Abstract
Mammalian natural killer (NK) cells are cytotoxic lymphocytes that express receptors specific for MHC class I molecules. The NK cell receptors belong to two structurally unrelated families, the killer cell Ig-like receptors and the killer cell C-type lectin receptors. We describe a cDNA clone derived from the bony (cichlid) fish Paralabidochromis chilotes and show that it encodes a protein related to the CD94/NK cell group 2 (NKG2) subfamily of the killer cell C-type lectin receptors. The gene encoding this receptor in a related species, Oreochromis niloticus, has a similar structure to the human CD94/NKG2 genes and is a member of a multigene cluster that resembles the mammalian NK cell gene complex. Thus, the CD94/NKG2 subfamily of NK cell receptors must have arisen before the divergence of fish and tetrapods and may have retained its function (possibly monitoring the expression of MHC class I molecules) for >400 million years.
Natural killer (NK) cells are ontogenetically related to lymphocytes but differ from the latter in that they do not express antigen-specific clonally distributed receptors. Instead, they display on their surfaces sets of glycoproteins called NK cell receptors, which recognize MHC class I or class I-like molecules on certain tumors and cells infected with viruses, intracellular bacteria, or other intracellular parasites (1, 2). Via their receptors, NK cells monitor the expression of class I molecules on the target cells and respond when it is lowered by tumorigenesis, infection, or some other incursion. The response can be either stimulatory (3) or inhibitory (4). The stimulatory response initiates an intracellular signaling pathway that leads to cytokine production and activation of cytotoxic mechanisms directed at the target cell. The inhibitory response generates negative signals that keep the cytotoxic mechanisms on a leash. Because individual NK cells possess both stimulatory and inhibitory receptors, the ultimate response of a given cell depends on the balance between positive and negative signals.
Structurally, the NK cell receptors fall into two broad evolutionarily unrelated classes, the killer cell Ig-like receptors (KIRs) and the killer cell C-type lectin receptors (KLRs). The KIRs are type I transmembrane glycoproteins containing one to three tandem Ig-like domains in their extracellular part. The KLRs are type II transmembrane homo- or heterodimers possessing an extracellular C-type lectin-like domain, which, however, lacks Ca2+-binding sites characteristic of true C-type lectins (5). Both KIRs and KLRs can be of either stimulatory or inhibitory type. Stimulatory receptors possess a positively charged residue (Lys or Arg) within their transmembrane regions, which interacts with a negatively charged residue (Asp) of an adaptor molecule such as DAP, CD3ζ, or FcRγ. Adaptor molecules contain an immunoreceptor tyrosine-based activating motif, which, when phosphorylated at the tyrosine residue, displays a docking site for protein tyrosine kinases and thus initiates the positive signaling pathway. Inhibitory receptors contain one or two immunoreceptor tyrosine-based inhibitory motifs, which, on phosphorylation of the tyrosine residue, recruit an src-homology domain containing tyrosine phosphatase and thus initiate the negative signaling pathway.
The genes encoding human killer cell Ig-like receptors are clustered in the leukocyte receptor complex on chromosome 19q13.42, together with related Ig-like transcript and leukocyteassociated inhibitory receptor genes (6). Similarly, the human KLR-encoding genes are clustered in the NK cell gene complex (NKC) on chromosome 12p12-p13. The NKC includes a family of genes denoted KLRs or NK cell group 2 (NKG2) receptors, with the individual members distinguished by the letters A, C, D, E, and F. In addition to these two clusters, other structurally diverse NK cell receptors are encoded in genes scattered over other human chromosomes.
At the time this study was submitted for publication, NK cell receptors had been identified in mammals only. A group of novel immune-type receptor genes has been described in teleost fishes (7, 8). The individual members of this group appear to be distantly related to some of the mammalian receptor genes outside of the leukocyte receptor complex (LRC) and NKC, but it is unclear whether they are bona fide NK cell receptors. Here we present evidence for the existence in teleostean fishes of genes homologous to the mammalian KLR genes.
Materials and Methods
Molecular Methods. RNA extraction, preparation of a cDNA library, PCR, and RT-PCR were carried out as described elsewhere (9). Total RNA was extracted from the head part of the teleostean fish Paralabidochromis chilotes (family Cichlidae, order Perciformes) with the RNeasy Midi Kit (Qiagen, Hilden, Germany) and used to prepare a directed cDNA library with the SMART cDNA Library Construction Kit (BD Biosciences CLONTECH, Heidelberg, Germany). PCR products obtained by the amplification of the cDNA library were purified and sequenced by the custom sequencing service of Agowa (Berlin) by using the primer pTripl5Seq2 (5g′-GAAGCGCGCCATTGTGTT-3′), which anneals next to the 5′ end of the insert cDNA. Insert sequences were subjected to a blastx search against the nonredundant protein database at the National Center for Biotechnology Information. A candidate clone showing sequence similarity with human CD94 was then subjected to further analysis. SuperScript One-Step RT-PCR with Platinum TaqDNA polymerase (Invitrogen) was carried out according to the manufacturer's instructions.
Screening of Bacterial Artificial Chromosome (BAC) Library. A BAC library with an average insert size of 150 kb was prepared by Katagiri et al. (10) from sperm cells of Oreochromis niloticus, another cichlid fish, related to P. chilotes. A total of 56,540 colonies was plated, and Hybond N+ nylon membranes (Amersham Pharmacia Biosciences) were then prepared from each plate and hybridized with a 32P-labeled full-length cDNA clone used as a probe.
Separation of Red and White Blood Cells. Blood (500–1,000 μl) was drawn from the ventral aorta of P. chilotes into a syringe containing the anticoagulant citrate dextrose (Sigma). On addition of the same volume of Percoll (Sigma), the blood sample was centrifuged at 1,700 × g for 4 min at 4°C. The supernatant containing the white blood cells was transferred into a buffer solution, centrifuged at 1,800 × g for 15 min at 4°C, and the pellet representing the white blood cell fraction was collected (11). The erythrocyte fraction was washed once with buffer before use.
Sequence and Phylogenetic Analysis. Sequences were subjected to blastx and blastp searches against the nonredundant protein database at the National Center for Biotechnology Information. Evolutionary relationships were evaluated by using P distances or Poisson correction distances between amino acid sequences, and phylogenetic trees were drawn by the neighbor-joining algorithm (12) by using the PAUP4* program (13). Unalignable parts of the sequences and positions with indels were excluded from the analysis. The reliability of the branching order was determined by 1,000 bootstrap replications. Transmembrane regions were predicted by using the TMpred server at www.ch.embnet.org/software/TMPRED_form.html; the TMHMM server, Ver. 2.0, at www.cbs.dtu.dk/services/TMHMM-2.0; and the TopPred server at http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html. Secondary structures were predicted by using the FUGUE version 2.s.05 program at www-cryst.bioc.cam.ac.uk/∼fugue (14).
Results
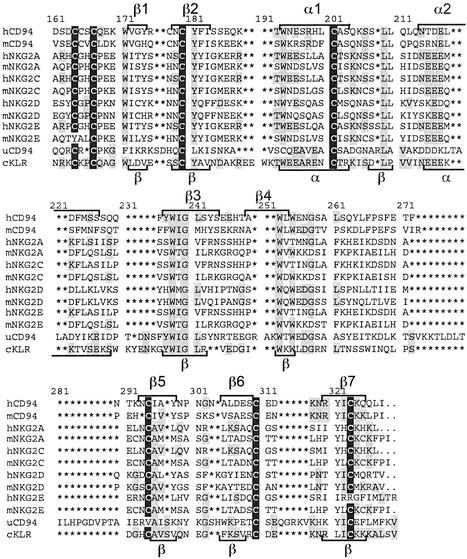
Characterization of a cDNA Clone. An expressed sequence tag was obtained from a cDNA library prepared from the cichlid fish P. chilotes. The sequence represented the full-length transcript of a gene encoding a protein homologous to a mammalian KLR. The 1,580-bp-long cDNA clone contained an ORF of 726 bp, potentially specifying a 242-residues-long polypeptide (Fig. 1). Hydrophilicity analysis (15) of the predicted sequence identified a putative transmembrane region near the N terminus (residues 24–45), suggesting that the gene encodes a type II integral membrane protein. An alignment of this protein with representative mammalian NK cell receptors of the KLR family (Fig. 2) and a phylogenetic tree based on this alignment (Fig. 3) revealed the cichlid protein to be most closely related to the mammalian CD94 and NKG2 group of receptors. Because the orthology of the cichlid protein to a particular member of the mammalian KLRs cannot be determined (see below and Discussion), we refer to it as cichlid cKLR. Secondary structure prediction (14) combined with comparison to the crystal structure of CD94 (16) and NKG2D (17, 18) suggested a division of the cKLR into the following regions (numbers in parentheses give length in amino acid residues of the cKLR and human CD94 proteins): cytoplasmic (23, 10), transmembrane (22, 24), stalk (64, 30), and carbohydrate-recognition domain (CRD; 123, 125). The CRD is predicted to contain seven β strands and two α helices arranged in the order β1, β2, α1, α2, β3, β4, β5, β6, β7, with the individual elements separated by loops (Fig. 2). The putative stalk region of the cKLR is unusually long (64 residues) compared with the human CD94 (26 residues) and NKG2A (22–24 residues) receptors.
Fig. 1.
Nucleotide sequence of a cDNA clone isolated from a P. chilotes library. UTR sequences are displayed without gaps, whereas the coding sequence is divided into codons and is translated into amino acid residues using the International Union of Pure and Applied Chemistry single-letter code. The transmembrane region (TMR) is highlighted, and the beginning of the CRD is indicated by an arrow. (Here we place the beginning of the CRD to coincide with the beginning of exon 5; some authors place it at Trp-120.) Exon–intron borders are marked by inverted filled triangles. The putative polyadenylation signal is underlined.
Fig. 2.
Amino acid alignment of P. chilotes cKLR sequence with human (h), mouse (m), and urochordate (u) CD94 and NKG2 sequences (CRD only). Asterisks indicate gaps introduced to optimize the alignment. Predicted (cKLR) or experimentally determined (other sequences; see refs. 17–19) elements of secondary structure are indicated by brackets (α, α helices; β, β strands). Conserved cysteine residues are highlighted in black; residues that cKLR shares with other sequences are highlighted in gray. The hNKG2E and uCD94 sequences are truncated at the C terminus. GenBank accession nos. are as follows: hCD94, Q13241; mCD94, NP_034784; hNKG2A, P26715; mNKG2A, AAD40221; hNKG2C, P26717; mNKG2C, NP_034783; hNKG2D, P26718; mNKG2D, XP_132877; hNKG2E, LAA04923; mNKG2E, AAF24982; and uCD94, AAO19649.
Fig. 3.
Phylogenetic (neighbor-joining) tree of human, mouse (m), fowl (f; chicken), baboon (b), urochordate (u), and cichlid (c) CRD sequences of C-type lectins. Numbers at nodes are bootstrap values obtained in 1,000 replications (only values >50% are indicated). C-type lectin groups are indicated by brackets and roman numbers. Sequence designations and accession codes are: Brevican, NP_068767; Versican, NP_004376; Aggrecan, P16112; Neurocan, NP_004377; CD23, NP_001993; mKupffer cell receptor, NP_058031; LANGERIN, NP_056532; fHepatic lectin, P02707; mCollectin 12, NP_569716; mMgl1, NP_034926; mMgl2, AAM52097; HML2, NP_006335; ASGR1, NP_001662; ASGR2, NP_001172; MINCLE, NP_055173; DCIR, NP_057268; DLEC, NP_569708; mDectin-2, NP_064385; mMCL, NP_034949; CLEC6, NP_525126; CD209, NP_066978; CD209L, NP_055072; MBL2, NP_000233; SELE, NP_000441; PRG3, NP_006084; NKG2A, P26715; NKG2C, P26717; NKG2E, Q07444; CD94, Q13241; KLRG1, AAC32200; KLRB1, AAA21605; KLRF1, NP_057607; mLy49 (Klra1), NP_057868; bLy49, AAK26161; CLL1, NP_612210; CLEC2, NP_057593; CLEC1, NP_057595; OLR1, NP_002534; DECTIN1, AAK20144; NKG2D, NP_031386; MDL1, NP_037384; uCD94, AAO19649; AICL, NP_005118; LLT1, NP_037401; and CD69, NP_001772.
The length of the stalk is largely responsible for the greater total length of the cKLR (242 residues) in comparison to most of the human receptors (179–240 residues). No immunoreceptor tyrosine-based inhibitory motifs characteristic of the inhibitory NK cell receptors could be identified in the cytoplasmic region, suggesting that the cichlid receptor might associate with an immunoreceptor tyrosine-based activating motif-bearing adaptor molecule. Consistent with this proposition is the presence of a positively charged residue (Arg-42) in the transmembrane region (in addition to the conserved Lys-23 at the transition from the cytoplasmic to the transmembrane region). The cichlid protein contains nine cysteine residues, of which six can be expected (in analogy with the human CD94/NKG2 proteins; see ref. 16) to form three pairs of intrachain disulfide bonds. The pairs correspond to the Cys-61–Cys-72, Cys-89–Cys-174, and Cys-152–Cys-166 pairs of the human CD94 protein. The seventh conserved cysteine residue of the fish sequence corresponds to Cys-58 of the human CD94 receptor and thus presumably participates in interchain bond formation of a dimeric molecule. The remaining two Cys residues (one in the cytoplasmic and the other in the transmembrane regions) are conserved in some but not all human receptors. The mammalian CD94 molecule possesses a fourth Cys pair (Cys-59–Cys-70), involved in intrachain bond formation (16), which is absent in the cichlid protein (and in mammalian NKG2 receptors). Thus, the cKLR possesses characteristics of both the mammalian CD94 and NKG2 proteins.
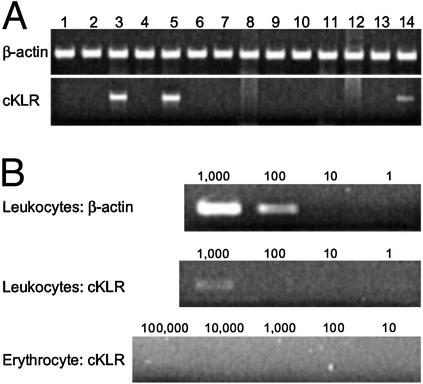
Expression of cKLR Genes. RNAs were extracted from the following frozen and homogenized tissues of an adult P. chilotes individual: brain, eye, pharyngeal jaw, heart, gill, stomach, liver, spleen, intestine, gas bladder, kidney, testes, bladder, and blood. The RNAs were subjected to RT-PCR by using pairs of primers specific for different segments of the cKLR gene. Amplification with primers specific for β-actin were included in each experiment as a positive control. In each pair, the two primers were complementary to different exons, so that the products of genomic DNA and cDNA amplifications could be distinguished by their size. Of the 14 different tissues examined, only blood and pharyngeal region tissues (gills and pharyngeal jaw) tested positive (Fig. 4A). When blood cells were separated into erythrocyte and leukocyte fractions, only the latter tested positive (Fig. 4B). In a serial dilution experiment, aliquots containing 1,000 white blood cells were positive by RT-PCR, whereas aliquots containing 100 cells were negative (but positive for β-actin transcripts). The identity of the amplification products was determined by cloning and sequencing. Two types of cKLR sequences were found in the products: one was identical with the original expressed sequence tag clone, and the other showed 95% nucleotide sequence identity with this clone. These results indicate that the cKLR gene is expressed most strongly in a subpopulation of blood leukocytes. The positivity of the gill and pharyngeal jaw RNA extracts might also be attributed to leukocytes in these tissues, lymphatics being particularly abundant in this part of a fish body.
Fig. 4.
(A) The results of RT-PCR with RNA extracts from brain (lane 1), eye (lane 2), pharyngeal jaws (lane 3), heart (lane 4), gill (lane 5), stomach (lane 6), liver (lane 7), spleen (lane 8), intestine (lane 9), gas bladder (lane 10), kidney (lane 11), testes (lane 12), bladder (lane 13), and blood (lane 14). (B) The result of RT-PCR with blood leukocytes and erythrocytes. β-Actin amplification is shown as a positive control. Numbers of cells are given above individual lanes. Fragment sizes are 685 and 296 bp for cKLR and β-actin, respectively. The RT-PCR primers were ND21 (GGACAAGGAAGCTGCACGTGAAG) and ND4 (CTTCGTTTTGGACAGAGACTGCACA) for cKLR, and BA3 (CCCAAAGTTCAGCCATGGAAGATG) and BA4 (CAGCTCGTTGTAGAAGGTGTGATG) for β-actin.
Gene Structure. To characterize the cKLR-encoding gene, we used a BAC library prepared from a related species, O. niloticus. Screening the library with a probe encompassing the entire coding part of the cDNA clone yielded six positive clones, of which three turned out to be identical. DNA isolated from one of the four different clones (no. 6) was used to prepare a shotgun library, which was then screened with a P. chilotes KLR exons 3 and 5 probe. Positive subclones were sequenced, and the exons were identified by comparison with the cDNA sequence.
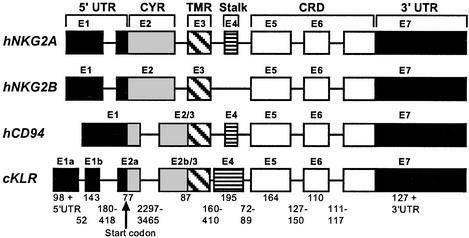
The cKLR gene consists of eight exons (Fig. 5). Exons 1a and 1b together encode the bulk of the 5′ UTR; they correspond to exon 1 of the human NKG2A gene (19). Exon 2a specifies the remainder of the 5′ UTR and part of the cytoplasmic region. Exon 2b/3 encodes the rest of the cytoplasmic region and corresponds to exons 2/3 of the human CD94 gene (ref. 20; in the human NKG2A gene, the entire cytoplasmic region, together with part of the 5′ UTR, is encoded in exon 2, and the transmembrane region is encoded in exon 3; see Fig. 5 and ref. 19). Exon 4 specifies the stalk and exons 5–7 the CRD (exon 7 also specifies part of the 3′ UTR). Hence, compared with human CD94, the cKLR-encoding gene has an extra intron that splits exon 2, whereas intron 2 of the NKG2A gene is eliminated in both the cichlid and human CD94 genes.
Fig. 5.
Exon–intron organization of cichlid (c) KLR, human (h) NKG2, and human (h) CD94 genes. Exons (E) are indicated by numbered boxes and introns by intervening lines. Lengths of exons are approximately proportional to the widths of the boxes; those of introns are not. Lengths (in base pairs) of cKLR exons and introns are as specified. The exon parts specifying different domains of the protein are distinguished by shading: CYR, cytoplasmic region; TMR, transmembrane region. The organization of the human genes is taken from refs. 20 and 21.
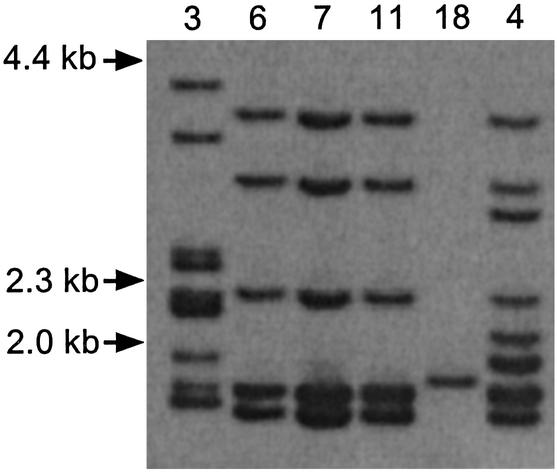
Clustering of cKLR Loci. To obtain information about other cKLR loci, the four distinct cKLR-bearing clones were digested with the MspI endonuclease, the digest was separated by electrophoresis, blotted according to the standard Southern blotting protocol, and the blots were hybridized with the cKLR exon 5-specific probe. The number of hybridizing bands ranged from one (clone 18) to nine (clone 3) (Fig. 6), indicating that the O. niloticus genome contains a region comprised of at least nine cKLR loci.
Fig. 6.
Southern blot hybridization of DNA extracted from O. niloticus BAC clones (numbers at the top), digested with MspI, and hybridized with a cKLR exon 5 probe. Sizes in kilobases are indicated (Left). The lower part of the gel showing a 0.6-kb band has been left out to save space.
DNA isolated from clones 3, 4, 6, and 18 was then subjected to PCR by using degenerate primers to amplify the conserved segment extending from exons 5 to 7 and including the short intervening introns (see Fig. 5). The amplification products were subcloned, the inserts of the individual clones were reamplified, the products of these secondary PCRs were separated by electrophoresis on a single-stranded conformational polymorphism gel (21, 22), and fragments identified as distinct were sequenced. Twenty-five different sequences were obtained (Fig. 7). Because 10 unique sequences were yielded by single clones (nos. 4 and 6), this means that at least 10 KLR-like loci are clustered on a single BAC encompassing ≈150 kb of sequence. The actual number of KLR loci in the cichlid genome is probably higher, because three of the four different BAC clones overlapped partially (only clone 18 did not overlap with these three; unpublished data), and they yielded different sequences. Apparently the KLR region of O. niloticus is at least as complex as the mammalian NKC.
Fig. 7.
Amino acid alignment of cKLR carbohydrate-recognition domain sequences obtained from indicated BAC clones (C, followed by a number) of O. niloticus. Simple majority consensus sequence is shown at the top; identity with the consensus is indicated by a dash (–). Dots indicate absence of information; asterisks, gaps introduced to optimize the alignment; and exclamation marks, stop codons. The segment shown corresponds to amino acid residues 173–285 in the numbering system of Fig. 2.
Discussion
C-type lectins are currently classified into seven groups, distinguished, in addition to sequence differences, by the number of CRDs, the types of domains they possess in addition to the CRDs, the presence or absence of Ca2+-binding sites (and hence their ability to bind carbohydrates), the orientation of the polypeptide chain relative to the plasma membrane, the presence or absence of immunoreceptor tyrosine-based inhibitory motifs and immunoreceptor tyrosine-based activating motifs, the organization of the lectin-encoding genes, and other features (23). On the basis of these criteria, the cKLR sequences clearly do not belong to groups I, III, IV, VI, or VII. This leaves groups II and V open for consideration. Lectins in these two groups are characterized by being type II integral membrane proteins with an intracellular N terminus and extracellular C terminus, by the possession of a single CRD separated from the transmembrane region by a stalk (neck) of variable length and by the lack of other domains (except the transmembrane and cytoplasmic regions). The two groups differ in that the group II proteins possess two Ca2+-binding sites and hence have carbohydrates as their ligands, whereas group V proteins have lost these sites. However, the situation is complicated by the existence of a set of molecules that fall between groups II and V in terms of their properties. These intermediates are closer to group II than to group V in that they possess at least one of the two Ca2+-binding sites. Comparison of the cKLR sequences with those of other C-type lectins reveals that the residues associated with the Ca2+-binding sites are not conserved in any of those identified thus far (not shown). Hence the cKLRs are group V lectins, and this assignment is supported by phylogenetic analysis of their sequences (Fig. 3). Group V contains all of the known C-type lectin NK cell receptors, but it also contains several other genes that have other functions. These genes (with one exception) are members of a cluster located in a single region, the NKC, on chromosome 12 in humans and chromosome 6 in the mouse (3, 4). The two categories of genes differ in their expression, the NK cell receptor genes being expressed predominantly on blood NK cells (i.e., lymphocytes), whereas the other genes are active in a variety of other cells (macrophages, monocytes, dendritic cells, and endothelial cells). The expression of the cKLR genes on a subfraction of blood leukocytes and their nondetectability in spleen and liver place them in the NK cell category of genes.
This placement is also supported by their exon–intron organization: the group V NK cell receptor genes tend to consist of more than six exons, and the cKLR genes each consist of seven exons. Most importantly, however, the assignment of the cKLRs into the category of group V NK cell receptors is supported by phylogenetic analysis. Although the bootstrap support for this assignment is low to moderate, the grouping emerges consistently in trees drawn by different methods (not shown) and does not strongly depend on the type of sequences included in or excluded from the analysis. We conclude, therefore, that all of the known cKLR genes are most closely related to the mammalian genes encoding group V NK cell receptors. Whether the products of these genes really function as NK cell receptors can be answered only by functional tests.
Although the phylogenetic relationship of the cKLRs to the mammalian CD94/NKG2 set of receptors is strongly indicated by our data, orthology to a particular receptor is not. The analysis of our data suggests that the fish and mammalian clusters of NK cell receptor genes arose independently by duplications from different ancestral genes. If so, all of the phylogenetic relationships between the fish and human genes in these two clusters are of the paralogous rather than orthologous type. While this manuscript was under review, an article appeared (24) in which the authors claim to have cloned a urochordate ortholog of the human CD94 gene. The inclusion of their sequence in the tree containing the fish sequence reveals, however, that although it is a group V C-type lectin, the urochordate protein is not orthologous to the human CD94 protein (Fig. 3).
The driving force behind the instability of the NKC is believed to be coevolution of the receptors and their ligands (6). In mammals, the ligands of the CD94/NKG2 receptors are the nonclassical class Ib MHC molecules (25). In the case of the CD94/NKG2A dimer, the ligands are HLA–E in humans (26–28) and Qa1b in mice (27, 29). These two molecules are not orthologous to each other and are not particularly similar in their sequence. Nevertheless, they are related functionally in that both bind peptides derived from the signal sequence of proteins encoded in many HLA-A, -B, -C, and -G alleles in the case of HLA–E (30) and the H2D and -L alleles in the case of Qa1b (31). Because cell surface expression of class I molecules depends on the presence of peptides attached to their peptide-binding regions, it has been suggested (32) that the NK cell recognition of HLA–E and Qa1b molecules loaded with MHC class I-derived signal peptides is a mechanism for monitoring the expression of class I molecules. It has been speculated that the interaction of human CD94 molecules with HLA-E is mediated by a cluster of charged amino acid residues, including Glu-123, Glu-124, Glu-145, Lys-150, Asp-163, Glu-164, Glu-167, Asp-168, Lys-169, and Arg-171 (16). Several of these residues are conserved at equivalent positions in the cKLR molecules. Hence, if CD94/NKG2 receptors indeed have a class I-monitoring function, one might speculate that the function emerged early in the evolution of the MHC molecules. This hypothesis could explain why the CD94/NKG2 subfamily of NK receptors has retained sequence similarity among taxa as distant as mammals and fishes, whereas other subfamilies have apparently not been conserved even between different mammalian orders (6). It will therefore be interesting to determine the homologies of the other genes in the cichlid KLR cluster.
Acknowledgments
We thank Dr. T. Aoki (Laboratory of Genetics and Biochemistry, Tokyo University of Fisheries, Tokyo) for giving us access to the O. niloticus BAC library; Dr. Naoko Takezaki for help with phylogenetic analysis; Sabine Rosner and Nadine Heckmann for technical assistance; and Jane Kraushaar for help with the preparation of the manuscript.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BAC, bacterial artificial chromosome; CRD, carbohydrate-recognition domain; KLR, killer cell C-type lectin receptor; cKLR, cichlid KLR; NK, natural killer; NKC, NK cell gene complex; NKG2, NK cell group 2 receptors.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY297060–AY297086).
References
- 1.Biassoni, R., Cantoni, C., Falco, M., Pende, D., Millo, R., Moretta, L., Bottino, C. & Moretta, A. (2000) Mol. Immunol. 37, 1015-1024. [DOI] [PubMed] [Google Scholar]
- 2.Borrego, F., Kabat, J., Kim, D.-K., Lieto, L., Maasho, K., Pena, J., Solana, R. & Coligan, J. E. (2001) Mol. Immunol. 38, 637-660. [DOI] [PubMed] [Google Scholar]
- 3.Lanier, L. L. (2001) Nat. Immunol. 2, 23-27. [DOI] [PubMed] [Google Scholar]
- 4.Ravetch, J. V. & Lanier, L. L. (2000) Science 290, 84-89. [DOI] [PubMed] [Google Scholar]
- 5.Drickamer, K. & Taylor, M. E. (1993) Annu. Rev. Cell Biol. 9, 237-264. [DOI] [PubMed] [Google Scholar]
- 6.Barten, R., Torkar, M., Haude, A., Trowsdale, J. & Wilson, M. J. (2001) Trends Immunol. 22, 52-57. [DOI] [PubMed] [Google Scholar]
- 7.Strong, S. J., Mueller, M. G., Litman, R. T., Hawke, N. A., Haire, R. N., Miracle, A. L., Rast, J. P., Amemiya, C. T. & Litman, G. W. (1999) Proc. Natl. Acad. Sci. USA 96, 15080-15085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Litman, G. W., Hawke, N. A. & Yoder, J. A. (2001) Immunol. Rev. 181, 250-259. [DOI] [PubMed] [Google Scholar]
- 9.Mayer, W. E., Uinuk-ool, T., Tichy, H., Klein, J. & Cooper, M. D. (2002) Proc. Natl. Acad. Sci. USA 99, 14350-14355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katagiri, T., Asakawa, S., Minagawa, S., Shimizu, N., Hirono, I. & Aoki, T. (2001) Anim. Genet. 32, 200-204. [DOI] [PubMed] [Google Scholar]
- 11.Overath, P., Ruoff, J., Stierhof, Y.-D., Haag, J., Tichy, H., Dyková, I. & Lom, J. (1998) Parasitol. Res. 84, 343-347. [DOI] [PubMed] [Google Scholar]
- 12.Saitou, N. & Nei, M. (1987) Mol. Biol. Evol. 4, 406-425. [DOI] [PubMed] [Google Scholar]
- 13.Swofford, D. L. (2002) paup*: Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA), Ver. 4.0b 10.
- 14.Shi, J., Blundell, T. L. & Mizuguchi, K. (2001) J. Mol. Biol. 310, 243-257. [DOI] [PubMed] [Google Scholar]
- 15.von Heijne, G. (1992) J. Mol. Biol. 225, 487-494. [DOI] [PubMed] [Google Scholar]
- 16.Boyington, J. C., Riaz, A. N., Patamawenu, A., Coligan, J. E., Brooks, A. G. & Sun, P. D. (1999) Immunity 10, 75-82. [DOI] [PubMed] [Google Scholar]
- 17.Radaev, S., Rostro, B., Brooks, A. G., Colonna, M. & Sun, P. D. (2001) Immunity 15, 1039-1049. [DOI] [PubMed] [Google Scholar]
- 18.Li, P., Morris, D. L., Willcox, B. E., Steinle, A., Spies, T. & Strong, R. K. (2001) Nat. Immunol. 2, 443-451. [DOI] [PubMed] [Google Scholar]
- 19.Plougastel, B., Jones, T. & Trowsdale, J. (1996) Immunogenetics 44, 286-291. [DOI] [PubMed] [Google Scholar]
- 20.Rodríguez, A., Carretero, M., Glienke, J., Bellón, T., Ramírez, A., Lehrach, H., Francis, F. & López-Botet, M. (1998) Immunogenetics 47, 305-309. [DOI] [PubMed] [Google Scholar]
- 21.Orita, M., Iwahana, H., Kanazawa, H., Hayashi, K. & Sekiya, T. (1989) Proc. Natl. Acad. Sci. USA 86, 2766-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagl, S., Tichy, H., Mayer, W. E., Takahata, N. & Klein, J. (1998) Proc. Natl. Acad. Sci. USA 95, 14238-14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drickamer, K. (1993) Curr. Opin. Struct. Biol. 3, 393-400. [Google Scholar]
- 24.Khalturin, K., Becker, M., Rinkevich, B. & Bosch, T. C. G. (2003) Proc. Natl. Acad. Sci. USA 100, 622-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanier, L. L. (1998) Annu. Rev. Immunol. 16, 359-393. [DOI] [PubMed] [Google Scholar]
- 26.Borrego, F., Ulbrecht, M., Weiss, E. H., Coligan, J. E. & Brooks, A. G. (1998) J. Exp. Med. 187, 813-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Braud, V. M., Allan, D. S. J., O'Callaghan, C. A., Söderström, K., D'Andrea, A., Ogg, G. S., Lazetic, S., Young, N. T., Bell, J. I., Phillips, J. H., et al. (1998) Nature 391, 795-799. [DOI] [PubMed] [Google Scholar]
- 28.Lee, N., Llano, M., Carretero, M., Ishitani, A., Navarro, F., López-Botet, M. & Geraghty, D. E. (1998) Proc. Natl. Acad. Sci. USA 95, 5199-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vance, R. E., Kraft, J. R., Altman, J. D., Jensen, P. E. & Raulet, D. H. (1998) J. Exp. Med. 188, 1841-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braud, V. M., Jones, E. Y. & McMichael, A. J. (1997) Eur. J. Immunol. 27, 1164-1169. [DOI] [PubMed] [Google Scholar]
- 31.Aldrich, C. J., DeCloux, A., Woods, A. S., Cootter, R. J., Soloski, M. J. & Forman, J. (1994) Cell 79, 649-658. [DOI] [PubMed] [Google Scholar]
- 32.Braud, V. M., Allan, D. J., Wilson, D. & McMichael, A. J. (1997) Curr. Biol. 8, 1-10. [DOI] [PubMed] [Google Scholar]