Abstract
We report a technique, named targeted gene methylation (TAGM), for identifying in vivo protein-binding sites in chromatin. M.CviPI, a cytosine-5 DNA methyltransferase recognizing GC sites, is fused to a DNA-binding factor enabling simultaneous detection of targeted methylation, factor footprints, and chromatin structural changes by bisulfite genomic sequencing. Using TAGM with the yeast transactivator Pho4, methylation enrichments of up to 34- fold occur proximal to native Pho4-binding sites. Additionally, significant selective targeting of methylation is observed several hundred nucleotides away, suggesting the detection of long-range interactions due to higher-order chromatin structure. In contrast, at an extragenic locus lacking Pho4-binding sites, methylation levels are at the detection limit at early times after Pho4 transactivation. Notably, substantial amounts of methylation are targeted by Pho4-M.CviPI under repressive conditions when most of the transactivator is excluded from the nucleus. Thus, TAGM enables rapid detection of DNA–protein interactions even at low occupancies and has potential for identifying factor targets at the genome-wide level. Extension of TAGM from yeast to vertebrates, which use methylation to initiate and propagate repressed chromatin, could also provide a valuable strategy for heritable inactivation of gene expression.
The interaction of proteins with chromosomal target sites, either directly or through recruitment by DNA-bound factors, is central to many processes, including transcriptional activation and repression, replication and repair of DNA, recombination, and chromosome segregation. Therefore, strategies are needed that can efficiently identify specific chromosomal sites at which factors act. Few techniques are capable of demonstrating these interactions in the context of native chromatin in living cells, and these methods have limitations (1). For example, with footprinting techniques, protection against chemical (e.g., dimethyl sulfate) or enzymatic probes expressed in cells, e.g., DNA methyltransferases (MTases) (2–6) or DNase I (7), requires close proximity of the interacting factor to DNA sites that are modified or cleaved by the footprinting agent. Footprinting methods also require that the factor resists displacement by the enzymatic or chemical probe. Moreover, because many proteins can exclude probe access, a footprint does not provide an unequivocal identity of the bound protein (8). To circumvent this latter problem, proteins have been fused to an endonuclease (9); however, the resulting DNA damage alters chromatin structure and activates checkpoint controls. Another method, chromatin immunoprecipitation (ChIP), uses in situ fixation with formaldehyde followed by immunoselection of DNA-bound complexes. The requirements for large numbers of cells and highly specific antibodies as well as low fixation efficiencies (≈0.1–0.5%) (10, 11) present distinct disadvantages of ChIP analysis. The approach of tethering chromatin proteins to the Dam MTase, which methylates GATC sites near their sites of chromosomal association, overcomes the above problems (12). This method has been used to detect factors bound at chromosomal regions containing multiple factor binding sites, e.g., 14 Gal4 (12) and 112 TetR sites (13); however, it is not known whether it can detect a factor bound at a single binding site. In addition, sensitive quantification of methylation frequencies can be performed for only one dam site at a time and requires real-time PCR analysis.
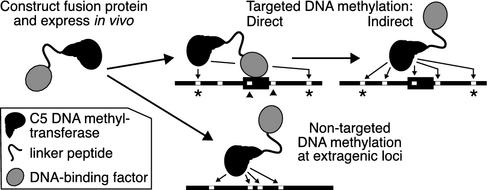
We report the specific targeting of cytosine methylation to promoters in living eukaryotic cells. Our strategy (Fig. 1), named TAGM, capitalizes on fusing chromatin-associating factors to M.CviPI, a cytosine-5 DNA MTase (C5 MTase) that methylates the C of a 2-bp GC site. This short specificity provides a M.CviPI recognition site, on average, once every 27 bp, increasing the frequency of MTase sites at least 10-fold over MTases that recognize 4-bp sites. Bisulfite genomic sequencing is used to provide a positive display of 5-methylcytosine (5meC) levels at many GC sites on a standard sequencing gel. We find that fusion of M.CviPI to a DNA-binding factor leads to substantial increases in, or targeting of, 5meC proximal to factor-binding sites that are accessible in chromatin. Moreover, 5meC is selectively targeted distal to the site of the bound factor, suggesting detection of higher-order chromatin structure. Thus, TAGM is sensitive, requiring small numbers of cells to monitor the interaction of a factor with a single native binding site. Because DNA methylation is a primary signal for establishing and maintaining repressive chromatin structures in vertebrates (14), our demonstration of targeting 5meC in a eukaryote is a critical step toward achieving heritable methylation-dependent gene silencing in such organisms.
Fig. 1.
The TAGM strategy for identifying DNA–protein interactions in vivo. Hypothetical sites protected against methylation (arrowheads) or directly methylated (asterisks) are indicated.
Materials and Methods
Yeast Strains, Plasmid Construction, and Growth Conditions. All yeast strains used in TAGM analyses have the S288C background and were derived from YPH500ΔL (MATα ade2-101 ura3-52 his3-Δ200 leu2-Δ1 trp1-Δ63 lys2-Δ1) (5). Zif268 (mut Zif), which contains a single amino acid mutation (H58E) (15) that abolishes DNA binding, was cloned as an in-frame fusion to M.CviPI into pMPK1 under the control of the GAL1 promoter and integrated at LYS2 as described (5). M.CviPI and mut Zif are separated by a linker peptide, GS(G)4SG4SG3LGST (16). Pho4-M.CviPI was constructed by tagging Pho4 with 3H A-(his)6-GPGS(G)6(SGG)2GLGST (linker)-M.CviPI at its C terminus under control of the constitutively expressed, endogenous PHO4 promoter (17). URA3, which was flanked by Zygosaccharomyces rouxii recombinase sites, was then deleted by homologous recombination (18).
For ChIP analysis, strains LFY2152 (S288C; MATα leu2-Δ0 lys2-Δ0 ura3-Δ0 pho3Δ) with the endogenous PHO4 locus tagged at its N terminus with a triple myc epitope or ADY2398 with wild-type PHO4 (no tag control) were used. Both strains also contain a mutated copy of the PHO5 promoter [deletion of both upstream activating sequences (UASs), from -401 to -352 and -258 to -209] integrated at the extragenic CAN1 locus.
Strains were pregrown in minimal media [2% raffinose/20 mM Mes, pH 5.5/14 mM L-glutamine/0.7 g of yeast nitrogen base without (NH4)2SO4, phosphate, or amino acids (Bio101)] brought to 13.4 mM KH2PO4. Cells were then washed and resuspended to an OD600 of 0.2 with the same minimal media containing either 13.4 mM KH2PO4 (+Pi, repressive conditions) or 13.4 mM KCl (-Pi, activating conditions) that also contained 2% galactose.
Bisulfite Genomic Sequencing. Genomic DNA was rapidly isolated and analyzed by bisulfite genomic sequencing (19, 20) as modified (5). PCR products amplified from bisulfite-deaminated DNA using JumpStart Taq DNA polymerase (Sigma) were purified and subjected to primer extension as described (5), except that the final concentrations of dNTPs (A, C, T) and ddGTP were 50 and 150 μM, respectively. Exclusion of dGTP from the PCR product primer extension reactions yields high termination efficiencies (>96%) (5) at template cytidines (5meC residues in vivo). Absolute frequencies of site methylation are calculated by dividing the intensity of a given band by all summed product intensities, including the run-off product at the top of the gel generated by extension on nonmethylated templates. Oligonucleotides used for the bisulfite genomic sequencing analysis of 5meC levels are listed in Table 1. A more detailed protocol is available on request.
Table 1. Bisulfite genomic sequencing primers.
| Oligonucleotides for PCR amplification
|
32P-end-labeled oligonucleotides for primer extension
|
||||
|---|---|---|---|---|---|
| Primer* | Sequence | Fig. | Primer | Sequence | Fig. |
| CAR1b1-60 | CCATTTaAaaaACTCaaaAC AATaTaaaAC | 2D | CAR1b1-60 | CCATTTaAaaaACTCaaaAC AATaTaaaAC | 2D |
| CAR1b2-61 | TAtGGAATTAGAGtttTtAA TGGAtGAG | 2D | PHO5b1-751 | TaTTTTCTCATaTAAaCaaA CaTCaTCT | 2A, B Upper, and C |
| PHO5a1-22 | CCAAATaaaTATATaCCTTa CCAAaTAAaaTaACC | 3 | PHO5b1-969 | AACaCAACTaCACAATaCCA A | 2B Lower |
| PHO5a2-21 | TAtAtATtGGAtTGATAAGT TAtTAtTGtAtATTGG | 3 | PHO5a1-22 | CCAAATaaaTATATaCCTTa CCAAaTAAaaTaACC | 3 (UASp1) |
| PHO5b1-922 | TTCAATTaCTAAATACAATa TTCCTTaaT | 2 and 4 | PHO5a1-20 | aaCTAaTTTaCCTAAaaaAA TaaTACCTaCATTaaCC | 3 (UASp2) |
| PHO5b2-924 | GAAAAtAGGGAttAGAATtA TAAATTTAGTtT | 2 and 4 | PHO5b1-768 | ATATATCTCGAGGACTAATAa AAaAAAACAAaAaACTCCa | 4 |
| PHO8b1-246 | ATAACCaCACCTaCAATaAC aaTA | 5A | PHO8b1-248 | AaAATCAAaTAAaACCTCAA aA | 5A |
| PHO8b2-247 | TtGAGTtAGATttAGGAAtA AGAtGT | 5A | PHO84a1-918 | ATaTTACCACCTTCaaTAAa aTaTTCTTTATaAA | 5B |
| PHO84a1-918 | ATaTTACCACCTTCaaTAAa aTaTTCTTTATaAA | 5B | |||
| PHO84a2-920 | AGATGAtTTtAAAtGAtTtG GTATAtTtTG | 5B | |||
Nucleotides in lowercase represent either G-to-a or C-to-t transitions
Pairs of “a” (a1 and a2) or “b” (b1 and b2) primers are used to PCR amplify the upper and lower DNA strands, respectively, from bisulfite-treated DNA
ChIP Analysis. Strains LFY2152 (3Myc-PHO4) and ADY2398 (PHO4) were grown for 4 h in the above minimal medium (2% glucose) containing the indicated concentrations of Pi before treatment with 1% paraformaldehyde for 15 min at room temperature. ChIP analysis was performed as described (21) by using 2 μl of rabbit A-14 anti-Myc antibody (Santa Cruz Biotechnologies). Two microliters of immunoselected and input DNA (1:2,000 dilution) were amplified in the presence of 10 μCi [α32P]dCTP by quantitative competitive PCR with primers ADO236 (CATGTAAGCGGACGTC) and LFO740 (GCCTTGCCAAGTAAGGTGAC), which simultaneously amplify both the wild-type (298-bp product) and mutant (198-bp product) copies of the PHO5 promoter. Radiolabeled PCR products were analyzed by 4% native PAGE.
Results
Targeting of Cytosine Methylation by Pho4 in Vivo. 5meC has been selectively targeted to oligonucleotides in vitro by fusing C5 MTases to heterologous DNA-binding factors (16, 22). To date, however, attempts to reproduce this capability in vivo have been unsuccessful (22). As a first step toward targeting C5 DNA methylation in vivo, we tested whether a native yeast protein could specifically target a C5 MTase and hence increase 5meC levels at promoters in the tractable eukaryote, Saccharomyces cerevisiae (Fig. 1). Yeast does not have detectable endogenous 5meC, and foreign expression of C5 MTases neither is deleterious nor has known effects on gene expression (5, 6). The sequences coding M.CviPI (23) were integrated at the end of the PHO4 gene, such that the MTase is fused to the C terminus of Pho4 and the fusion protein is constitutively expressed from the endogenous PHO4 promoter (17). Pho4 is a basic helix-loop-helix transactivator that induces expression of the PHO gene cluster after binding as a homodimer to E boxes (CACGTG or CACGTT) when Pi is limiting (24). The factor to which M.CviPI is fused is designated the targeting factor. Acid phosphatase activity is increased in PHO4-M.CviPI strains at least 25-fold after 6 h of Pi starvation, as has been observed for wild-type strains and those expressing other Pho4 C-terminal fusions (25, 26). Because fusing foreign proteins to MTases can decrease the affinity of the MTase for its recognition site (16), as a control, we expressed M.CviPI tethered to a mutated version of the zinc-finger protein, Zif268, that is severely impaired for DNA-binding activity (mut Zif). This “free” nontargeted MTase controls for the extent of GC methylation due to MTase site preferences in protein-free DNA and accessibility in chromatin (2–6).
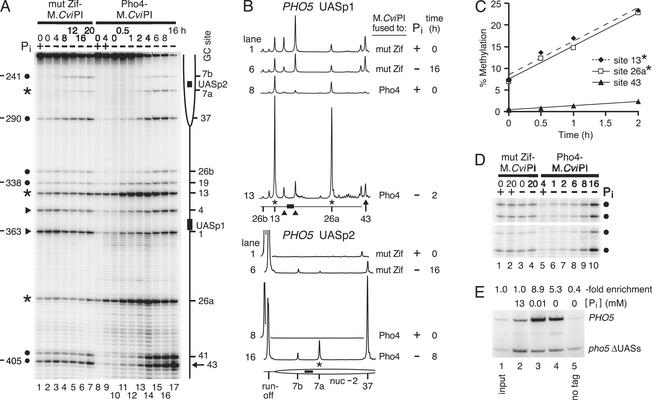
We investigated the Pho4-dependent targeting of M.CviPI to the PHO5 promoter, a well-studied locus of Pho4 binding, in a PHO4-M.CviPI/PHO4 strain. The use of a heterozygote rigorously tests whether Pho4 can target the MTase in the presence of wild-type Pho4, and more closely approximates the experimental conditions likely to be used if TAGM were used in vertebrate cells. Relative methylation frequencies at multiple GC sites were determined by bisulfite genomic sequencing (5, 19, 20), where the extent of primer extension termination is directly proportional to the level of 5meC at a given GC site. PHO4 expression is constitutive (17); in high Pi medium, Pho4 is phosphorylated by the nuclear cyclin-, cyclin-dependent kinase Pho80-Pho85 and is exported to the cytoplasm, thereby leading to the repression of PHO genes (25). Consistent with the predominantly cytoplasmic localization of Pho4 under conditions of high Pi, on the lower DNA strand of the nucleosome-free region of the PHO5 promoter (27), C5 methylation by Pho4-M.CviPI of six GC sites (sites 1, 4, 19, 26b, 41, and 43) is at background levels (Fig. 2A, lanes 8 and 9).
Fig. 2.
Pho4 specifically targets M.CviPI to the PHO5 promoter. (A) Time course of targeted methylation. Cultures expressing Pho4-M.CviPI and Pho4 (lanes 8–17) or mut Zif-M.CviPI as a free MTase control (lanes 1–7) were grown under repressive conditions in high Pi medium (lanes 1, 8, and 9; only these +Pi samples from the full time course are shown, because all others for both MTase fusions were identical), then washed and transferred to Pi-free medium to activate PHO genes (lanes 2–7 and 11–17). Genomic DNA isolated from cells removed at the indicated times was analyzed for 5meC levels at GC sites on the lower strand of the PHO5 promoter by a modification (5) of bisulfite genomic sequencing (19, 20). The locations of the two known Pho4-binding sites (filled bars), the UASp1 E box (CACGTT), and UASp2 E box (CACGTG), as well as positioned nucleosome –2 (27) (partial ellipse), are shown. The distance (base pairs) of each GC site from the respective proximal edge of UASp1 in the nuclease hypersensitive region (27) (GC sites from –405 to –331 relative to the PHO5 ATG) or UASp2 (GC sites from –290 to –241) are also indicated on the right. The same number of total counts was loaded in each lane. In strains expressing either MTase fusion, the ratios of 5meC between several sites (•) in a given lane were similar, identifying sites to which methylation is nontargeted or targeted indirectly (see Fig. 1). Normalization of 5meC levels to an accessible histone-free site remote from UASp1, site 43 (←), enables lane-to-lane comparisons (see B) and demonstrates protection against methylation (▸) as well as efficient targeting of M.CviPI to three GC sites (*) by bound Pho4. Selective targeting of 5meC to these latter three sites is highly reproducible, as evidenced in lanes 9–17 and in five additional experiments analyzing one +Pi and a 4-h -Pi sample. Note that, after 2 h, high levels of methylation in the Pho4-M.CviPI samples lead to considerable departure from single-hit kinetics and underestimation of signal intensity from –344 to –241. (B) Quantitative scans of bisulfite genomic sequencing data. (Upper) Selected lanes (as indicated) in A are scanned (PHO5 UASp1). Methylation levels can be normalized to that at site 43. (Lower) Scans (PHO5 UASp2) were obtained by reextension of the same PCR products used in the analysis in A with primer PHO5b1–969 that anneals between sites 26b and 37. The same number of total counts was loaded in each lane. Symbols are as in A.(C) Initial rates of methylation are linear. Quantification of absolute 5meC frequencies (percentage of total summed product intensities) of the indicated sites from the data in A, lanes 10–13. (D) M.CviPI is specifically targeted by Pho4 to PHO5 and not to CAR1 at early times after PHO activation. CAR1 sequences (+159 to +558) were amplified from a subset of the bisulfite-treated samples analyzed in A and analyzed for 5meC levels. Only four GC sites at CAR1 are shown; the ratios among eight additional sites are also identical. (E) TAGM detects Pho4 binding more sensitively than ChIP analysis. Immunoselected (lanes 2–5) and nonimmunoselected (lane 1, input) samples from either wild-type PHO4 (lane 5, no tag) or 3Myc-PHO4 (lanes 2–5) strains that contain a wild-type PHO5 promoter and a mutated promoter (pho5 ΔUASs) were analyzed by competitive PCR. The folds of enrichment (PHO5:pho5 ΔUASs), normalized to the input ratio, are given.
During a time course of PHO transactivation (Fig. 2 A, –Pi, lanes 11–17), methylation at most nucleosome-free sites (sites 13, 19, 26a, 26b, 41, and 43) in the PHO5 promoter increased over time in the Pho4-M.CviPI strain, in agreement with the well-known nuclear accumulation of Pho4 under these conditions (25). In contrast, in the mut Zif-M.CviPI control strain (Fig. 2 A, lanes 1–7), methylation remained rather constant at most of these sites in this histone-free region, except at sites 1 and 4 adjacent to UASp1, which are probably protected against methylation by bound Pho4 (5, 6). Closer analysis of 5meC levels during 0–2 h after activation (Fig. 2 A, lanes 10–13) indicates that Pho4 predominantly targets M.CviPI to PHO5 sites 13 and 26a (asterisks), achieving enrichments of up to 20- and 34-fold, respectively, over mut Zif-M.CviPI. Directly targeted methylation is readily identifiable by inspecting for peak areas that are altered relative to other peaks in a given lane with Pho4-M.CviPI compared with mut Zif-M.CviPI (Fig. 2B). Further, in the PHO4-M.CviPI strain, methylation frequencies of sites 13 and 26a increase linearly from 7% to 23% between 0 and 2 h (Fig. 2 A, lanes 10–13; Fig. 2C) and plateau at 4 h after induction (Fig. 2 A, lane 14). By comparison, from 0–2 h activation, 5meC accumulates at an 8-fold slower rate at site 43 than at sites 13 and 26a (Fig. 2C). In addition, similar ratios of 5meC levels among GC sites in a given lane at an extragenic locus (CAR1), which lacks Pho4 sites, demonstrate that the enhanced methylation of sites 13 and 26a at PHO5 is due to site-specific DNA binding by Pho4 (Fig. 2D). These results suggest that the frequency of targeted 5meC parallels the increase in Pho4 binding to UASp1 that occurs when cells are starved for Pi (28, 29). We conclude that M.CviPI is efficiently and directly targeted (see Fig. 1) to C residues of GC sites 13 and 26a on the lower strand of the PHO5 promoter, which agrees well with the optimal distance range of 10–40 bp observed for targeting DNA MTases to oligodeoxynucleotide substrates in vitro (16, 22). It is likely that the MTase can reach sites within this distance range when the targeting factor (i.e., Pho4) is specifically bound to its UAS. Interestingly, other sites, e.g., site 19, are not selectively modified by Pho4-M.CviPI (Fig. 2 A and B; see Discussion).
Indirect targeting of M.CviPI, Pho4-dependent accumulation of 5meC that occurs locally when Pho4 dissociates from its UAS, is also observed (see Fig. 1). For instance, methylation at sites 41 and 43 increases abruptly at 4 h –Pi and continues to rise for the remainder of the time course (Fig. 2 A, lanes 14–17). Moreover, while 5meC amounts introduced by Pho4-M.CviPI at PHO5 (e.g., sites 41 and 43) surpass those attained with the free MTase (Fig. 2 A, compare lanes 14–17 to 3–7), the converse occurs at the extragenic CAR1 locus at all times until 16 h postinduction (Fig. 2D, compare lanes 3 and 4 to 6–10). This demonstrates that, at early times after induction, Pho4 preferentially targets M.CviPI to PHO promoters (see Fig. 5) and, at extragenic loci, at least 4 h more is required to accumulate high levels of 5meC. Therefore, at ≥4 h in Pi-free medium (Fig. 2 A, lanes 14–17), the significant increases in methylation at PHO5 sites 41 and 43 are due to indirect targeting of M.CviPI; Pho4-M.CviPI dissociating from either UAS creating a local region of 5meC.
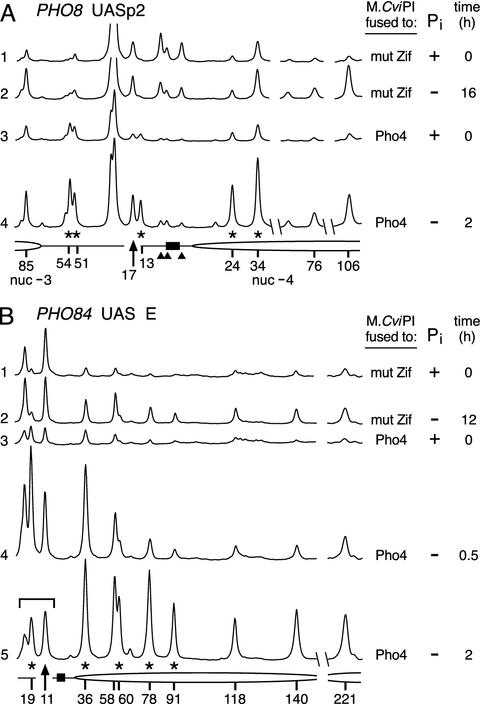
Fig. 5.
M.CviPI is targeted by Pho4 to the PHO8 (A) and PHO84 (B) promoters. 5meC levels were determined at PHO8 and PHO84 from cells expressing either mut Zif- or Pho4-M.CviPI grown in the presence (+) and absence (-) of Pi, as indicated. Shown are the quantitative scans of the phosphorimage obtained from the gel (same total counts per lane). GC sites to which M.CviPI directly targeted methylation (*), GC sites protected against methylation (▴), and Pho4-binding sites (filled bars), are labeled. 5meC levels can be compared with the sites marked with arrows. The positions of nucleosomes (nuc -3 and nuc -4, partial ellipses), previously mapped at PHO8 (41), are shown. From the data in B, we infer the disruption of two nucleosomes in the analyzed PHO84 region (increased methylation on activation at seven GC sites, 36–221 bp from UAS E; compare scan 2 to scan 1 in B). To augment peak heights, quantification of the run-off products has been omitted. A region in scan 5 where the signal is underestimated due to departure from single-hit kinetics is bracketed.
Between 2 and 16 h of induction, Pho4-M.CviPI also increased 5meC levels substantially at PHO5 sites 7a, 7b, and 37 located in positioned nucleosome –2 (27) (Fig. 2 A, lanes 13–17). Because nucleosomes block accessibility of MTases (4–6), the increased methylation of these sites by both M.CviPI fusion proteins is indicative of nucleosomal disruption concomitant with PHO5 activation (27). Methylation by mut Zif-M.CviPI at site 37 in the presence of Pi (Fig. 2 A, lane 1) occurs because MTases can access two helical turns of DNA that enter and exit nucleosomes (4–6). Note that methylation levels at sites 7a, 7b, and 37 in the Pho4-M.CviPI samples (Fig. 2 A, lanes 13–17) are substantially underestimated due to high levels of primer extension termination at sites closer to the primer (i.e., the analysis does not satisfy single-hit kinetics at the most primer distal sites). Thus, extension with a primer annealing just downstream of site 26b demonstrates that Pho4-M.CviPI methylates sites 7a, 7b, and 37 more efficiently than mut Zif-M.CviPI (Fig. 2B Lower). The extensive methylation of these sites by Pho4-M.CviPI is consistent with the high level of indirect targeting of methylation to the UASp1 region that occurs ≥4 h Pi starvation. In addition, the marked increase in 5meC at site 7a relative to 7b with Pho4-M.CviPI, and not with mut Zif-M.CviPI, strongly suggests that Pho4 targets the MTase to site 7a after binding UASp2 and/or from a distance when bound at UASp1. Therefore, 5meC is targeted to the central region of nucleosomes, which is inaccessible to MTases, only when they have been disrupted.
Strikingly, methylation is targeted directly to several GC sites when the majority of Pho4-M.CviPI is expected to be excluded from the nucleus (25). This is evidenced by the significant level of methylation present at sites 13 and 26a in the presence of Pi (Fig. 2B, PHO5 UASp1; compare scan 8 to scans 1 and 6). After normalization of 5meC levels to a histone-free site, site 43, >20-fold enrichments in targeting of M.CviPI to sites 13 and 26a by Pho4 is observed, compared with the free MTase, mut Zif-M.CviPI. Significant methylation is also targeted under repressive conditions to the opposite strand of the PHO5 promoter (Fig. 3, scan 3) as well as the PHO8 (Fig. 5A, scan 3) and PHO84 promoters (Fig. 5B, scan 3). A possible explanation for targeted methylation under repressive conditions is that the MTase fusion impairs the ability of Pho80-Pho85 to phosphorylate Pho4 and hence increases the nuclear retention of Pho4-M.CviPI. This is unlikely, because acid phosphatase expression is not derepressed in the Pho4-M.CviPI strain. Nevertheless, we tested this possibility further by comparing the rate at which PHO5 transcript levels decrease in wild-type and Pho4-M.CviPI strains after adding Pi back to cultures subjected to 10 h of Pi starvation. For both strains, PHO5 transcript levels decreased by 90% within 20 min of Pi addition, indicating that Pho4 and Pho4-M.CviPI are regulated similarly (data not shown). Thus, TAGM detects Pho4 binding, even under repressive conditions where its nuclear concentration is low (25), and therefore promoter occupancy by Pho4 is very low. Repeated attempts to detect Pho4 binding in the presence of Pi by ChIP analysis were unsuccessful (Fig. 2E, lane 2); significant immunoselection of Pho4 crosslinked to PHO5 was detected only on transactivation (Fig. 2E, lanes 3 and 4).
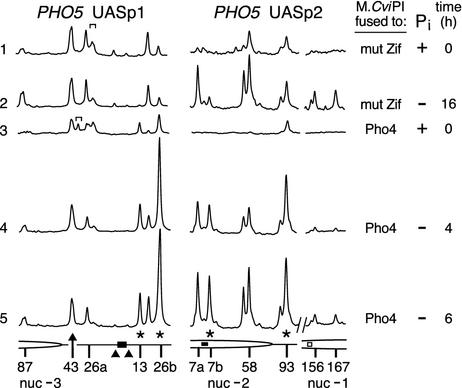
Fig. 3.
Targeting of C5 methylation by Pho4-M.CviPI to the upper strand of the PHO5 promoter. The same bisulfite-treated samples used in the analysis in Fig. 2 A were used in the PCR amplification. Scans of the phosphorimage of the gel that was loaded with the same number of total counts per lane are shown. (Left) The brackets above scans 1 and 3 (PHO5 UASp1) indicate a nonspecific primer extension pause that occurred in samples 1–5 or only sample 3, respectively. See the legend to Fig. 2 A for symbol definitions.
We also analyzed 5meC levels on the upper strand of the PHO5 promoter (Fig. 3). After transactivation, methylation is enhanced at several GC sites near UASp1 (site 87) and UASp2 (sites 7a, 52, 58, 154, 156, and 167), as expected with the increased access of both MTase fusion proteins that accompanies nucleosome disruption (27). Methylation amounts are significantly altered at sites 13, 26b, 7b, and 93 (asterisks) relative to other sites in cells expressing Pho4-M.CviPI (scans 3–5) compared with the control, mut Zif-M.CviPI (scans 1 and 2), indicating Pho4-dependent targeting of M.CviPI. Interestingly, despite the high level of 5meC targeted to sites 7a and 26a on the lower strand (Fig. 2), M.CviPI is not directly targeted to these sites on the upper strand. The reason for this strand-specific, targeting of 5meC to pairs of GC sites that symmetrically flank each Pho4 binding site (7 or 26 bp away) is not understood.
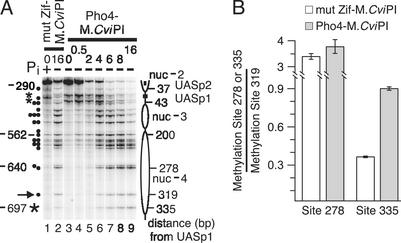
Pho4 Targets M.CviPI at a Distance. In Fig. 3, the marked methylation of site 93 compared with other sites on the upper strand of the PHO5 promoter suggests that M.CviPI is targeted at distances (93 bp from UASp2 and 202 bp from UASp1), well beyond the optimal targeting distance of 10–40 bp observed in vitro (16, 22). To investigate this possibility further, we determined methylation levels at GC sites farther upstream in the PHO5 promoter (Fig. 4). 5meC levels at a number of GC sites increased at the positions of two additional nucleosomes (-3 and -4) that are known to be perturbed on promoter activation (27) (Fig. 4A, compare lane 2 to 1 and lanes 6–9 to 3). 5meC was reproducibly enriched at a GC site located 335 bp from UASp1, in the PHO4-M.CviPI compared with the mut Zif-M.CviPI strain, suggesting the formation of long-range interactions stemming from higher-order chromatin folding (Fig. 4, compare lanes 7–9 to 2). A DNA-bound homodimer of Pho4 similarly targets M.CviPI distally (60, 78, and 91 bp) to a low-affinity Pho4-binding site (UAS E) in the PHO84 promoter (30) (Fig. 5B). The Gal4 DNA-binding domain- and TetR-Dam MTase, bound at 14 and 112 sites, respectively, can also distally target a tethered Dam MTase (12, 13). Thus, in comparison to a free MTase control, TAGM can discern activation-dependent perturbations in nucleosome structure and preferential MTase targeting at a distance.
Fig. 4.
Pho4 targets M.CviPI at a distance. (A) Determination of 5meC levels upstream of PHO5 UASs. PHO5 sequences were amplified from a subset of the bisulfite-treated samples analyzed in Fig. 2 A to assay for 5meC levels. The two asterisks at the top of the gel indicate sites 13 and 26a that are directly targeted by Pho4-M.CviPI near UASp1. Symbols are as in Fig. 2A, except that double (••) and triple GC sites (•••) that did not resolve during electrophoresis are also indicated. Site 319 used for normalization in B is marked as well (→). (B) Quantification of preferential targeting of M.CviPI by Pho4 to site 335, but not to site 278. The mean ± standard error of 5meC levels for the indicated sites (normalized to site 319) for mut Zif-M.CviPI (n = 3) and Pho4-M.CviPI (n = 6) is shown.
Pho4 Targets M.CviPI to Additional PHO Promoters. Pho4 targeted M.CviPI directly to several GC sites at the PHO8 and PHO84 promoters (Fig. 5). For example, in contrast to mut Zif-M.CviPI cells, yeast expressing Pho4-M.CviPI exhibited significantly higher levels of 5meC at PHO8 sites 13, 51, and 54 compared with site 17, and at PHO84 sites 19 and 36 relative to site 11 (compare the relative peak areas of scans 3 and 4 in Fig. 5A or scans 3–5 in Fig. 5B to those of 1 and 2). Pho4 also significantly targets M.CviPI to each of these sites under repressive conditions when Pho4 binding is very low (scan 3). In addition, after starving PHO4-M.CviPI cells for Pi (scan 4 in Fig. 5A), amounts of methylation at PHO8 sites 24 and 34 in disrupted nucleosome –4 surpass those at site 17. Although M.CviPI targeting was evident near UASp2 of PHO8, none of four GC sites located 11–42 bp from the putative UASp1 is targeted in the repressed or activated promoter (data not shown), indicating further that UASp2 is the only functional Pho4-binding site in the PHO8 promoter (31). After 2 h of activation, methylation at PHO84 sites 60, 78, and 91 exceeds that at neighboring sites 118, 140, and 221 with Pho4-M.CviPI, but not mut Zif-M.CviPI (Fig. 5B, compare scan 5 to 2), suggesting that bound Pho4 directly targets M.CviPI to distal PHO84 sites 60, 78, and 91. We conclude that the native transcription factor Pho4 can efficiently target M.CviPI to each of the endogenous single-copy PHO promoters that we have tested.
Discussion
We have demonstrated that TAGM is a highly effective and sensitive technique for detecting DNA–protein interactions and activation-dependent changes chromatin structure in vivo. The method provides several distinct advantages over other available approaches (1) (see introduction), including: (i) identification of sites of factor interaction at relatively high resolution in living cells; (ii) high sensitivity, requiring only small amounts of cells and detecting factor binding even at single native sites; and (iii) the ability to monitor nucleosomal rearrangements kinetically. In vitro, the ability to target 5meC is primarily related to the distance between a particular MTase site and the factor-binding site, which is likely related to the length and nature of the peptide separating the targeting factor and the MTase (16, 22). In addition to these constraints, our results demonstrate that, in chromatin, the efficiency of targeting 5meC to a given site is determined by its accessibility, its rotational orientation relative to the factor-binding site, and/or higher-order chromosome structure.
Taken together, at PHO5, our data suggest that a homodimer of Pho4, initially binding to the accessible UASp1 E box, preferentially targets M.CviPI to sites 13 and 26a (28). Subsequently, disruption of nucleosome –2, presumably mediated by the recruitment of coactivators such as histone acetyltransferases (32, 33) and ATP-dependent remodelers (34–36) to PHO5, facilitates Pho4 binding at the high-affinity UASp2 site (28, 29). Increases in the local MTase concentration due to cooperative binding of Pho4-M.CviPI that accompanies chromatin perturbation may account for the accumulation of high levels of methylation at sites to which the MTase is indirectly targeted (e.g., sites 41 and 43). In that Pho4 targets M.CviPI at a distance (Figs. 3, 4, 5), it is interesting to speculate that it can also do so with recruited coactivators and hence disrupt distal nucleosomes (37).
Previous studies have suggested that residual levels of Pho4 are present in the nucleus in high Pi (38–40), despite its predominant cytoplasmic localization under these conditions (25). The presence of marked targeted 5meCat PHO5, PHO8, and PHO84 provides direct evidence of a low level of Pho4 binding in the presence of Pi. This binding occurs either before phosphorylation of Pho4 by Pho80-Pho85 or after its modification and before subsequent nuclear export. The sensitivity of TAGM is underscored by this result because, in the presence of Pi, Pho4 binding is not detectable by ChIP analysis (36) (Fig. 2E) or genomic footprinting (28). Thus, TAGM is a powerful and complementary alternative to existing technologies.
In addition to the use of TAGM with Pho4 fused to M.CviPI presented here, we have also targeted M.CviPI as well as M.SssI, acting on CG sites, both near and several hundred nucleotides from single Zif268-binding sites (C.D.C., R. L. Parr, G.-L. Xu, and M.P.K., unpublished work). Thus, the successful application of TAGM for these two factors tested thus far, each at three different loci validates its efficacy in targeting C5 methylation and hence detection of factor interactions. We are currently extending TAGM to additional transcription factors and coactivators. It may also be possible to apply TAGM in other organisms that lack cytosine methylation or perhaps in vertebrate cells at regions that do not have an abundance of 5meC (e.g., CpG islands). Further, our observation that substantially more methylation by Pho4-M.CviPI at PHO5 vs. CAR1 occurs at early times after PHO activation is promising for using TAGM in genome-wide identification of targets for Pho4 and other transcription factors. Finally, because C5 DNA methylation is a primary signal leading to the formation of repressive chromatin structures in vertebrates (14), the ability to specifically target such methylation could provide a strategy for inactivating gene expression.
Acknowledgments
We thank Shiv Grewal for training L.B.F. in ChIP analysis. This work was supported by awards from the American Heart Association–Texas Affiliate, the Texas Higher Education Coordinating Board, and the National Cancer Institute (CA095525).
Abbreviations: 5meC, 5-methylcytosine; ChIP, chromatin immunoprecipitation; MTase, methyltransferase; Pi, inorganic orthophosphate; TAGM, targeted gene methylation; mut Zif, mutated Zif268; C5, cytosine-5; UAS, upstream activating sequence.
References
- 1.Simpson, R. T. (1999) Curr. Opin. Genet. Dev. 9, 225-229. [DOI] [PubMed] [Google Scholar]
- 2.Singh, J. & Klar, A. J. S. (1992) Genes Dev. 6, 186-196. [DOI] [PubMed] [Google Scholar]
- 3.Gottschling, D. E. (1992) Proc. Natl. Acad. Sci. USA 89, 4062-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kladde, M. P. & Simpson, R. T. (1994) Proc. Natl. Acad. Sci. USA 91, 1361-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kladde, M. P., Xu, M. & Simpson, R. T. (1996) EMBO J. 15, 6290-6300. [PMC free article] [PubMed] [Google Scholar]
- 6.Xu, M., Simpson, R. T. & Kladde, M. P. (1998) Mol. Cell. Biol. 18, 1201-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang, X. & Simpson, R. T. (2001) Nucleic Acids Res. 29, 1943-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rigaud, G., Roux, J., Pictet, R. & Grange, T. (1991) Cell 67, 977-986. [DOI] [PubMed] [Google Scholar]
- 9.Lee, J. S., Lee, C. H. & Chung, J. H. (1998) Proc. Natl. Acad. Sci. USA 95, 969-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka, T., Cosma, M. P., Wirth, K. & Nasmyth, K. (1999) Cell 98, 847-858. [DOI] [PubMed] [Google Scholar]
- 11.Reid, J. L., Iyer, V. R., Brown, P. O. & Struhl, K. (2000) Mol. Cell 6, 1297-1307. [DOI] [PubMed] [Google Scholar]
- 12.van Steensel, B. & Henikoff, S. (2000) Nat. Biotechnol. 18, 424-428. [DOI] [PubMed] [Google Scholar]
- 13.Lebrun, E., Fourel, G., Defossez, P.-A. & Gilson, E. (2003) Mol. Cell. Biol. 23, 1498-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bird, A. (2002) Genes Dev. 16, 6-21. [DOI] [PubMed] [Google Scholar]
- 15.Nardelli, J., Gibson, T. J., Vesque, C. & Charnay, P. (1991) Nature 349, 175-178. [DOI] [PubMed] [Google Scholar]
- 16.Xu, G. L. & Bestor, T. H. (1997) Nat. Genet. 17, 376-378. [DOI] [PubMed] [Google Scholar]
- 17.Legrain, M., De Wilde, M. & Hilger, F. (1986) Nucleic Acids Res. 14, 3059-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roca, J., Gartenberg, M. R., Oshima, Y. & Wang, J. C. (1992) Nucleic Acids Res. 20, 4671-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frommer, M., MacDonald, L. E., Millar, D. S., Collis, C. M., Watt, F., Grigg, G. W., Molloy, P. L. & Paul, C. L. (1992) Proc. Natl. Acad. Sci. USA 89, 1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark, S. J., Harrison, J., Paul, C. L. & Frommer, M. (1994) Nucleic Acids Res. 22, 2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hecht, A. & Grunstein, M. (1999) Methods Enzymol. 304, 399-414. [DOI] [PubMed] [Google Scholar]
- 22.McNamara, A. R., Hurd, P. J., Smith, A. E. & Ford, K. G. (2002) Nucleic Acids Res. 30, 3818-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu, M., Kladde, M. P., Van Etten, J. L. & Simpson, R. T. (1998) Nucleic Acids Res. 26, 3961-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oshima, Y., Ogawa, N. & Harashima, S. (1996) Gene 179, 171-177. [DOI] [PubMed] [Google Scholar]
- 25.O'Neill, E. M., Kaffman, A., Jolly, E. R. & O'Shea, E. K. (1996) Science 271, 209-212. [DOI] [PubMed] [Google Scholar]
- 26.Komeili, A. & O'Shea, E. K. (1999) Science 284, 977-980. [DOI] [PubMed] [Google Scholar]
- 27.Almer, A., Rudolph, H., Hinnen, A. & Hörz, W. (1986) EMBO J. 5, 2689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venter, U., Svaren, J., Schmitz, J., Schmid, A. & Hörz, W. (1994) EMBO J. 13, 4848-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svaren, J., Schmitz, J. & Hörz, W. (1994) EMBO J. 13, 4856-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogawa, N., Saitoh, H., Miura, K., Magbanua, J. P., Bun-ya, M., Harashima, S. & Oshima, Y. (1995) Mol. Gen. Genet. 249, 406-416. [DOI] [PubMed] [Google Scholar]
- 31.Münsterkötter, M., Barbaric, S. & Hörz, W. (2000) J. Biol. Chem. 275, 22678-22685. [DOI] [PubMed] [Google Scholar]
- 32.Gregory, P. D., Schmid, A., Zavari, M., Lui, L., Berger, S. L. & Hörz, W. (1998) Mol. Cell 1, 495-505. [DOI] [PubMed] [Google Scholar]
- 33.Galarneau, L., Nourani, A., Boudreault, A. A., Zhang, Y., Heliot, L., Allard, S., Savard, J., Lane, W. S., Stillman, D. J. & Côté, J. (2000) Mol. Cell 5, 927-937. [DOI] [PubMed] [Google Scholar]
- 34.Santisteban, M. S., Arents, G., Moudrianakis, E. N. & Smith, M. M. (1997) EMBO J. 16, 2493-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sudarsanam, P., Iyer, V. R., Brown, P. O. & Winston, F. (2000) Proc. Natl. Acad. Sci. USA 97, 3364-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steger, D. J., Haswell, E. S., Miller, A. L., Wente, S. R. & O'Shea, E. K. (2003) Science 299, 114-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, Y. & Clark, D. J. (2002) Proc. Natl. Acad. Sci. USA 9, 15381-15386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han, M., Kim, U. J., Kayne, P. & Grunstein, M. (1988) EMBO J. 7, 2221-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han, M. & Grunstein, M. (1988) Cell 55, 1137-1145. [DOI] [PubMed] [Google Scholar]
- 40.Wechser, M. A., Kladde, M. P., Alfieri, J. A. & Peterson, C. L. (1997) EMBO J. 16, 2086-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barbaric, S., Fascher, K. D. & Hörz, W. (1992) Nucleic Acids Res. 20, 1031-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]