between cells from patients in M0 or M1 stage of the disease. To do so, we performed a feature-ranking analysis of the chromosomal aberrations to determine the information that a specific aberration carries about clinically evident metastatic disease. Thereafter, several classifiers were trained to select the subset of loci that predict metastatic disease with highest accuracy. The final subset of features contained 8q, 18q, 17qcen-21.3, 17p, and 12q. These five mutations were used to assign probability values of being isolated from a metastatic patient to each individual cell. Using these probabilities, we defined three groups (Fig. 3). Cluster 1 grouped cells with little probability (P < 0.22) to be isolated from a clinically metastatic patient (displaying a genotype here called “M0-like”), whereas cluster 3 comprised cells from all but two M1 patients (“M1-like genotype;” P > 0.75). Cases with intermediate probabilities (0.22 < P < 0.75) were grouped in cluster 2. CK-positive cells of 92% of the M1 patients were grouped together in cluster 3 irrespective of the anatomical site of the manifest metastasis. Apparently, they can be regarded as representatives of clinically evident metastasis anywhere in the body (for details of bioinformatic evaluation, see Supporting Text).
Fig. 3.
Identification of M0 and M1 genotypes. All cells were grouped on the basis of their probability of being isolated from a metastatic patient, ranging from 0.006 (Left) to 1.000 (Right). The five cluster-defining regions on top are shown in the order of the amount of information they provided to the classification (8q >18q >17qcen-21.3 > 17p >12q). All other genomic regions did not contribute to the classification and are ranked according to the feature-ranking analysis (see text). Gains and losses are indicated as green and red squares, respectively; balanced chromosomal regions are in black color. Chromosomal regions are shown (Right). Identifiers of metastastic patients (nos. 101–123) are depicted above the clusters, identifiers of nonmetastatic patients below (nos. 001–030). When several cells were isolated from one patient, they were labeled by an additional number.
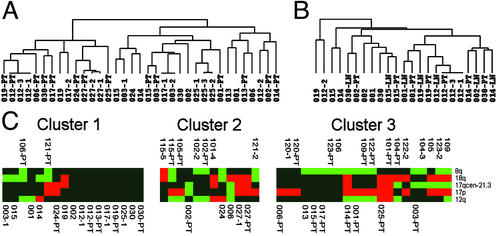
Comparison of Disseminated Cells with Their Matched Primary Tumor. We further asked whether the clear genomic differences of M0 and M1 cells reflect the genomic organization of their matched primary tumors or whether the primary tumors of our collective generally have an M0-like genotype. Therefore, areas with malignant morphology were histologically identified in primary tumors (n = 27) and positive regional lymph nodes (n = 7). After microdissection, DNA was prepared by the same global amplification method as for single cells (18). The primary tumors displayed the typical CGH abnormalities of breast cancer, demonstrating that the relevant areas had been microdissected (Fig. 2C). We then applied unsupervised hierarchical cluster analysis to determine the genomic relationship of the M0 cells with their primary tumors (26). Only two (nos. 012 and 027) of 14 matched pairs showed a high degree of similarity between disseminated cells and primary tumors (Fig. 4A). The distinctive genotypes of M0 tumor cells that were disseminated by way of blood became even more evident when we compared them with regional lymph node metastasis and their matched primary tumors. In four of seven cases, the lymph node samples were by far more closely related to the primary tumor than the cells isolated from the bone marrow (nos. 001, 014, 015, and 019; Fig. 4B). In addition, we applied the same classifier that established the separation of M0 and M1 cells to compare primary tumors and their disseminated cells. Again, this analysis showed that in most cases, the pattern of genomic aberrations in the primary tumor does not reflect the genomic aberrations of the disseminated tumor cells (Fig. 4C). From both groups of patients, approximately one-half of the primary tumors were grouped into cluster 3. Finally, we noted that M0 cells displayed significantly fewer genomic aberrations per cell than the microdissected areas of the primary tumors (5.9 vs. 10.4; P < 0.008).
Fig. 4.
Comparison of disseminated tumor cells with their matched primary tumors and lymph node metastasis. (A) Hierarchical clustering of primary tumors and their disseminated cells from patients in clinical stage M0. The applied algorithm (in complete linkage mode) organizes the CGH data on the basis of overall similarity in their genomic aberration patterns. The relationships are summarized in a dendrogram, in which the pattern and length of the branches reflect the relatedness of the samples. (B) Samples from seven patients in stage M0 and positive lymph nodes were analyzed for relatedness of primary tumors, lymph node metastasis, and disseminated tumor cells. (C) Twenty-four primary tumors and their descendent tumor cells were grouped in three clusters using the same classifier and probability thresholds as in Fig. 3. Identifiers of metastastic patients are depicted above the clusters, identifiers of nonmetastatic patients below. Patient identifiers for the disseminated cells are as in Fig. 3; primary tumors and lymph nodes samples are labeled PT and LN, respectively, after the patient identifier.
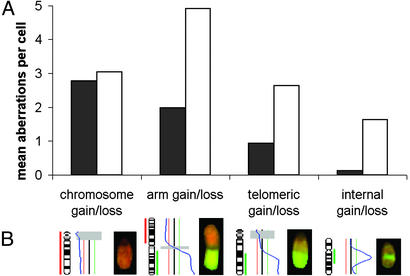
Time Point of Dissemination. Because these findings contradict a simple accumulation of imbalances in M0 cells in addition to those found in the primary tumor, we intended to define the time point of dissemination. We examined whether the cells left the primary tumor before or after the occurrence of a process that results from telomere shortening leading to characteristic chromosomal rearrangements and that has been termed crises (27, 28). We evaluated the CGH profiles for numerical genomic aberrations affecting either whole chromosomes or chromosome arms and defined intrachromosomal and telomeric regions on the other side (Fig. 5B). These changes indicate the passage of the cells through crisis, at least when they comprise the telomeric regions, because they often result from telomere shortening, chromosome fusion, and breakage (28). In contrast, gains or losses of whole chromosomes are caused by different mechanisms (29). Interestingly, the majority of chromosomal aberrations present in M0 cells consisted in aneuploidy, whereas M1 cells showed a significant increase in all changes involving chromosomal breaks (Fig. 5A; P < 0.0001). Thus, although we detected signs of crisis in M1 cells, their absence in M0 cells suggests that many tumor cells disseminate before they undergo crisis.
Fig. 5.
Types of chromosomal aberrations. (A) Mean number of chromosomal gains and losses per cell that affect the whole chromosome, a chromosome arm, a telomeric, or an internal fragment for M0 (black) and M1 (white) cells. (B) Example for each type of chromosomal aberration (on chromosome 14, 1, 13, and 17, from left to right) showing the ideogram and the respective hybridization picture.
Apparently chromosomal aberrations do not disclose the clonal relationship between the primary tumor and its seed, because many tumor cells seem to acquire them later at the distant anatomical site. However, shared mutations would not only reveal such a clonal relationship but would also define mutations that are present before dissemination. Unfortunately, the somatic mutations initiating sporadic breast cancer are not known. We had started our search by sequencing TP53, the most commonly mutated gene in human cancer (30). However, TP53 mutations were not found in any of 10 M0 breast cancer patients (31), disqualifying TP53 mutations as a marker for clonal relatedness and necessitating a search for another early somatic mutation.
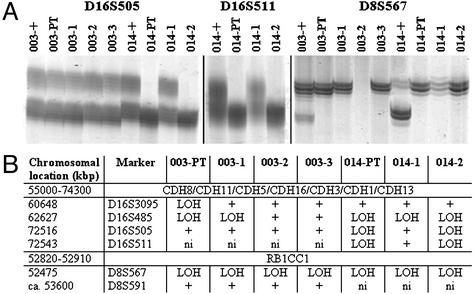
Because loss of cell–cell adhesion might be an initiating event both of oncogenesis and dissemination, we tested LOH in the proximity of CDH1 at 16q22. This region encodes E-cadherin and displays LOH in 30–40% of primary breast cancers (32). However, we had to exclude all matched pairs showing loss of the region by CGH either in the primary tumor sample or in the disseminated cells, because such large CGH-detectable aberrations result from a mutational event other than subchromsomal deletions. Six pairs of tumors and disseminated cells could be tested, and LOH at 16q22 was observed in two primary tumors by at least one of the four markers tested (Fig. 6 A and B). Whenever a disseminated cell had lost one allele, it was the same as in the primary, although not all cells displayed the LOH found in the primary tumor. In patient 014, the pattern of LOH was identical between the primary and one of the two disseminated cells for three markers. It is noteworthy that this cell belonged to the group of CK-positive cells without CGH aberrations, demonstrating that at least some of these cells have a malignant origin.
Fig. 6.
LOH analysis of disseminated M0 cells and their primary tumors. LOH analysis for markers mapping within the cadherin cluster region or close to RB1CC1. (A) Polyacrylamide gel electrophoresis of the markers D16S505, D16S511, and D8S567 (“patient identifier”+, positive control from normal cells). (B) Summary of results obtained for all markers (+, informative marker and both alleles present; ni, marker not informative).
Recently, RB1CC1 has been described as a tumor suppressor gene that is mutated in 20% of primary breast cancers. It is thought to be an activator of the retinoblastoma tumor suppressor gene (19). Inactivation of the retinoblastoma pathway is believed to be a very early genetic event in oncogenesis. Eleven pairs of primary tumors and M0 cells had no CGH loss at 8q11, the RB1CC1 region, and were therefore analyzed for LOH. Two primary tumors, the same that already had LOH at 16q22, displayed LOH at one of the two markers tested (Fig. 6 A and B). The disseminated cells from both tumors displayed LOH at D8S567, the marker closest to RB1CC1, including the cell displaying a balanced CGH (014-2) profile. Thus, LOH of RB1CC1 apparently occurred before dissemination.
Defining the Extent of Systemic Disease by Single-Cell Analysis. Breast cancer cells that disseminate in a less progressed stage must therefore acquire the characteristic chromosomal aberrations of M1 cells at the distant anatomical site if they are to grow into metastasis. The assumption of metastatic growth from such precursor cells depends on the premise of whether the relevant cells were analyzed. If, however, the genome of a single cell picked from thousands to millions of undetectable tumor cells elsewhere in the body allows a correct assessment of the patient's clinical status, this single cell should indeed be a representative of the actual systemic disease. Therefore, we calculated the predictive value of the single-cell genome for the presence and absence of clinical metastasis, including only cells for which the malignant nature was proven by CGH. As classification threshold, 0.5 was chosen, i.e., the probability of more or less than 50% of a cell being derived from a patient with a clinically metastasized tumor.
From some patients, we had isolated several cells for which the classifier had calculated different probabilities. Hence, the predictive power of disseminated tumor cells was determined twice, using either the cells with the highest or those with the lowest probability, which is reflected in the best and worst predictive power, respectively. Thereby the range of the sampling error can be determined. To judge the data from single cells, we compared them with those obtained by clinical routine parameters such as tumor size, lymph node involvement, estrogen and progesteron receptor status, and the Her-2/neu positivity. The analysis was performed on a subset of cases (M0, n = 29; M1, n = 10) for which clinical variables were available. For comparison, both classifiers were trained on this subset of 39 samples. The accuracy of prediction (i.e., the number of correctly predicted M1 and M0 patients divided by all cases) for clinical routine parameters was 69%, applying the best combination of clinical variables as input, which in our collective was the T and N stage, whereas the classification performance of single disseminated cells reached 80–85% (worst–best prediction; P < 0.002). As for sensitivity (the number of correctly predicted M1 patients divided by the total number of predicted M1 patients), and specificity (the number of cases correctly classified as M0 patients divided by the total number of patients classified as M0), the classifiers based on disseminated cells demonstrate the superior prediction. Although the sensitivity to correct identification of a metastatic patient was 70–80% (worst–best) for the genomic aberrations of disseminated cells vs. 10% for clinical variables, patients without metastatic disease were classified correctly with similar specificity [83–86% (worst–best) vs. 90%].
Discussion
Here we show that human breast cancer cells disseminate much earlier in genomic development than expected from a sequential model of cancer progression. The observation that, despite complete resection of their primary tumor, patients with initial stage M0 also relapse demonstrates that seed cells of distant metastasis must have spread before surgery or even first diagnosis. Because these cells harbor fewer and different aberrations from the primary tumor and do not display signs of telomeric crisis, our data do not support the view that dissemination starts from the most advanced clone within the primary tumor. Of course, we cannot formally exclude the possibility that advanced cells hide at anatomical sites other than bone marrow. However, the large number of analyzed samples (n = 371) minimizing the effect of potential sampling errors has yielded significant differences between the analyzed groups. Moreover, both the strong prognostic impact of CK-positive cells of breast cancer patients for both skeleton metastasis and overall survival (10, 12) and the correct identification of the clinical status of the patient from a single-cell genome underscore the relevance of the analyzed cells.
Previous cytogenetic analyses of breast cancer have not verified the postulated gradual acquisition of genomic changes, because in situ carcinomas already display chromosomal aberrations very similar to invasive carcinomas (33, 34). Likewise, similar genetic alterations were observed in primary tumors and synchronous regional lymph node metastasis (35, 36). In contrast, asynchronous distant metastases often differ extensively from the corresponding matched primary tumors (37). This finding makes a simple descent model very unlikely. From the synoptic analysis of cytogenetic data from thousands of solid tumors, it was deduced that the number of cytogenetic imbalances per tumor reflects to some extent the biological age of the tumor (38). Hence, the relatively small number of chromosomal aberrations in many M0 cells suggests they left the primary site early and the further accumulation of imbalances may have been decelerated, perhaps by environmental constraints. The numerous M0 cells without detectable aberrations by CGH analysis are of particular interest. At present, there is no evidence whatsoever that they are unrelated to the primary tumor. On the contrary, applying a high-resolution technique, microsatellite PCR, we found that at least some of these cells share identical subchromosomal deletions (e.g., 16q22 and 8q11) with the primary tumor, suggesting their malignant origin. The further development of M0 cells into metastasis, and hence M1 cells, apparently is a matter of mutation and selection, leading to a more plausible explanation for tumor dormancy. In this interpretation, dormancy reflects the time needed for M0 cells to acquire the full capacity of unrestrained growth. On the other hand, the rare detection of disseminated cancer cells displaying the typical aberrations of the primary tumors might indicate that cells of the advanced clones are selected for growth within the local environment. Consequently, cancer of unknown primary syndrome may be the infrequent situation that an early disseminated cell acquires an advantageous mutation at a distant site and evolves faster than the progenitor cell at the primary site.
Our findings may not affect only the current view on progression of systemic breast cancer but may also have some important clinical implications. First, all adjuvant therapies that do not target genetic or epigenetic events occurring early during tumorigenesis are unlikely to eradicate minimal residual disease, because disseminated cancer cells may not uniformly share mutations that are acquired later on. Because M0 cells are genetically very heterogeneous on a chromosomal level (17), reflecting the extent of genetic instability of human cancer (39), the search for shared aberrations must exploit high-resolution techniques. Second, because disseminated cells progress independently from the primary tumor, a simple extrapolation from primary tumor data to disseminated cancer cells is impossible. In conclusion, our study underscores the need to validate potential cellular targets for adjuvant and systemic therapies on disseminated cancer cells directly.
Supplementary Material
Acknowledgments
We thank Manfred Meyer, Simone Pentz, Susanne Ehnle, and Ines Heidrich for excellent technical assistance and Judith P. Johnson and Nikolas H. Stoecklein for help with the manuscript. This work was supported by Grant SFB 456 from the Deutsche Forschungsgemeinschaft and BioFuture Grant 0311884 from the German Federal Ministery for Education, Science, Research, and Technology.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CGH, comparative genomic hybridization; CK, cytokeratin; LOH, loss of heterozygosity.
References
- 1.Fearon, E. R. & Vogelstein, B. (1990) Cell 61, 759-767. [DOI] [PubMed] [Google Scholar]
- 2.van't Veer, L. J., Dai, H., van de Vijver, M. J., He, Y. D., Hart, A. A., Mao, M., Peterse, H. L., van der Kooy, K., Marton, M. J., Witteveen, A. T., et al. (2002) Nature 415, 530-536. [DOI] [PubMed] [Google Scholar]
- 3.Bernards, R. & Weinberg, R. A. (2002) Nature 418, 823. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan, D. & Weinberg, R. A. (2000) Cell 100, 57-70. [DOI] [PubMed] [Google Scholar]
- 5.Abbruzzese, J. L., Abbruzzese, M. C., Hess, K. R., Raber, M. N., Lenzi, R. & Frost, P. (1994) J. Clin. Oncol. 12, 1272-1280. [DOI] [PubMed] [Google Scholar]
- 6.Riethmuller, G. & Klein, C. A. (2001) Semin. Cancer Biol. 11, 307-311. [DOI] [PubMed] [Google Scholar]
- 7.Bernards, R. & Weinberg, R. A. (2002) Nature 419, 560 (lett.). [Google Scholar]
- 8.Edwards, P. A. (2002) Nature 419, 559-560 (lett.). [DOI] [PubMed] [Google Scholar]
- 9.Sherley, J. L. (2002) Nature 419, 560 (lett.). [DOI] [PubMed] [Google Scholar]
- 10.Braun, S., Pantel, K., Muller, P., Janni, W., Hepp, F., Kentenich, C. R., Gastroph, S., Wischnik, A., Dimpfl, T., Kindermann, G., et al. (2000) N. Engl. J. Med. 342, 525-533. [DOI] [PubMed] [Google Scholar]
- 11.Pantel, K., Cote, R. J. & Fodstad, O. (1999) J. Natl. Cancer Inst. 91, 1113-1124. [DOI] [PubMed] [Google Scholar]
- 12.Gebauer, G., Fehm, T., Merkle, E., Beck, E. P., Lang, N. & Jager, W. (2001) J. Clin. Oncol. 19, 3669-3674. [DOI] [PubMed] [Google Scholar]
- 13.Oberneder, R., Riesenberg, R., Kriegmair, M., Bitzer, U., Klammert, R., Schneede, P., Hofstetter, A., Riethmuller, G. & Pantel, K. (1994) Urol. Res. 22, 3-8. [DOI] [PubMed] [Google Scholar]
- 14.Pantel, K., Schlimok, G., Kutter, D., Schaller, G., Genz, T., Wiebecke, B., Backmann, R., Funke, I. & Riethmüller, G. (1991) Cancer Res. 51, 4712-4715. [PubMed] [Google Scholar]
- 15.Muller, P., Weckermann, D., Riethmuller, G. & Schlimok, G. (1996) Cancer Genet. Cytogenet. 88, 8-16. [DOI] [PubMed] [Google Scholar]
- 16.Klein, C. A., Schmidt-Kittler, O., Schardt, J. A., Pantel, K., Speicher, M. R. & Riethmuller, G. (1999) Proc. Natl. Acad. Sci. USA 96, 4494-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein, C. A., Blankenstein, T. J., Schmidt-Kittler, O., Petronio, M., Polzer, B., Stoecklein, N. H. & Riethmuller, G. (2002) Lancet 360, 683-689. [DOI] [PubMed] [Google Scholar]
- 18.Stoecklein, N. H., Erbersdobler, A., Schmidt-Kittler, O., Diebold, J., Schardt, J. A., Izbicki, J. R. & Klein, C. A. (2002) Am. J. Pathol. 161, 43-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chano, T., Kontani, K., Teramoto, K., Okabe, H. & Ikegawa, S. (2002) Nat. Genet. 31, 285-288. [DOI] [PubMed] [Google Scholar]
- 20.Ragg, T., Menzel, W., Baum, W. & Wigbers, M. (2002) Neurocomputing 43, 127-144. [Google Scholar]
- 21.Ragg, T. (2002) in AI Commun. 15, 61-74. [Google Scholar]
- 22.Bishop, C. M. (1995) Neural Networks for Pattern Recognition (Oxford Univ. Press, Oxford).
- 23.Empson, M. B. (2001) Pathology 33, 93-95. [DOI] [PubMed] [Google Scholar]
- 24.Press, W. H., Flannery, B. P., Teukolsky, S. A. & Vetterling, W. T. (1993) Numerical Recipes in C (Cambridge Univ. Press, Cambridge, U.K.).
- 25.Kallioniemi, A., Kallioniemi, O. P., Piper, J., Tanner, M., Stokke, T., Chen, L., Smith, H. S., Pinkel, D., Gray, J. W. & Waldman, F. M. (1994) Proc. Natl. Acad. Sci. USA 91, 2156-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Artandi, S. E., Chang, S., Lee, S. L., Alson, S., Gottlieb, G. J., Chin, L. & DePinho, R. A. (2000) Nature 406, 641-645. [DOI] [PubMed] [Google Scholar]
- 28.Maser, R. S. & DePinho, R. A. (2002) Science 297, 565-569. [DOI] [PubMed] [Google Scholar]
- 29.Pihan, G. A. & Doxsey, S. J. (1999) Semin. Cancer Biol. 9, 289-302. [DOI] [PubMed] [Google Scholar]
- 30.Greenblatt, M. S., Bennett, W. P., Hollstein, M. & Harris, C. C. (1994) Cancer Res. 54, 4855-4878. [PubMed] [Google Scholar]
- 31.Klein, C. A., Seidl, S., Petat-Dutter, K., Offner, S., Geigl, J. B., Schmidt-Kittler, O., Wendler, N., Passlick, B., Huber, R. M., Schlimok, G., et al. (2002) Nat. Biotechnol. 20, 387-392. [DOI] [PubMed] [Google Scholar]
- 32.Cheng, C. W., Wu, P. E., Yu, J. C., Huang, C. S., Yue, C. T., Wu, C. W. & Shen, C. Y. (2001) Oncogene 20, 3814-3823. [DOI] [PubMed] [Google Scholar]
- 33.Aubele, M., Mattis, A., Zitzelsberger, H., Walch, A., Kremer, M., Welzl, G., Hofler, H. & Werner, M. (2000) Int. J. Cancer 85, 82-86. [DOI] [PubMed] [Google Scholar]
- 34.Buerger, H., Otterbach, F., Simon, R., Poremba, C., Diallo, R., Decker, T., Riethdorf, L., Brinkschmidt, C., Dockhorn-Dworniczak, B. & Boecker, W. (1999) J. Pathol. 187, 396-402. [DOI] [PubMed] [Google Scholar]
- 35.Nishizaki, T., DeVries, S., Chew, K., Goodson, W. H., III, Ljung, B. M., Thor, A. & Waldman, F. M. (1997) Genes Chromosomes Cancer 19, 267-272. [DOI] [PubMed] [Google Scholar]
- 36.Pandis, N., Teixeira, M. R., Adeyinka, A., Rizou, H., Bardi, G., Mertens, F., Andersen, J. A., Bondeson, L., Sfikas, K., Qvist, H., et al. (1998) Genes Chromosomes Cancer 22, 122-129. [PubMed] [Google Scholar]
- 37.Kuukasjarvi, T., Karhu, R., Tanner, M., Kahkonen, M., Schaffer, A., Nupponen, N., Pennanen, S., Kallioniemi, A., Kallioniemi, O. P. & Isola, J. (1997) Cancer Res. 57, 1597-1604. [PubMed] [Google Scholar]
- 38.Hoglund, M., Gisselsson, D., Sall, T. & Mitelman, F. (2002) Cancer Genet. Cytogenet. 135, 103-109. [DOI] [PubMed] [Google Scholar]
- 39.Loeb, L. A., Loeb, K. R. & Anderson, J. P. (2003) Proc. Natl. Acad. Sci. USA 100, 776-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.