Abstract
A major concern in therapy of acute liver failure is protection of hepatocytes to prevent apoptosis and maintain liver function. Small interfering RNA (siRNA) is a powerful tool to silence gene expression in mammalian cells. To evaluate the therapeutic efficacy of siRNA in vivo we used different mouse models of acute liver failure. We directed 21-nt siRNAs against caspase 8, which is a key enzyme in death receptor-mediated apoptosis. Systemic application of caspase 8 siRNA results in inhibition of caspase 8 gene expression in the liver, thereby preventing Fas (CD95)-mediated apoptosis. Protection of hepatocytes by caspase 8 siRNA significantly attenuated acute liver damage induced by agonistic Fas (CD95) antibody (Jo2) or by adenovirus expressing Fas ligand (AdFasL). However, in a clinical situation the siRNAs most likely would be applied after the onset of acute liver failure. Therefore we injected caspase 8 siRNA at a time point during AdFasL- and adenovirus wild type (Adwt)-mediated liver failure with already elevated liver transaminases. Improvement of survival due to RNA interference was significant even when caspase 8 siRNA was applied during ongoing acute liver failure. In addition, it is of particular interest that caspase 8 siRNA treatment was successful not only in acute liver failure mediated by specific Fas agonistic agents (Jo2 and AdFasL) but also in acute liver failure mediated by Adwt, which is an animal model reflecting multiple molecular mechanisms involved in human acute viral hepatitis. Consequently, our data raise hope for future successful application of siRNA in patients with acute liver failure.
Acute liver failure (ALF) is a dramatic clinical syndrome with high mortality rates in which a previously normal liver fails within days or weeks. Three subgroups have been proposed: hyperacute, acute, and subacute liver failure (1). Patients with hyperacute disease have the most favorable prognosis, whereas survival of patients with acute and subacute liver failure is <15% (2). Worldwide the most frequent cause of ALF is viral hepatitis. Bioartificial liver support and hepatocyte transplantation are promising treatment options, but currently orthotopic liver transplantation remains the only treatment modality that has an impact on survival. Because of limitations of donor organs, new effective tools for the treatment of patients with ALF are needed urgently.
In ALF, signals released from the cell membrane of hepatocytes trigger suicide pathways, leading to the activation of caspase cascades that subsequently execute apoptotic death of hepatocytes. Mice injected with agonistic Fas (CD95) antibody rapidly develop ALF and die within hours (3). In animal models of acute viral hepatitis, apoptosis of hepatocytes is mediated by death receptors such as Fas (CD95), tumor necrosis factor (TNF)α, and TNF-related apoptosis-inducing ligand (4–6), and several studies in humans showed that apoptosis of hepatocytes in Wilson's disease, toxic liver damage, and viral hepatitis is triggered through death receptors (7, 8). Consequently an attractive strategy to treat patients with ALF would be to inhibit death receptor-mediated apoptosis to maintain liver function and save the organ.
Transfection of mammalian cells with synthetic small interfering RNAs (siRNAs) 21–23 nt in length specifically suppresses expression of endogenous and heterologous genes by RNA interference (RNAi) in cell culture (9, 10). Recently two reports suggested that siRNA could be delivered effectively into hepatocytes in vivo by rapid systemic injection of large volumes of physiological solutions (11, 12), and Song et al. (13) demonstrate protection against agonistic Fas (CD95) antibody (Jo2)-mediated liver failure and improvement of liver fibrosis in ConA-mediated hepatitis in mice by application of siRNA targeting the Fas (CD95) receptor. However, until now there are no studies demonstrating improvement of survival by siRNA treatment during ongoing ALF.
In the present study we demonstrate that systemic application of caspase 8 siRNA inhibits caspase 8 expression in the liver of mice and is capable of preventing Fas (CD95)-mediated apoptosis of hepatocytes. Protection of hepatocytes by caspase 8 siRNA results in reduced liver damage after application of activating anti-Fas (CD95) antibody or adenovirus expressing Fas ligand (AdFasL). Delayed treatment of mice with caspase 8 siRNA after administration of AdFasL or adenovirus wild type (Adwt) significantly improves the survival of the animals, demonstrating the therapeutic value of siRNA for acute liver diseases.
Methods
Cell Lines, siRNA Transfection, Adenoviral Infection, Measurement of Caspase 8 Activity, and Detection of Apoptosis in Cell Culture. HepG2 cells were obtained from the American Type Culture Collection. The cells were maintained in growth medium [DMEM/Glutamax (GIBCO/BRL) supplemented with 10% heat-inactivated FBS (GIBCO/BRL), 100 units/ml penicillin, and 100 μg of streptomycin] at 37°C in 5% CO2. Twenty-one-nucleotide RNA with 3′-dTdT overhangs was synthesized by Dharmacon Research (Lafayette, CO) in the “ready-to-use” option. The AA-N19 mRNA targets were caspase 8 target sequence 1 (5′-AAC CUC GGG GAU ACU GUC UGA-3′), caspase 8 target sequence 2 (5′-AAG AAG CUC UUC UUC CCU CCC U-3′), and scrambled sequence (5′-AAU CGC AUA GCG UAU GCC GUU-3′). Transfection of siRNA was performed by using Oligofectamine (Invitrogen) and Opti-MEM (Invitrogen) media according to manufacturer recommendations. HepG2 cells grown to a confluency of 40–50% in 24-well plates were transfected with 60 pmol of siRNA per well. After 48 h cells were infected with AdFasL and AdGFP at a multiplicity of infection of 15 in DMEM supplemented with 2% FBS. Caspase 8 activity was measured by using the ApoAlert caspase fluorescent-assay kit (CLONTECH) according to manufacturer instructions. Cells were harvested 8 h after infection, and 1 × 106 cells were resuspended in 50 μl of the provided lysis buffer. For detection of apoptosis in hepatoma cells after adenoviral application, cells were harvested 12 h after infection for photometric histone ELISA. Histone ELISA was performed with the cell-death detection ELISA Plus kit (Roche Diagnostics) according to the manufacturer's instructions.
Animal Experiments, Jo2 Application, and Detection of Apoptosis and Organ Damage. Pathogen-free male BALB/c- and C57BL/6J-TgN(MtnlacZ) mice (age 6–8 weeks) were obtained from the Animal Research Institute of the Medizinische Hochschule Hannover (Hannover, Germany). In vivo delivery of siRNA was performed either via the tail vein by high-volume injection or via portal vein by using either 0.6 (AdFasL- and Adwt-mediated liver damage) or 0.45 (Jo2-mediated liver damage and experiments with C57BL/ 6J-TgN(MtnlacZ) mice) nmol of siRNA per gram of body weight. For tail-vein injection siRNA was applied in a total volume of 2.5 ml (0.5 ml of dH20/2.0 ml of 0.9% NaCl) for experiments with Jo2-mediated liver damage or 2.0 ml (0.35 ml of dH20/1.65 ml of 0.9% NaCl) for experiments with adenoviral-mediated liver damage. Injection time was 4–10 sec. For portal-vein application animals were anesthetized with ketamine/Rompune. A 0.3 × 12-mm injection cannula (Braun, Melsungen, Germany) was inserted into the portal vein, and 0.45 nmol of siRNA were delivered in a 1-ml volume containing 10% Lipiodol (0.5 ml of dH20/0.4 ml of 0.9% NaCl). Injection time was ≈10 sec. Before removing the cannula an adhesive solution of Fibrine/Thrombine (Beriplast, Aventis, Marburg, Germany) was spread on the portal vein to prevent bleeding.
Fas (CD95)-activating antibody Jo2 (Becton Dickinson Biosciences) was applied i.v. in a dose of 0.5 μg/g of body weight. Livers were harvested for caspase 8 assays and terminal deoxynucleotidyltransferase-mediated dUTP end labeling (TUNEL) staining 6 h after Jo2 application and for the investigation of histopathology 10 h after Jo2 treatment. For TUNEL-staining, 7-μm cryosections were prepared and fixed in PBS-buffered paraformaldehyde solution (4%) for 30 min at room temperature. Staining with the in situ cell-death detection kit (Roche) was performed following the manufacturer's instructions.
For investigation of histopathology hematoxylin/eosin-stained sections of paraffin-embedded tissue were used. Hematoxylin/eosin-stained sections were examined by an experienced pathologist (P.F.).
Biochemical markers of liver injury were assessed by measuring serum alanine aminotransferase and serum aspartate transaminase by standard procedures.
Adenovirus Preparation and in Vivo Infection Experiments. To generate high-titer viral stocks of Adwt and AdFasL, 293 packaging cells were infected at a multiplicity of infection of 5–10 plaque-forming units (pfu) per cell. Adenovirus preparation and viral plaquing were performed as described (5, 6). AdFasL was administered via tail vein in a dose of 6.25 × 107 pfu/g of body weight. Adwt was administered in a dose of 5.5 × 108 pfu/g of body weight.
Statistical Analysis. Cumulative survival of mice after induction of ALF was assessed by Kaplan–Meier analysis. Statistical significance of serum alanine aminotransferase and serum aspartate transaminase for each time point was calculated by using the t test. P values <0.05 were considered to indicate statistical significance. All statistical analyses were performed with the use of SPSS 11.0.1 software (SPSS, Chicago).
Determination of siRNA Uptake into the Liver. For in vivo experiments with siRNA targeted against the Escherichia coli lacZ gene two different procedures were performed. For experiments with siRNA lacZ sequence 1 (5′-AAG CAA AAC ACC AGC AGC AGU-3′), MT-lacZ transgene C57BL/6J-TgN(MtnlacZ) mice were first injected i.p. with 1 mg of Cd2+ per kg of body weight 20 and 6 h before siRNA delivery to induce lacZ gene expression. siRNA was applied via tail vein as described above, and 48 h later livers were harvested for lacZ whole-liver staining and β-galactosidase (β-gal) assay. For experiments with siRNA lacZ sequence 2 (5′-AAU UUA ACC GCC AGU CAG GCU-3′) mice were treated with siRNA 24 h before triggering lacZ gene expression. Induction of lacZ gene expression by i.p. injection of 1 mg of Cd2+ per kg of body weight was performed 20 and 6 h before harvesting of the organs for whole-liver staining and β-gal assay. Whole-liver staining with β-gal substrate 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-gal) was performed as described elsewhere (14). For determination of the proportion of hepatocytes with effective uptake of siRNA, siRNA was administered 24 h before triggering lacZ gene expression, and 24 h later the hepatocytes were isolated by collagenase perfusion and cultured as described elsewhere (6). Twelve hours after seeding the cells, the proportion of hepatocytes with LacZ gene expression was determined by X-gal staining.
For the β-gal assay, 0.5 g of liver tissue was homogenized in extraction buffer containing 125 mM Tris-PO4 (pH 7.8), 10 mM EDTA, 10 mM DTT, 50% glycerol, and 5% Triton. The liver homogenate was centrifuged at 20,000 × g for 20 min. The supernatant then was assessed for o-nitrophenyl-β-D-galactopyranoside (ONPG) turnover in a buffer containing 60 mM Na2HPO4, 39 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 2 mM DTT, and 1 mg/ml ONPG. Extinction was measured at 405 nm.
Measurement of Caspase 8 Activity in Vivo, Isolation of mRNA from Liver Tissue, and Northern Blot Analysis. For determination of caspase 8 activity in the mouse liver, freshly harvested organs were subjected to a buffer containing 25 mM Hepes, 5 mM MgCl2,5mM EDTA, 2 mM DTT, 0.1% CHAPS, 0.5 mM Pefabloc, 0.1 mg/ml leupeptin, and 0.1 mg/ml pepstatin in a ratio of 1:3 (wt/vol). For preparation of whole-cell extracts, tissue was homogenized on ice and subsequently centrifuged at 12,000 × g for 10 min (4°C). Total protein (100 μg) from the supernatant was used to measure caspase 8 activity by the ApoAlert caspase fluorescent-assay kit (CLONTECH).
For isolation of mRNA from mouse livers, mTRAP Midi mRNA isolation kit (Active Motif, Carlsbad, CA) was used according to manufacturer instructions.
mRNA (8 μg) was separated on a 1% agarose formaldehyde gel and transferred to a Hybond N membrane (Amersham Pharmacia Biotech). Mouse caspase 8 and GAPDH cDNA probes were generated by RT-PCR by using oligo(dT)15 primers (Promega) in combination with an Omniscript kit (Qiagen, Valencia, CA) to produce single-stranded cDNA. Total RNA (2 μg) was used for each reaction. Amplification of cDNA probes was performed by using a PCR with 15 sec of denaturation at 92°C, annealing for 30 sec at 55°C, and elongation for 80 sec at 72°C. The sequence of the primers for the caspase 8 probe were forward (5′-tgccctcaagttcctgtgcttggac-3′) and reverse (5′-ggatgctaagaatgtcatctcc-3′). Primers for GAPDH were forward (5′-tgcatcctgcaccaccaact-3′) and reverse (5′-aacacggaaggccatgccag-3′). Caspase 8 and GAPDH cDNA probes were labeled with [α-32P]CTP by using Rediprime II random prime labeling kit (Amersham Pharmacia Biotech) and hybridized at 62°C overnight. Membranes were exposed on x-ray films for 7 days (caspase 8) or 1 day (GAPDH). The ratio of caspase 8 mRNA to GAPDH mRNA was quantified by using PCBAS 2.09f software (Raytest, Straubenhardt, Germany).
Results
Inhibition of Caspase 8 by siRNA Prevents Fas (CD95)-Mediated Apoptosis. RNAi targeted against death receptor-mediated apoptosis would be an important intervention to protect hepatocytes and maintain liver function. Caspase 8 is presumed to be the apex of the death receptor-mediated apoptosis pathways and seems to be a key enzyme in FasL-, TNF-related apoptosis-inducing ligand-, and TNFα-induced apoptosis (15, 16). Therefore we designed specific siRNA against two target sequences of the human and murine caspase 8 mRNA.
We transfected human HepG2 hepatoma cells with caspase 8 siRNA and subsequently treated the cells with AdFasL. For a negative control we used a GFP-expressing adenoviral vector. Both caspase 8 siRNA sequences strongly inhibit caspase 8 activity and prevent effectively FasL-mediated apoptosis (as shown in Fig. 1). These results indicate that caspase 8 siRNA silences an essential enzyme in the Fas (CD95) pathway and thus may be an interesting tool for the assessment of the therapeutic utility of siRNA in animal models.
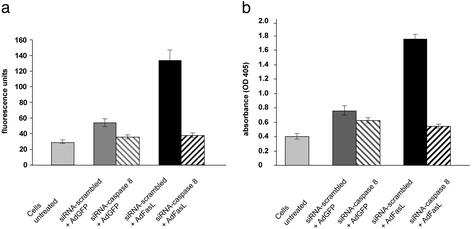
Fig. 1.
Transfection of siRNA targeted against caspase 8 mRNA prevents FasL-mediated apoptosis of HepG2 hepatoma cells. HepG2 cells were transfected with either a nonsense siRNA or a specific siRNA targeted against caspase 8. After 48 h cells were infected with AdFasL or AdGFP. (a) Caspase 8 assay (caspase 8 sequence 1). Cells pretreated with caspase 8 siRNA show ≈2.5-fold reduced caspase 8 activity compared with cells pretreated with scrambled siRNA 8 h after adenoviral infection. (b) Histone ELISA (caspase 8 sequence 1). Caspase 8 siRNA treatment inhibits Fas (CD95)-mediated apoptosis as shown by photometric histone ELISA 12 h after infection. The figure shows representative data obtained from three independent experiments.
siRNA Is Delivered Effectively into Hepatocytes by Systemic Large-Volume Injection and Inhibits Liver Gene Expression. To investigate the efficacy of systemically administered siRNA for silencing hepatic gene expression, we used two siRNAs targeted against the E. coli lacZ gene to inhibit lacZ gene expression in a transgenic mouse strain (14). LacZ siRNA was injected into the tail vein in a dose of 0.45 nmol/g of body weight, and β-gal expression was monitored by β-gal staining of the whole liver and isolated hepatocytes and by β-gal assay from liver cell lysates (see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). For controls we used mice receiving non-sense siRNA (scrambled siRNA). We show that application of lacZ-siRNA either before or after induction of lacZ gene expression by Cd2+ leads to 3- to 4-fold-reduced β-gal activity in MT-lacZ transgene C57BL/6J-TgN(MtnlacZ) mice. Approximately 70% of all hepatocytes in the liver take up siRNA after systemic hydrodynamic application as shown by the reduced number of x-gal-positive primary hepatocytes after application of LacZ-siRNA.
Caspase 8 siRNA Prevents Fas (CD95)-Mediated Apoptosis of Hepatocytes. To determine whether systemic caspase 8 siRNA treatment is capable of down-regulating caspase 8 gene expression in the liver we injected siRNA into the tail vein of mice and subsequently harvested the livers for mRNA isolation and Northern blot analysis. As shown in Fig. 2a, systemic delivery of caspase 8 siRNA results in >3-fold-reduced caspase 8 mRNA expression in the liver.
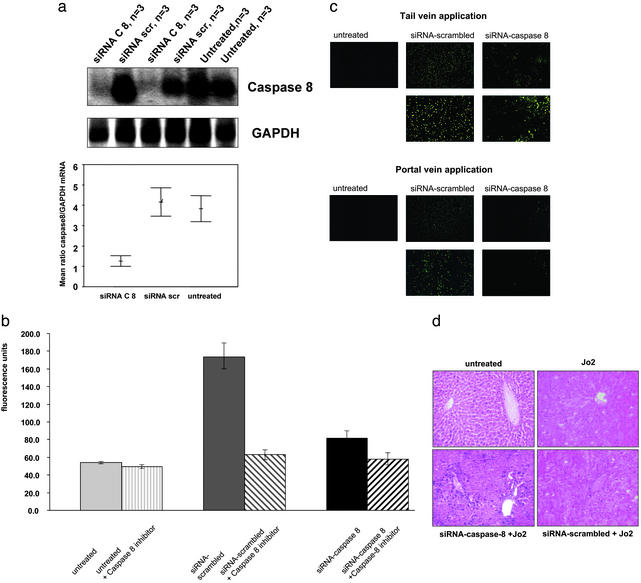
Fig. 2.
Caspase 8 siRNA inhibits caspase 8 gene expression in the liver and prevents Jo2-mediated apoptosis of hepatocytes. (a) Mice were treated with 0.45 nmol of siRNA per g of body weight into the tail vein. After 48 h livers were harvested for mRNA isolation and Northern blot analysis. Compared with animals treated with scrambled (scr) siRNA or control animals without prior siRNA treatment, caspase 8 siRNA-treated animals show >3-fold-reduced mRNA expression in Northern blot analysis. Each lane contains mRNA extracted from pooled tissues of three animals. (b) For analysis of caspase 8 activity, mice were treated with Fas (CD95)-activating antibody Jo2 48 h after siRNA application. Six hours after Jo2 application, livers were harvested for measurement of caspase 8 activity. Extracts of pooled liver tissue from three animals in each group were prepared. Pretreatment with caspase 8 siRNA significantly inhibits up-regulation of caspase 8 activity after Jo2 application. (c) Mice treated systemically with caspase 8 siRNA show heterogenous and overall reduced apoptosis in TUNEL staining 6 h after Jo2 application compared with mice treated with scrambled-siRNA. Inhibition of Jo2-mediated liver cell apoptosis can be improved by portal-vein application compared with tail-vein application. (Magnification, ×100 and ×200.) Shown are representative data from three independent experiments. (d) Hematoxylin/eosin-stained tissue sections were investigated for histopathology to show the morphological effects of caspase 8 siRNA treatment. Unprotected livers were totally destroyed, whereas 10–20% of hepatocytes from mice pretreated with caspase 8 siRNA were still alive 10 h after Jo2 treatment. (Magnification, ×200.)
To evaluate the impact of the silenced caspase 8 mRNA expression on caspase 8 activity and apoptosis of hepatocytes in vivo, we used a mouse model of Fas (CD95)-mediated ALF. Forty-eight hours after administration of siRNA, Fas (CD95)-mediated apoptosis was induced by systemic injection of 0.5 μg/g of body weight of the Fas (CD95)-agonistic antibody Jo2. Treatment with caspase 8 siRNA prevents almost completely up-regulation of caspase 8 activity in the liver and inhibits significantly Jo2-mediated apoptosis of hepatocytes as shown by TUNEL staining (Fig. 2 b and c).
In humans with acute liver diseases, regional therapy by portal-vein injection may be more feasible than systemic injection of high volumes. Consequently we investigated whether administration of smaller volumes of siRNA into the portal vein would also reduce Jo2-mediated apoptosis of hepatocytes. As shown in Fig. 2c inhibition of apoptosis seemed more effective after regional treatment compared with systemic injection (Fig. 2c). However, both approaches led to heterogenous TUNEL staining patterns in mice treated with caspase 8 siRNA, whereas mice in the control groups treated with scrambled siRNA showed homogeneous intense apoptosis throughout the liver. Some areas in livers of mice treated with caspase 8 siRNA were completely negative for TUNEL staining, indicating complete protection from Jo2-mediated apoptosis.
We next investigated the histopathology of the livers 10 h after application of Jo2. According to the results obtained with TUNEL staining, ≈10–20% of the hepatocytes remained alive due to systemic caspase 8 siRNA treatment, whereas the livers of mice treated with scrambled siRNA and Jo2 or with Jo2 alone were destroyed completely (Fig. 2d).
To confirm protection of Fas (CD95)-mediated apoptosis of hepatocytes by caspase 8 siRNA we used another mouse model of liver failure. In contrast to Jo2, application of FasL-expressing adenoviral vector leads to liver failure not within a few hours but within 2 days. Forty-eight hours after systemic application of siRNA, 6.25 × 107 pfu/g of body weight of AdFasL were injected into the tail vein of mice. Liver injury was assessed by measuring transaminases and investigating liver histopathology 32 and 40 h after adenoviral application. Delivery of caspase 8 siRNA results in significant prevention of AdFasL-mediated liver injury even more effectively than in Jo2-mediated liver failure (Fig. 3). These data underline the essential contribution of caspase 8 in Fas (CD95)-mediated apoptosis of hepatocytes.
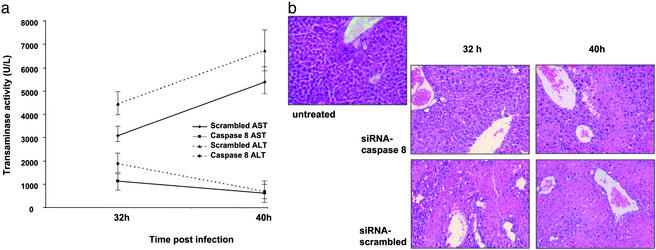
Fig. 3.
Caspase 8 siRNA prevents liver damage in a model of AdFasL-induced liver failure. Delivery of caspase 8 siRNA 48 h before application of AdFasL is capable of preventing liver damage as shown by significantly reduced serum transaminases (a) and distinctly improved histopathologies (b; magnification, ×200) in caspase 8 siRNA-protected mice compared with mice treated with scrambled siRNA 32 and 40 h after adenoviral application. The difference between the siRNA scrambled- and caspase 8 siRNA-treated animals for serum alanine aminotransferase and serum aspartate transaminase at each time point was highly significant (P < 0.001). Furthermore, the increase for siRNA scrambled-treated animals and decrease for caspase 8 siRNA-treated animals in time course was also significant (P < 0.001 and 0.05, respectively).
Systemic Fas (CD95) engagement induced by the agonistic antibody Jo2 has been reported to cause apoptosis in hepatocytes (3, 7), lymphocytes (17), endothelial cells (18, 19), and alveolar epithelial cells of the lung (20) as well as glomerular and mesangial cells in the kidney (21). Accordingly, 10 h after application of Jo2 we observed an increase in caspase 8 activity in the kidneys and lungs of the mice. Up-regulation of caspase 8 activity in kidneys and lungs was inhibited by pretreatment with caspase 8 siRNA, but in contrast to the liver, we did not observe any inhibition of TUNEL staining in kidneys or lungs due to caspase 8 siRNA treatment (data not shown). Histology sections showed no differences in morphology of the kidneys and lungs 10 h after Jo2 application, indicating that Jo2-mediated injury of these organs has no significant impact on the survival of the animals in our experiments (data not shown).
Systemic Application of Caspase 8 siRNA Prevents Fas (CD95)-Mediated ALF and Improves Survival of Mice with Adenoviral Hepatitis. Next we investigated the effect of caspase 8 siRNA on survival of animals with ALF. Scrambled siRNA- and caspase 8 siRNA-treated mice as well as untreated control animals were injected with 0.5 μg/g Jo2 or 6.25 × 107 pfu/g AdFasL. Systemic pretreatment with caspase 8 siRNA protected the mice against Jo2- and AdFasL-induced ALF and significantly improved the survival of the animals as shown in Fig. 4a.
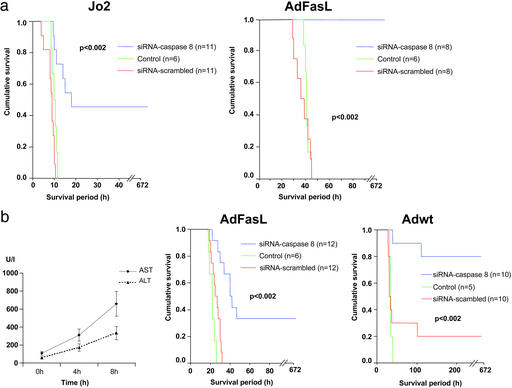
Fig. 4.
Application of caspase 8 siRNA leads to significantly improved survival in different models of ALF. Prevention of Fas (CD95)-mediated apoptosis by caspase 8 siRNA was investigated by application of caspase 8 siRNA or scrambled siRNA 48 h before administration of Jo2 or AdFasL. Control animals received Jo2 or AdFasL without siRNA treatment. (a) Whereas all animals with unprotected livers died, prevention of apoptosis by caspase 8 siRNA led to 100% survival in AdFasL-mediated liver failure and 45% survival in Jo2-mediated ALF. (b) Therapeutic intervention by RNAi in an ongoing ALF was investigated by application of caspase 8 siRNA after viral liver infection with AdFasL or Adwt. Increase of transaminases (serum alanine aminotransferase and serum aspartate transaminase) demonstrates liver injury already 4 and 8 h after application of Adwt. Treatment with caspase 8 siRNA 8 h after application of AdFasL or Adwt significantly improves survival of animals. Control animals received AdFasL or Adwt without siRNA treatment.
In a clinical situation caspase 8 siRNA would be applied after the onset of liver injury. Therefore, we assessed the therapeutic efficacy of siRNA in mouse models resembling the clinical situation. Because mice injected with Jo2 die rapidly within a few hours, Jo2-mediated liver failure is not a suitable model to evaluate therapeutic interventions in ongoing liver damage. Therefore we used AdFasL and Adwt to induce ALF in mice. Compared with Jo2, these viruses cause a more delayed liver injury and represent better agents to simulate the natural course of human ALF.
To determine the beginning of liver injury after viral infection we measured serum transaminases 4 and 8 h after adenoviral application (Fig. 4b). To evaluate whether siRNA treatment would be effective after the onset of liver injury we selected a time point for siRNA application when liver transaminases were already elevated due to the beginning liver failure. Even in this setting caspase 8 siRNA treatment was capable of improving significantly the survival of the animals in AdFasL- and Adwt-mediated ALF, demonstrating the therapeutic value of RNAi after the onset of virally induced liver failure (Fig. 4b).
Discussion
The discovery that siRNA could be delivered effectively into the liver of mammals has raised the possibility that selective intervention in hepatic gene regulation might be feasible for the treatment of liver diseases (11, 12). While our article was in preparation, Song et al. (13) published a study demonstrating successful treatment of Jo2- and ConA-mediated hepatitis by effective delivery of siRNA targeting Fas (CD95) into hepatocytes in mice. In contrast to Song et al., we used a downstream target of all known death receptors (caspase 8), which is essential in Fas (CD95) as well as in TNFα and TNF-related apoptosis-inducing ligand-mediated apoptosis of hepatocytes. Because TNFα and TNF-related apoptosis-inducing ligand are important mediators of apoptosis in human acute viral hepatitis (6, 22), caspase 8 seems to be the more suitable target compared with Fas (CD95) to achieve future therapeutic success in humans with ALF. However, the results in our experiments with Jo-2 were a little less dramatic than in the study from Song et al. (13), which may be explained by the more effective delivery of siRNA into the hepatocytes due to repeated application. In the study from Song et al., 89% of the hepatocytes took up siRNA after three repeated injections, whereas single application of siRNA in our study resulted in transduction of ≈70% of hepatocytes.
Prevention of hepatocyte apoptosis in our study was not only evident after systemic high-volume application of caspase 8 siRNA but also after intraportal injection of low volumes of a Lipiodol/ caspase 8 siRNA solution, which seems to be more feasible for clinical application in humans. Recently Lewis et al. (12) demonstrated that systemic coinjection of luciferase reporter plasmids with siRNA luciferase results in strong inhibition of luciferase activity not only in the liver but also in kidney, spleen, lung, and pancreas of postnatal mice, suggesting that siRNA is delivered effectively into many organs after systemic application in mammals. In our study we observed an inhibition of caspase 8 up-regulation in kidneys and lungs of mice treated with caspase 8 siRNA and Jo2, confirming the uptake of siRNA into these organs after systemic application. However, in contrast to the liver, Fas (CD95)-mediated apoptosis was not inhibited in lungs or kidneys by caspase 8 siRNA. Because Jo2 antibodies do not change the morphology of kidneys and lungs, our experiments were not suitable to give an answer to the question of whether systemic treatment with siRNA is capable of providing therapeutic effects in these organs. Other animal models should be used to determine the value of systemic siRNA treatment for extrahepatic diseases.
In our study significant therapeutic effects were obtained in ALF induced by specific Fas (CD95) agonistic agents such as Jo2 and AdFasL as well as in acute liver damage mediated by Adwt. Because Adwt-mediated liver failure is an animal model reflecting multiple molecular mechanisms involved in human acute viral hepatitis, our results indicate a clinical relevance for caspase 8 gene silencing in human hepatitis. In addition, it is of considerable interest that improvement of survival was not only obtained in mice protected from liver failure by siRNA application before viral infection but also in animals treated after the onset of liver injury. The effective therapeutic intervention after viral liver infection highlights the possibility of future successful clinical application of siRNA in patients with ALF. In consideration of the distinctly longer time course of human ALF one can even expect a prolonged therapeutic time slot in patients compared with the animals in our experiments.
The ability to apply siRNA in the liver of mammals will be of outstanding interest not only for ALF but also for many other human liver diseases. T lymphocytes have the ability to kill Fas (CD95)-bearing hepatocytes in viral and autoimmune hepatitis, and there is evidence that Fas (CD95)-mediated liver injury plays a critical role in alcoholic liver disease. The present study raises the hope of new therapeutic approaches to inhibit apoptosis of hepatocytes, thereby maintaining the liver function of patients. Inhibition of inflammation and fibrosis and improvement of liver regeneration are also as-yet-unsolved clinical problems of great relevance. In addition to caspase 8 other key molecules in various signal transduction pathways may also be attractive targets for therapeutic RNAi in the liver. Recently studies revealed that siRNA targeted to viral RNA sequences can effectively protect human cells against RNA viruses such as HIV (23–25), poliovirus (26), or hepatitis C virus (27). These findings, in combination with the results of our study, suggest that RNAi provides an attractive therapeutic strategy for patients with hepatitis C infection, one of the most important viral diseases of humans.
Our study demonstrates successful improvement of survival by therapeutic application of siRNA in mammals during ongoing ALF. The possibility to target siRNA against any appropriate mRNA sequence promises the clinical application of RNAi as a powerful tool for molecular therapy of many liver diseases.
Supplementary Material
Acknowledgments
This research was supported by grants from the Deutsche Forschungsgemeinschaft Sonderforschungsbereich 265, Projekt C4A, and Medizinische Hochschule Hannover, Hochschulinterne Leistungsfōrderung.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ALF, acute liver failure; TNF, tumor necrosis factor; siRNA, small interfering RNA; RNAi, RNA interference; FasL, Fas ligand; AdFasL, adenovirus expressing FasL; Adwt, adenovirus wild type; TUNEL, terminal deoxynucleotidyltransferase-mediated dUTP end labeling; pfu, plaque-forming unit(s); β-gal, β-galactosidase; X-gal, 5-bromo-4-chloro-3-indolyl-β-d-galactoside.
References
- 1.O'Grady, J. G., Schalm, S. W. & Williams, R. (1993) Lancet 342, 273-275. [DOI] [PubMed] [Google Scholar]
- 2.Plevris, J. N., Schina, M. & Hayes, P. C. (1998) Aliment. Pharmacol. Ther. 12, 405-418. [DOI] [PubMed] [Google Scholar]
- 3.Ogasawara, J., Watanabe-Fukunaga, R., Adachi, M., Matsuzawa, A., Kasugai, T., Kitamura, Y., Itoh, N., Suda, T. & Nagata, S. (1993) Nature 364, 806-809. [DOI] [PubMed] [Google Scholar]
- 4.Kondo, T., Suda, T., Fukuyama, H., Adachi, M. & Nagata, S. (1997) Nat. Med. 3, 409-413. [DOI] [PubMed] [Google Scholar]
- 5.Kuhnel, F., Zender, L., Paul, Y., Tietze, M. K., Trautwein, C., Manns, M. & Kubicka, S. (2000) J. Biol. Chem. 275, 6421-6427. [DOI] [PubMed] [Google Scholar]
- 6.Mundt, B., Kühnel, F., Zender, L., Paul, Y., Tillmann, H., Trautwein, C., Manns, M. P. & Kubicka, S. (2003) FASEB J. 17, 94-96. [DOI] [PubMed] [Google Scholar]
- 7.Galle, P. R., Hofmann, W. J., Walczak, H., Schaller, H., Otto, G., Stremmel, W., Krammer, P. H. & Runkel, L. (1995) J. Exp. Med. 182, 1223-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strand, S., Hofmann, W. J., Grambihler, A., Hug, H., Volkmann, M., Otto, G., Wesch, H., Mariani, S. M., Hack, V., Stremmel, W., et al. (1998) Nat. Med. 4, 588-593. [DOI] [PubMed] [Google Scholar]
- 9.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- 10.Caplen, N. J., Parrish, S., Imani, F., Fire, A. & Morgan, R. A. (2001) Proc. Natl. Acad. Sci. USA 98, 9742-9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCaffrey, A. P., Meuse, L., Pham, T. T., Conklin, D. S., Hannon, G. J. & Kay, M. A. (2002) Nature 418, 38-39. [DOI] [PubMed] [Google Scholar]
- 12.Lewis, D. L., Hagstrom, J. E., Loomis, A. G., Wolff, J. A. & Herweijer, H. (2002) Nat. Genet. 32, 107-108. [DOI] [PubMed] [Google Scholar]
- 13.Song, E., Lee, S. K., Wang, J., Ince, N., Ouyang, N., Min, J., Chen, J., Shankar, P. & Lieberman, J. (2003) Nat. Med. 9, 347-351. [DOI] [PubMed] [Google Scholar]
- 14.Rhim, J. A., Sandgren, E. P., Degen, J. L., Palmiter, R. D. & Brinster, R. L. (1994) Science 263, 1149-1152. [DOI] [PubMed] [Google Scholar]
- 15.Ashkenazi, A. & Dixit, V. M. (1998) Science 281, 1305-1308. [DOI] [PubMed] [Google Scholar]
- 16.Shi, Y. (2002) Mol. Cell 9, 459-470. [DOI] [PubMed] [Google Scholar]
- 17.Nishimura, Y., Hirabayashi, Y., Matsuzaki, Y., Musette, P., Ishii, A., Nakauchi, H., Inoue, T. & Yonehara, S. (1997) Int. Immunol. 9, 307-316. [DOI] [PubMed] [Google Scholar]
- 18.Wanner, G. A., Mica, L., Wanner-Schmid, E., Kolb, S. A., Hentze, H., Trentz, O. & Ertel, W. (1999) FASEB J. 13, 1239-1248. [DOI] [PubMed] [Google Scholar]
- 19.Janin, A., Deschaumes, C., Daneshpouy, M., Estaquier, J., Micic-Polianski, J., Rajagopalan-Levasseur, P., Akarid, K., Mounier, N., Gluckman, E., Socie, G., et al. (2002) Blood 99, 2940-2947. [DOI] [PubMed] [Google Scholar]
- 20.Matute-Bello, G., Winn, R. K., Jonas, M., Chi, E. Y., Martin, T. R. & Liles, W. C. (2001) Am. J. Pathol. 158, 153-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Cuadrado, S., Lorz, C., Garcia, d. M., O'Valle, F., Alonso, C., Ramiro, F., Ortiz-Gonzalez, A., Egido, J. & Ortiz, A. (1997) Kidney Int. 51, 1739-1746. [DOI] [PubMed] [Google Scholar]
- 22.Streetz, K., Leifeld, L., Grundmann, D., Ramakers, J., Eckert, K., Spengler, U., Brenner, D., Manns, M. & Trautwein, C. (2000) Gastroenterology 119, 446-460. [DOI] [PubMed] [Google Scholar]
- 23.Jacque, J. M., Triques, K. & Stevenson, M. (2002) Nature 418, 435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, N. S., Dohjima, T., Bauer, G., Li, H., Li, M. J., Ehsani, A., Salvaterra, P. & Rossi, J. (2002) Nat. Biotechnol. 20, 500-505. [DOI] [PubMed] [Google Scholar]
- 25.Novina, C. D., Murray, M. F., Dykxhoorn, D. M., Beresford, P. J., Riess, J., Lee, S. K., Collman, R. G., Lieberman, J., Shankar, P. & Sharp, P. A. (2002) Nat. Med. 8, 681-686. [DOI] [PubMed] [Google Scholar]
- 26.Gitlin, L., Karelsky, S. & Andino, R. (2002) Nature 418, 430-434. [DOI] [PubMed] [Google Scholar]
- 27.Randall, G., Grakoui, A. & Rice, C. M. (2003) Proc. Natl. Acad. Sci. USA 100, 235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.