Abstract
Cytolytic killing is a major effector mechanism in the elimination of virally infected and tumor cells. The innate cytolytic effectors, natural killer (NK) cells, and the adaptive effectors, cytotoxic T cells (CTL), despite differential immune recognition, both use the same lytic mechanism, cytolytic granule release. Using live cell video fluorescence microscopy in various primary cell models of NK cell and CTL killing, we show here that on tight target cell contact, a majority of the NK cells established cytoskeletal polarity required for effective lytic function slowly or incompletely. In contrast, CTLs established cytoskeletal polarity rapidly. In addition, NK cell killing was uniquely sensitive to minor interference with cytoskeletal dynamics. We propose that the stepwise NK cell cytoskeletal polarization constitutes a series of checkpoints in NK cell killing. In addition, the use of more deliberate progression to effector function to compensate for inferior immune recognition specificity provides a mechanistic explanation for how the same effector function can be used in the different functional contexts of the innate and adaptive immune response.
The immune system eliminates virally infected and tumor cells through cytolytic killing. Natural killer (NK) cells (1) are the cytolytic effectors of the innate immune system (2). They are present with great frequency, and their effector functions can be activated rapidly as a first line of immunological defense. However, their recognition specificity is limited to a small number of prototypical immune patterns, such as lack of MHC class I, the presence of stress-induced proteins, or MHC-related viral proteins (3–7). The cytolytic effectors of the adaptive immune system, cytotoxic T cells (CTLs), dominate an immune response after ≈6 d and recognize a practically unlimited number of small nonself peptides with high specificity (8). They exist in small precursor frequencies and require extensive proliferation and effector differentiation explaining their belated action. However, the precursor expansion allows for extensive controls against erroneous activation. Interestingly, NK cells and CTLs, despite their different immune recognition properties, use the same cytolytic mechanisms, directed release of cytotoxic granules and expression of ligands for death receptors. Here, we have investigated the cytoskeletal dynamics of innate and adaptive cytolytic effectors. We propose that the compensation of inferior immune recognition specificity, by more deliberate progression to effector function, provides a mechanistic understanding of how the same effector mechanism can be used under the different physiological conditions of the innate and adaptive immune response.
Experimental Procedures
Cells. Lymphokine-activated killer (LAK) cells were obtained by culturing B6 splenocytes with 1 ng/ml IL-12 and 100 ng/ml IL-18 for 5 days (9) and contained >90% NK cells (CD3–2B4+DX5+). Poly I/C-stimulated NK cells were prepared by isolating splenocytes from SCID mice that had been injected i.p. with 200 μg of polyinosinic:polycytidylic acid the day before (10). They contained 15% NK cells (CD3–NK1.1+). YAC-1 lymphoma intercellular adhesion molecule-1–GFP (11) transfected YAC-1, or CD48–GFP (similar to ref. 11) P815 cells were used as targets. CTLs from P14 TCR-transgenic mice (12) were obtained as described for 5C.C7 T cells (11). B6 CTLs were obtained in the same way from CD4+ cell-depleted lymph node suspensions but primed by using irradiated spleen plus 2 μg/ml anti-CD3ε antibody 2C11 (PharMingen). In some cultures, additional IL-12 and -18 was added (9). EL4 thymoma or H2-Db-transfected CH27 B lymphoma cells preincubated with 10 μM of the p33 LCMV peptide (KAVYNFATM) were used as targets for P14 CTLs; DC2.4 dendritic cells preincubated with 10 μg/ml 2C11 were used as targets for B6 CTLs.
Microscopy. NK cell–target cell couple formation is relatively rare. Large fields have to be imaged to allow the analysis of a significant number of interactions, thus limiting resolution. Only one to five interactions on average for different assays could be analyzed per experiment. To avoid bias in data interpretation, we used exactly defined analysis criteria as described below. The microscopy system and sample handling were the same as before (11), with the following modifications. Cells were loaded with dye by incubation with 0.4 or 4 μM SNARF-1-AM ester or 4 μM fura 2-AM ester (Molecular Probes). Effector-to-target cell interactions were followed for 60–90 min at 37°C by acquiring one differential interference contrast microscopy (DIC) bright-field image, one SNARF, and the two fura 2 images every 20 s by using a ×20 air (for NK cells) or a ×40 oil (for CTLs) objective. When intercellular adhesion molecule–GFP transfected YAC-1 or actin-GFP or tubulin-GFP transduced effectors were used, images were recorded every 20 s to 1 min, and an additional 13–21 GFP images with a z-distance of 1 μm were acquired at each time point.
Drug Treatments. For moderate interference with cytoskeletal dynamics, effectors were pretreated with the minimal effective concentration (0.5 μM) of Jasplakinolide (13) (Molecular Probes) at room temperature for 5 min, and the marginal concentration of 0.1 μM Jasplakinolide (13) was present during the effector–target cell interaction. Effectors were pretreated with 0.1 μM Nocodazole (Calbiochem) for 30 min at 37°C and 0.1 μM Nocodazole was present in the assay. For strong interference, 2 μM/1 μM Jasplakinolide and 10 μM/10 μM Nocodazole were used.
Interface Diameters, Actin–GFP, F-Actin Staining, and Tubulin–GFP. Effector–target cell interface diameters were measured in DIC images by using the METAMORPH software (Universal Imaging, Downingtown, PA). We defined an initial tight NK cell–target contact as a cellular interface with a diameter of at least two-thirds of the NK cell diameter in conjunction with tight membrane apposition. This criterion excluded random cell contacts, as seen in incidental target cell–target cell and NK cell–NK cell encounters. β-Actin–GFP and β-5 tubulin–GFP (14, 15) were expressed from a Moloney murine leukemia virus-derived retroviral expression vector (16) containing an additional internal phosphoglycerol kinase promoter (actin-GFP) or not (tubulin-GFP). T cell and NK cell transduction was performed as described (16). For analysis of 3D actin–GFP data at each time point, the average actin–GFP fluorescence intensity of the area of actin accumulation at the effector–target cell interface was measured (METAMORPH) and compared with the average intensity of an area of the same size at the most intense part of the rest of the cell (>40% of that is a strong accumulation), and at the least intense part of the cell, the cellular background (>40% of that is a partial accumulation). For analysis of the 3D tubulin–GFP data, a cell was counted as having a well defined microtubule organizing center (MTOC), when in a maximum projection (METAMORPH) the top one-third of the tubulin–GFP fluorescence was contained in <10% of the effector area. This was the case in >90% of the effectors (the remainder was excluded from the analysis). We defined the MTOC to be at the NK cell–target cell interface if it was located at the edge of the NK cell and not farther than one-quarter of the NK cell diameter away from the center of the interface.
Other Assays. Specific target lysis was determined in a standard chromium release assay, and Western blots for perforin were performed as before (17).
Results
Variable NK Cell–Target Cell Couple Fates. To study innate cytolysis, we investigated the interaction of live IL-12/IL-18-stimulated LAK cells (9) (>90% NK cells (Experimental Procedures) with susceptible YAC-1 target cells in real time by using video fluorescence microscopy. Killing in this interaction depends entirely on cytotoxic granule release (17). To visualize NK cell signaling, we used the calcium-sensitive dye fura 1. To visualize killing, we used the release of a membrane-impermeable inert dye, SNARF-1, from target cells on lysis.
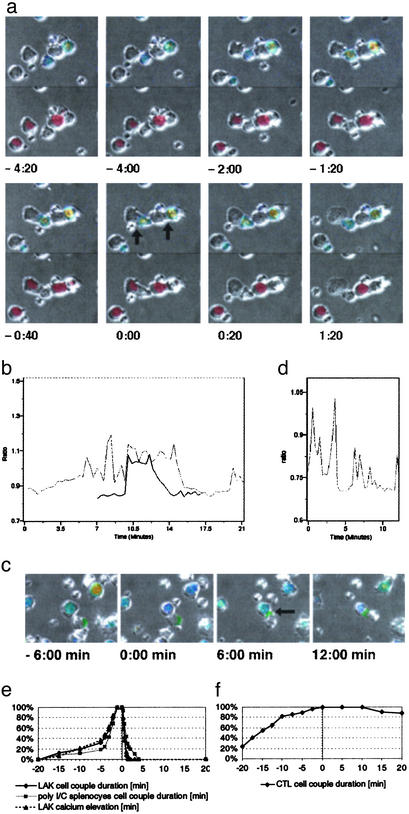
Target cell lysis was dramatic. All SNARF-1 was released and the target cell turned into a membrane ghost in <20 s (Fig. 1; Movie 1, which is published as supporting information on the PNAS web site, www.pnas.org). Target cell lysis could be brief. The minimal time between initial tight NK cell-target cell contact and target cell lysis was 2–5 min (Fig. 1, Movie 1). During this time, the NK cell intracellular calcium concentration was elevated (Fig. 1b, Movie 1), the two cells were closely apposed, and the target cell was lysed. This brief period of target lysis is designated here a lytic hit. In 47% of the lytic events (n = 72), the NK cell killed the target in such a manner within 5 min. We term this rapid killing. In the remaining 53% of the lytic events, the lytic hit was preceded by 9 ± 5 min of NK cell–target cell contact. We term this delayed killing. During the time preceding the final lytic hit, the NK cell intracellular calcium concentration fluctuated, often returning to baseline (Fig. 1b). We term such a phase of an NK cell–target cell interaction tentative. Next, we determined whether all initial tight contacts (Experimental Procedures) (n = 151) resulted in target cell lysis. Only 48% did, as characterized above. The remainder persisted without target cell lysis throughout the observation period of 1h or until the NK cell detached (Fig. 5, which is published as supporting information on the PNAS web site). We term this a nonkilling interaction. NK cell binding to target cells was strong, as judged by the persistence of the cell couples during target cell movement (Fig. 1c, Movie 2, which is published as supporting information on the PNAS web site), including occasional abrupt changes of direction (data not shown). Signaling was tentative, because the elevations of the NK cell intracellular calcium concentration were intermittent and fluctuating (Fig. 1d).
Fig. 1.
Three target cell fates in NK cell killing, rapid killing, delayed killing, and nonkilling. (a) The interaction of two LAKs with YAC-1 target cells (black arrows at t = 0:00 min) is shown in panels from Movie 1. DIC bright-field images have been duplicated. The top is overlaid with a rainbow color scale representation of the NK cell intracellular calcium concentration (blue for low to red for high), which also identifies NK cells. The bottom is overlaid with an image of the red fluorescent dye SNARF that is released on target cell lysis and also identifies target cells. The left couple displays a rapid killing event. The tight interface forms at -1:20 min, lysis (SNARF release and target cell swelling) occurs between 0:00 and 0:20 min, and detachment occurs at 1:20 min. The right cell couple displays a delayed killing event. Persistent tight cell apposition is visible, but target cell lysis can be seen only in Movie 1. An additional small NK cell and the NK cell of the left couple touch the target cell of the right couple but do not form a tight interface and are, therefore, highly unlikely to contribute to the lysis of that cell. (b) Traces of the NK cell intracellular calcium concentration (as the ratio of the emission intensities of the calcium-sensitive dye fura 2) for the two cell couples shown in a are given. The dark and light traces correspond to the NK cells in the left and right couples, respectively. Because of NK cell movement, one trace is shown for only part of the experiment. Target cell lysis coincides with the decline in the calcium concentration at 13 and 15 min, respectively. At 20 min, an elevation of the intracellular calcium concentration of the right NK cell as triggered by new target cell contact can be seen. (c) A nonkilling interaction of a LAK with a polarized YAC-1 target cell (arrow at t = 6:00 min) is shown in panels from Movie 2. The NK cell has been overlaid with a representation of the intracellular calcium concentration similar to a. The target cell has been overlaid with the intensity of the intercellular adhesion molecule (ICAM)-1–GFP fluorescence in green as a marker of polarity. High ICAM-1 concentration marks the posterior end of the target cell. Interface formation was at t = 0:00 min. As seen here and better in Movie 2, only a narrow interface was maintained. (d) The trace of the intracellular calcium concentration similar to b is shown for the NK cell from c.(e) The cumulative percentage of the indicated type of NK cells in killing interactions bound to their YAC-1 target cells or showing an elevation of their intracellular calcium concentration before (negative time values) and after (positive time values) lysis (t = 0 min) is shown. Thirty to 40 cell couples from at least six independent experiments were analyzed per condition. (f) The cumulative percentage of P14 CTLs in killing interactions bound to their target cells before (negative time values) and after (positive time values) the onset of blebbing (t = 0 min) is shown. Data for EL4 and H2-Db-transfected CH27 target cells were pooled. Thirty-four cell couples from five independent experiments were analyzed.
Three principal target cell fates (a statistical analysis of the reliability of their distinction can be found in Supporting Text, which is published as supporting information on the PNAS web site) were thus observed after the formation of an initial tight NK cell–target cell interface. (i) The target cell was rapidly lysed in an immediate lytic hit (23% of all cell couples), called rapid killing. (ii) The target cell was belatedly lysed in a final lytic hit after an extended period of tentative cell couple maintenance (25%), called delayed killing. (iii) The target cell was spared, persisting in a tentative cell couple (52%), called nonkilling. The tentative nature of many NK cell–target cell couples suggests the existence of regulatory checkpoints in the progression from cell couple formation to target cell lysis. Such variable dynamics were not restricted to LAK–YAC-1 interactions, because they were also seen in the killing of YAC-1 target cells by splenic NK cells from SCID mice stimulated with polyinosinic/polycytidylic acid (Experimental Procedures) (Fig. 1e), in the killing of P815 tumor cells by LAKs (Fig. 5B), and in the interaction of the rat-derived RNK-16 NK cell leukemia line with susceptible target cells (18). These data suggest that variable dynamics are a general feature of NK cell killing. Potential explanations for this variability are discussed in Supporting Text.
Decisive CTL–Target Cell Couple Maintenance. To investigate adaptive cytolysis, we used primary in vitro primed CTLs from P14 T cell receptor transgenic mice (12) that effectively lyse EL4 thymoma and H2-Db-transfected CH27 B cell lymphoma target cells incubated with the lymphocytic choriomeningitis virus gp33 peptide (12) (Fig. 6A, which is published as supporting information on the PNAS web site). Anti-FasL antibodies did not block this lysis (Fig. 6A), suggesting (10) that target cell death depended primarily on cytotoxic granule release. The CTLs and NK cells used here thus rely on the same effector mechanism.
Cellular events in CTL-mediated target cell lysis controlled by recognition of allo-MHC have previously been characterized (19, 20). Using peptide-MHC recognition, we extend these data here (Supporting Text and Fig. 6; see also Fig. 7 and Movies 3 and 4, which are published as supporting information on the PNAS web site). As our main observations, we found that CTL–target cell contact was persistently tight, and the elevation of the CTL intracellular calcium concentration was sustained. In contrast to the tentative nature of many NK cell–target cell couples, CTL cell couple maintenance was thus decisive. However, target cell lysis was slower (Supporting Text). The differential cell couple maintenance of CTLs and NK cells suggests that often-tentative progression from cell couple formation to target cell lysis is unique for NK cells.
Stepwise Cytoskeletal Polarization in NK Cell–Target Cell Couples. To better understand these distinguishing cell couple characteristics, we investigated effector cytoskeletal polarization by measuring interface diameters, by visualizing actin distribution as a likely mediator of interface maintenance, and by visualizing MTOC orientation as an indicator of cell polarity.
First, we measured interface diameters. The mostly spherical interface geometry and the availability of the whole interface for receptor/ligand binding (21–23) make the determination of an interface diameter based on bright-field data a reasonable approximation for the extent of effector/target cell contact. In NK cell–target cell interactions, the initial interface remained tight in rapid or delayed killing interactions. Its diameter was 87 ± 15% of the NK cell diameter (Fig. 8, which is published as supporting information on the PNAS web site). In contrast, the initial tight interface could not be maintained in nonkilling NK cell–target cell interactions despite persistent cell contact, because the interface diameter dropped significantly (P < 0.001) from 79 ± 11% to 49 ± 9% of the NK cell diameter (Fig. 8, Movie 2). These data suggest that the maintenance of a large contact area may be required for efficient NK cell killing, possibly because a wider interface is likely to allow increased receptor–ligand engagement.
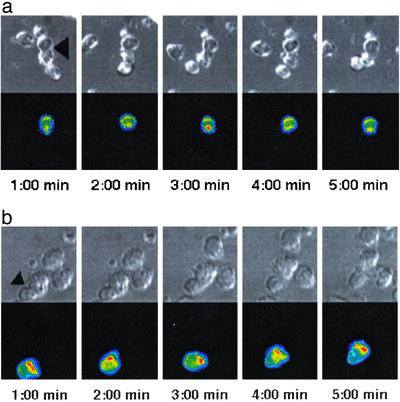
To characterize NK cell actin dynamics, we used a retrovirally expressed (16) actin–GFP fusion protein (14) in combination with 3D live cell video fluorescence microscopy (Fig. 2a, Movie 5, which is published as supporting information on the PNAS web site). As quantified in more detail in the Supporting Text, we made three principal observations. (i) Actin accumulated at the interface during the lytic hit in all cell couples (Fig. 9, which is published as supporting information on the PNAS web site). (ii) In tentative cell couples, actin accumulated at the NK cell–target cell interface only intermittently (Figs. 2a and 9 and Movie 5). Analyzing one time point per minute for the first 5 min after cell couple formation, 60% of the delayed (before the lytic hit) and 89% of the nonkilling cell couples did not show any strong (Experimental Procedures) actin accumulation (Fig. 9), corroborating the tentative nature of such couples. (iii) These data also demonstrate that more interface actin accumulation occurred in delayed killing interactions before the lytic hit than in nonkilling interactions, thus corroborating the differential interface maintenance described above. We corroborated the only occasional occurrence of actin accumulation at the NK cell–target cell interface by F-actin staining of fixed NK cell–target cell couples [data not shown; as also described earlier (24–26)], and in a much less extensive analysis of live cell actin accumulation in the anti-2B4 redirected lysis of P815 targets (data not shown). In summary, we found moderate actin accumulation at the NK cell–target cell interface, consistent with the often-tentative nature of the NK cell–target cell couples.
Fig. 2.
Actin accumulates differentially at the NK cell- and CTL-target cell interface. (a) A nonkilling interaction of an actin-GFP transduced LAK with a YAC-1 target cell is shown in panels from Movie 5. (Upper) A DIC image. (Lower) A projection of the 3D actin–GFP fluorescence data in a rainbow color scale (increasing from blue to red). The interface is marked with an arrow (Left). The NK cell is on top, and the target cell is on the bottom. A strong actin accumulation with an extended NK cell lamellopod can be seen in the third panel (3:00 min). One additional cell each is bound to the NK and the target cell. These interactions were not productive, because the cellular interfaces never reached two-thirds of the NK cell diameter. (b) A persistent interaction of an actin–GFP-transduced P14 CTL with an EL4 target cell is shown in panels from Movie 8, similar to a. The interface is marked with an arrow (Left); the CTL is on the bottom, the target cell on top.
Next, we investigated NK cell polarity by visualizing the MTOC with a tubulin-GFP fusion protein (15) similar to the actin–GFP approach (Fig. 3a and Movies 6 and 7, which is published as supporting information on the PNAS web site) (n = 54). As expected (27, 28), the MTOC was at the interface (Experimental Procedures) during the lytic hit (Fig. 3a, Movies 6 and 7). In rapid killing, it moved there in 1.0 ± 1.1 min after interface formation (Fig. 10A, which is published as supporting information on the PNAS web site). In delayed killing, MTOC reorientation was significantly (P = 0.001) delayed, occurring only 5.4 ± 4.4 min after interface formation (Fig. 10A), corroborating the tentative nature of the initial cell couples in delayed killing. In nonkilling interactions, a reorientation of the MTOC occurred in only 14% of the cell couples (Supporting Text), revealing a predominant lack of NK cell polarization. In summary, MTOC localization at the NK cell–target cell interface was also moderate, being mostly limited to the later stages of lytic interactions. Interestingly, it was not sufficient for target cell lysis, because the MTOC was located at the interface in several nonkilling interactions. In this context, we observed that the actin accumulation at the NK cell–target cell interface that accompanied target cell lysis occurred significantly (P < 0.05) (1.9 ± 2.6 min before target cell lysis) later than the MTOC orientation (4.9 ± 6.0 min) (Fig. 10B), suggesting that, after successful MTOC reorientation, an additional actin cytoskeletal polarization step might be required for the delivery of the lethal hit.
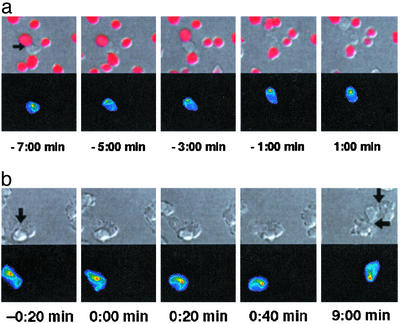
Fig. 3.
The MTOC orients toward the center of the interface differentially in NK cell– and CTL–target cell interactions. (a) A delayed killing interaction of a tubulin—GFP-transduced LAK with a YAC-1 target cell is shown in panels from Movie 6. (Upper) A DIC image is overlaid with a red intensity scale of the SNARF fluorescence to visualize membrane permeability. (Lower) A projection of the 3D tubulin–GFP fluorescence data is shown in a rainbow color scale (increasing from blue to red). The interface is marked with an arrow (Left). Target cell lysis is set to t = 0:00 min. NK cell–target cell contact without MTOC localization at the interface can be seen (Left). The MTOC reorients 5 min before target cell lysis (t = –5:00 min), remains at the interface until target cell lysis, and moves away from the interface after lysis (Right). (b) An interaction of a tubulin–GFP-transduced P14 CTL with an EL4 target cell is shown in sections derived from panels from Movie 9 similar to a but lacking the SNARF overlay. The cell couple is marked with an arrow (Left), the CTL is on the left, and the target cell is on the right. In the first four panels, a rapid reorientation of the CTL MTOC toward the center of the interface after interface formation (t = 0:00 min) can be seen. In the last frame, blebbing indicates substantial target cell damage; the MTOC, however, is still located at the center of the interface (bottom arrow). The CTL has formed a second interface (top arrow). A later reorientation of the MTOC toward the second interface can be seen in Movie 9.
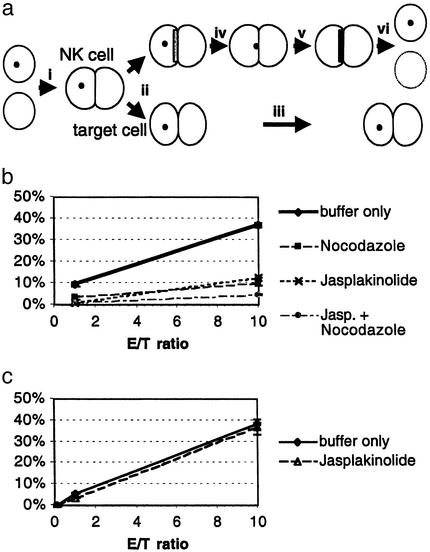
Table 1 summarizes how differential establishment of NK cell cytoskeletal polarity and signaling correlated with target cell fate. The more tentative the NK cell–target cell couple, the less likely the target cell was to be lysed. These data suggest that a series of sequential steps needs to be completed for successful progression of NK cell–target cell couples to target cell lysis (Fig. 4a). First, a tight interface (likely involving actin dynamics) needs to be maintained. Second, the MTOC has to orient toward the interface. Finally, actin accumulation at the interface occurs as part of the lytic hit. Although a requirement for MTOC orientation was expected (27, 28), it was surprising that it was only a single parameter in a series of cytoskeletal polarization steps. That the dynamics and extent of cytoskeletal polarization correlated with the effectiveness of target cell lysis strongly suggests that cytoskeletal polarization serves as a series of checkpoints in NK cell killing.
Table 1. More deliberate cytoskeletal polarization in models of NK cell vs. CTL killing.
| Assay | Lytic hit* | Tentative cell couple in delayed killing† | Tentative cell couple in nonkilling | CTL |
|---|---|---|---|---|
| Interface tightness | Tight | Tight | Narrow | Tight |
| Intracellular calcium | Elevated | Fluctuating | Fluctuating | Elevated |
| Actin accumulation | Always | Intermittent, more | Intermittent, less | Always |
| MTOC reorientation | Always | Belated | Rare | Always |
All of rapid and final phase of delayed killing
Applies to delayed killing prior to the lytic hit
Fig. 4.
Stepwise cytoskeletal polarization in effective NK cell killing. (a) A schematic representation of the stepwise polarization in NK cell killing is given. (i) NK cell killing begins with the formation of a tight NK cell–target cell couple. (ii) If the initial tight interface cannot be maintained (intermittent actin accumulation at the interface as depicted with a light gray bar is likely important for interface maintenance), (iii) the cell couple persists without target cell lysis. If a tight interface is maintained, (iv) the MTOC as depicted with a black dot can reorient toward the interface. (v) Actin accumulation at the interface as depicted with a dark gray bar always coincides with the lytic hit that (vi) leads to target cell lysis. (b) LAK-mediated YAC-1 killing was determined in a 1-h chromium-release assay (matching the duration of the corresponding microscopy experiments) as percent of specific lysis. For moderate interference with actin and microtubule dynamics, LAKs were treated with Jasplakinolide, Nocodazole, or both (Experimental Procedures). One representative of three assays is shown. (c) P14 CTL-mediated EL4 target cell killing was independent of a low concentration (Experimental Procedures)of Jasplakinolide, as in b.
Rapid Cytoskeletal Polarization in CTL–Target Cell Couples. Next, we investigated cytoskeletal polarization in CTL killing. In contrast to NK cells, the interface diameter in CTL-target cell couples was consistently large at 98 ± 10% of the T cell diameter (Figs. 6B, 7A, and 8 and Movies 3 and 4). Actin accumulated at the CTL–target cell interface immediately and remained there through most of the interface persistence (Supporting Text and Figs. 6B and 9; Movie 8, which is published as supporting information on the PNAS web site). Analyzing one time point per minute for the first 5 min after cell couple formation, all CTL–target cell couples showed strong actin accumulation at least once, 76% in more than half of the time points (Fig. 9). This decisive actin accumulation likely explains the superior ability of CTLs to maintain a tight effector–target cell interface. The CTL MTOC reoriented toward the CTL–target cell interface within 0.9 ± 0.7 min (n = 33) of cell couple formation and remained localized at the interface unless the CTL was moving to another target (Figs. 3B, 7, and 10A; Movie 9, which is published as supporting information on the PNAS web site). The rapid CTL polarization was seen over a wide range of concentrations of antigenic peptide and with different TCR transgenic T cell–APC combinations (data not shown and I. Tskvitaria-Fuller and C.W., unpublished results). To establish that decisive cell couple formation and cytoskeletal polarization are not limited to TCR transgenic models, we studied redirected lysis of DC2.4 dendritic cells by B6 CTLs (Experimental Procedures) by using an anti-CD3ε antibody. Killing as determined in a chromium-release assay was equal to P14 CTL-mediated target cell lysis and independent of FasL engagement (Fig. 6 and Fig. 11, which is published as supporting information on the PNAS web site). The elevation of the CTL intracellular calcium concentration on cell couple formation was immediate and sustained (data not shown). Ninety-five percent of the B6 CTL–DC2.4 target cell couples showed strong actin accumulation in at least one of five time points within 5 min of cell couple formation (Fig. 9). Although this percentage is slightly less than the 100% seen with P14 CTLs, it is substantially more than the 40% and 11% seen in NK cell–target cell interactions (Fig. 9). In our models of CTL–target cell as opposed to NK cell–target cell interactions, cell couple polarization was thus decisive (Table 1). These data are consistent with earlier reports on T cell actin and MTOC localization using fixed cell couples or modulated polarization microscopy (29–32).
NK Cell Killing Is Particularly Sensitive to Interference with Cytoskeletal Dynamics. The differential establishment of cytoskeletal polarity (stepwise in NK cells and immediate in CTLs) suggests that cytoskeletal polarization serves as a series of checkpoints in NK cell but not CTL killing. If this were true, a moderate interference with cytoskeletal dynamics should inhibit killing in NK cells but not CTLs. To test this hypothesis, we interfered with actin dynamics by using small concentrations of the inhibitor of actin depolymerization, Jasplakinolide (13, 33) (Experimental Procedures). As detailed in Supporting Text, this did not block but only slowed actin dynamics in both NK cells and CTLs, thus constituting a moderate interference.
As assayed in a chromium-release assay, this moderate Jasplakinolide treatment significantly reduced NK cell-mediated target cell lysis to 35 ± 18% of control (P < 0.05) (Fig. 4b and Fig. 12 A and B, which is published as supporting information on the PNAS web site), as consistent with reduced NK cell-mediated killing in humans that are deficient in the regulator of actin polymerization, Wiskott–Aldrich syndrome protein (26). The effect was of the same size as that a blocking MTOC reorientation with Nocodazole (Fig. 4b). A combination of Jasplakinolide and Nocodazole led to even less killing, consistent with a sequential limiting action of actin and microtubule dynamics (Figs. 4b and 12C). These data strongly suggest that the stepwise establishment of cytoskeletal polarity constitutes a series of checkpoints in a mechanism to regulate NK cell killing. In contrast, moderate interference with CTL cytoskeletal dynamics using Jasplakinolide or Nocodazole did not significantly impair CTL-mediated killing (Figs. 4c and 12 D and E), corroborating that cytoskeletal polarization is a robust feature of T cell activation. As expected, strong interference with cytoskeletal dynamics using high inhibitor concentrations substantially reduced both CTL and NK cell killing (Supporting Text and Fig. 12). The differential sensitivity of NK cell and CTL killing to moderate interference with cytoskeletal dynamics suggests that the establishment of cytoskeletal polarity is used as a series of checkpoints in NK cell but not CTL killing.
Discussion
In our experiments, NK cells established cytoskeletal polarity in a stepwise fashion, strongly suggesting a series of checkpoints. In contrast, the establishment of CTL polarity was rapid and robust. The majority of our data is based on the comparison of primary TCR transgenic CTLs (i.e., a homogeneous CTL population) with primary NK cells with more variable receptor expression. However, the consistent occurrence of the basic target cell variability in the three NK cell/target cell combinations studied here (Figs. 1e and 5 and Supporting Text) and in an additional clonal NK cell line (18) in combination with the corroboration of decisive CTL actin dynamics using B6 CTLs in redirected lysis (Fig. 9) suggest that the data are valid beyond the effector–target combinations investigated here. Fc receptor-mediated NK cell activation as an adaptive effector function in antibody-dependent cell-mediated cytotoxicity will need to be investigated separately. The stepwise establishment of NK cell polarity is reminiscent of the succession of different receptor/ligand couple patterns seen in the interaction of T cells with activating supported lipid bilayers (34).
The concept of stepwise cytoskeletal polarization as a series of NK cell checkpoints helps to understand NK cell cytolysis as part of the innate immune response. CTLs as part of the adaptive immune response are subject to a variety of cell proliferation and differentiation checkpoints to ensure that only those cells that specifically recognize antigen-presenting targets become part of the effector pool, thus effector availability is delayed by several days. NK cells as part of the innate immune response do not undergo such an education process. Therefore, their effector functions are available within hours after antigen exposure. However, immune recognition specificity is coarse and not double-checked in proliferation and differentiation. Effector CTLs thus can afford to establish cytoskeletal polarity in an immediate and robust manner without further checkpoints, because the chance of their responding vigorously to anything other than their intended target is slim. NK cells, however, have to be more restrained, requiring additional checkpoints after the formation of an NK cell–target cell couple to compensate for their coarse immune recognition specificity. Our data strongly suggest that stepwise cytoskeletal polarization provides such checkpoints. This could well be the consequence of signals from activation receptors being balanced by inhibitory ones (3–6). The strong role of inhibitory receptors in NK cell biology might thus reflect the need for balanced signaling in a series of checkpoints to prevent erroneous killing.
Supplementary Material
Acknowledgments
We thank Michael Bennett, David Farrar, and Iwona Stroynowski for helpful discussions; Pamela Verrett for expert technical assistance; Gina Costa and Gary Nolan for advice in establishing the retroviral transduction; Irina Tskvitaria-Fuller for help in the production of retroviral supernatants; Beat Imhof for the actin–GFP and tubulin–GFP constructs; and Nicolai van Oers for the breeding of P14 mice. This work was supported in part by the National Institutes of Health (to J.D.S.) and the Welch Foundation (to C.W.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NK, natural killer; CTL, cytotoxic T cell; LAK, lymphokine-activated killer; MTOC, microtubule organizing center; DIC, differential interference contrast.
References
- 1.Trinchieri, G. (1989) Adv. Immunol. 47, 187-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janeway, C. A., Jr., & Medzhitov, R. (2002) Annu. Rev. Immunol. 20, 197-216. [DOI] [PubMed] [Google Scholar]
- 3.Yokoyama, W. M. (1995) Curr. Opin. Immunol. 7, 110-120. [DOI] [PubMed] [Google Scholar]
- 4.Lanier, L. L. (1998) Annu. Rev. Immunol. 16, 359-393. [DOI] [PubMed] [Google Scholar]
- 5.Moretta, A., Biassoni, R., Bottino, C., Mingari, M. C. & Moretta, L. (2000) Immunol. Today 21, 228-234. [DOI] [PubMed] [Google Scholar]
- 6.Garni-Wagner, B. A., Purohit, A., Mathew, P. A., Bennett, M. & Kumar, V. (1993) J. Immunol. 151, 60-70. [PubMed] [Google Scholar]
- 7.Arase, H., Mocarski, E. S., Campbell, A. E., Hill, A. B. & Lanier, L. L. (2002) Science 296, 1323-1326. [DOI] [PubMed] [Google Scholar]
- 8.Germain, R. N. (2001) Science 293, 240-245. [DOI] [PubMed] [Google Scholar]
- 9.Lauwerys, B. R., Renauld, J. C. & Houssiau, F. A. (1999) Cytokine 11, 822-830. [DOI] [PubMed] [Google Scholar]
- 10.Kagi, D., Vignaux, F., Ledermann, B., Burki, K., Depraetere, V., Nagata, S., Hengartner, H. & Golstein, P. (1994) Science 265, 528-530. [DOI] [PubMed] [Google Scholar]
- 11.Wülfing, C., Sumen, C., Sjaastad, M. D., Wu, L. C., Dustin, M. L. & Davis, M. M. (2002) Nat. Immunol. 3, 42-47. [DOI] [PubMed] [Google Scholar]
- 12.Pircher, H., Burki, K., Lang, R., Hengartner, H. & Zinkernagel, R. M. (1989) Nature 342, 559-561. [DOI] [PubMed] [Google Scholar]
- 13.Cramer, L. P. (1999) Curr. Biol. 9, 1095-1105. [DOI] [PubMed] [Google Scholar]
- 14.Ballestrem, C., Wehrle-Haller, B. & Imhof, B. A. (1998) J. Cell Sci. 111, 1649-1658. [DOI] [PubMed] [Google Scholar]
- 15.Ballestrem, C., Wehrle-Haller, B., Hinz, B. & Imhof, B. A. (2000) Mol. Biol. Cell 11, 2999-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costa, G. L., Benson, J. M., Seroogy, C. M., Achacoso, P., Fathman, C. G. & Nolan, G. P. (2000) J. Immunol. 164, 3581-3590. [DOI] [PubMed] [Google Scholar]
- 17.Taylor, M. A., Bennett, M., Kumar, V. & Schatzle, J. D. (2000) J. Immunol. 165, 5048-5053. [DOI] [PubMed] [Google Scholar]
- 18.Eriksson, M., Leitz, G., Fallman, E., Axner, O., Ryan, J. C., Nakamura, M. C. & Sentman, C. L. (1999) J. Exp. Med. 190, 1005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanderson, C. J. (1981) Biol. Rev. Camb. Philos. Soc. 56, 153-197. [DOI] [PubMed] [Google Scholar]
- 20.Poenie, M., Tsien, R. Y. & Schmitt-Verhulst, A. M. (1987) EMBO J. 6, 2223-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis, D. M., Chiu, I., Fassett, M., Cohen, G. B., Mandelboim, O. & Strominger, J. L. (1999) Proc. Natl. Acad. Sci. USA 96, 15062-15067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eriksson, M., Ryan, J. C., Nakamura, M. C. & Sentman, C. L. (1999) Immunology 97, 341-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vyas, Y. M., Maniar, H. & Dupont, B. (2002) J. Immunol. 168, 3150-3154. [DOI] [PubMed] [Google Scholar]
- 24.Radosevic, K., van Leeuwen, M. T., Segers-Nolten, I. M., Figdor, C. G., de Grooth, B. G. & Greve, J. (1994) Cytometry 15, 320-326. [DOI] [PubMed] [Google Scholar]
- 25.Carpen, O., Virtanen, I., Lehto, V. P. & Saksela, E. (1983) J. Immunol. 131, 2695-2698. [PubMed] [Google Scholar]
- 26.Orange, J. S., Ramesh, N., Remold-O'Donnell, E., Sasahara, Y., Koopman, L., Byrne, M., Bonilla, F. A., Rosen, F. S., Geha, R. S. & Strominger, J. L. (2002) Proc. Natl. Acad. Sci. USA 99, 11351-11356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kupfer, A., Dennert, G. & Singer, S. J. (1983) Proc. Natl. Acad. Sci. USA 80, 7224-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sancho, D., Nieto, M., Llano, M., Rodriguez-Fernandez, J. L., Tejedor, R., Avraham, S., Cabanas, C., Lopez-Botet, M. & Sanchez-Madrid, F. (2000) J. Cell Biol. 149, 1249-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sedwick, C. E., Morgan, M. M., Jusino, L., Cannon, J. L., Miller, J. & Burkhardt, J. K. (1999) J. Immunol. 162, 1367-1375. [PubMed] [Google Scholar]
- 30.Kupfer, A. & Singer, S. J. (1989) Annu. Rev. Immunol. 7, 309-337. [DOI] [PubMed] [Google Scholar]
- 31.Kuhn, J. R. & Poenie, M. (2002) Immunity 16, 111-121. [DOI] [PubMed] [Google Scholar]
- 32.Stowers, L., Yelon, D., Berg, L. J. & Chant, J. (1995) Proc. Natl. Acad. Sci. USA 92, 5027-5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bubb, M. R., Senderowicz, A. M., Sausville, E. A., Duncan, K. L. & Korn, E. D. (1994) J. Biol. Chem. 269, 14869-14871. [PubMed] [Google Scholar]
- 34.Grakoui, A., Bromley, S. K., Sumen, C., Davis, M. M., Shaw, A. S., Allen, P. M. & Dustin, M. L. (1999) Science 285, 221-226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.