Abstract
Cell-cycle checkpoints controlling the orderly progression through mitosis are frequently disrupted in human cancers. One such checkpoint, entry into metaphase, is regulated by the CHFR gene encoding a protein possessing forkhead-associated and RING finger domains as well as ubiquitin–ligase activity. Although defects in this checkpoint have been described, the molecular basis and prevalence of CHFR inactivation in human tumors are still not fully understood. To address this question, we analyzed the pattern of CHFR expression in a number of human cancer cell lines and primary tumors. We found CpG methylation-dependent silencing of CHFR expression in 45% of cancer cell lines, 40% of primary colorectal cancers, 53% of colorectal adenomas, and 30% of primary head and neck cancers. Expression of CHFR was precisely correlated with both CpG methylation and deacetylation of histones H3 and H4 in the CpG-rich regulatory region. Moreover, CpG methylation and thus silencing of CHFR depended on the activities of two DNA methyltransferases, DNMT1 and DNMT3b, as their genetic inactivation restored CHFR expression. Finally, cells with CHFR methylation had an intrinsically high mitotic index when treated with microtubule inhibitor. This means that cells in which CHFR was epigenetically inactivated constitute loss-of-function alleles for mitotic checkpoint control. Taken together, these findings shed light on a pathway by which mitotic checkpoint is bypassed in cancer cells and suggest that inactivation of checkpoint genes is much more widespread than previously suspected.
Keywords: DNA methylation, DNA methyltransferase, mitotic checkpoint, histone deacetylation
Evidence now indicates that the majority of human cancers exhibit genetic instability, and that this instability exists on two levels (1). On one level, the expression or sequence of human mitotic-checkpoint genes are altered. For example, decreased expression of hMAD2 has been documented in breast cancers; haploinsufficiency of this gene causes chromosomal instability (CIN) in both mouse cells and human cancer cells harboring stable karyotypes (2). In addition, a small fraction of colorectal cancers have been shown to contain somatic mutations of either hBUB1 or hBUBR1 (3). Mutations in BUB1 can function in a dominant-negative manner in both mouse and human cells, conferring an abnormal spindle checkpoint when expressed endogenously. On a second level, defects in the pathway downstream of the spindle checkpoint have been noted in some cancers. For example, genetic inactivation of hSecurin in human cancer cells leads to nondisjunction of sister chromatids during prolonged anaphase and a concomitant CIN phenotype (4).
Recent evidence also suggests the presence of another checkpoint in mammalian cells, upstream of the spindle checkpoint. This checkpoint delays chromosome condensation in response to mitotic stress and is regulated by the expression of a protein designated CHFR (checkpoint with FHA and RING finger). Normal primary cells and cancer cell lines that express CHFR exhibit delayed entry into metaphase after treatment with microtubule inhibitors (5). By contrast, cancer cell lines lacking CHFR entered metaphase without delay, and ectopic expression of CHFR was necessary and sufficient to restore the cell-cycle delay (5). Recently, inactivation of CHFR was shown to be caused by DNA methylation in several tumors (6, 7). However, the causal role of DNA methylation in gene silencing and the functional consequences of inactivation of CHFR in tumorigenesis are not yet fully understood.
In the present study, we show that the loss of CHFR expression in human tumors is predominantly mediated via an epigenetic pathway involving DNA methylation and histone modification and that maintenance of CHFR in a silenced state requires DNA methyltransferase 1 (DNMT1) and DNMT3B. We also showed that tumor cells in which CHFR has been inactivated are functionally null for early mitotic checkpoint control. These findings broaden the spectrum of pathways by which mitotic checkpoints are known to be bypassed in cancer cells and suggest that inactivation of checkpoint genes is much more widespread than previously thought.
Materials and Methods
Cell Lines and Tumor Specimens. DNA was prepared from colorectal cancer cell lines (Caco2, RKO, SW48, HCT116, DLD-1, LoVo, SW837, HT29, Colo205, Colo201, BM314 SW480, and Colo320DM), hepatocellular cancer cell lines (CHC4, CHC20, CHC32, HLE, and HuH7), pancreatic cancer cell lines (Panc1 and Miapaca2), lung cancer cell lines (H249, H157, H209, and H1299), myeloma cell lines (HO3238, KMS-12PE, RPMI8226, KR12, HS-Sultan, RPMI1788, and IM9), acute lymphoblastic leukemia (ALL) cell lines (BALL1, Jurkat, CEM, HSB2, Malt4, SupT1, PEER, and TALL), one chronic myeloid leukemia cell line (K562), malignant lymphoma cell lines (Daudi, SCC3, and Raji), one adult T cell leukemia cell line (ATL2), head and neck cancer cell lines (HSC3, HSC4, Ca9–22, OSC-20, OSC-19, OSC30, OSC-70, SAS, KOSC3, and OSC40), one osteosarcoma cell line (Saos2), and one glioblastoma cell line (T98G). All cell lines were obtained from the American Type Culture Collection or the Japanese Collection of Research Bioresources (Tokyo). HCT116 cells with genetic disruptions in DNMT1 and/or DNMT3b loci have been described (8, 9). Isolation and characterization of specimens from 63 colorectal cancers and 51 colorectal adenomas have been described (10, 11). Genomic DNA was extracted with phenol chloroform. Total RNA was extracted by using Isogen (Nippon Gene, Toyama, Japan). To analyze restoration of CHFR expression, cell lines were incubated for 96 h with 0.2 μM or 1 μM 5-aza-2′-deoxycytidine (5-aza-dC) (Sigma) and/or for 24 h with 300 nM trichostatin A (TSA), a histone deacetylase inhibitor (WAKO, Tokyo), after which they were harvested and their RNA was extracted for further analysis.
Methylation Analysis. DNA treatment and bisulfite PCR were performed as described (12). Briefly, bisulfite PCR was carried out with primers that amplify both methylated and unmethylated alleles. After amplification, the PCR products were digested with restriction enzymes that cleave exclusively methylated CpG sites (Fig. 8, which is published as supporting information on the PNAS web site, www.pnas.org). Bisulfite-treated PCR products were then sequenced as described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site, using primers CHFRGM1F and CHFRGM1R cloned into pCR4 vector by using a TOPO-cloning kit (Invitrogen). The primer sequences and cycling parameters are included in Tables 2 and 3, which are published as supporting information on the PNAS web site.
Gene Expression Analysis. cDNA was synthesized by using SuperscriptII (Invitrogen) according to the manufacturer's instructions. PCR was carried out in solution containing 1× PCR buffer (Takara, Tokyo), 200 μM of each deoxynucleotide triphosphate, 2.5 pmol of each primer, 1 unit of ExTaq polymerase (Takara), and 5% (vol/vol) DMSO. The oligonucleotide sequences and specific amplification/reaction conditions can be found in Tables 2 and 3.
Chromatin Immunoprecipitation. Chromatin immunoprecipitation assays were carried out as described (13). Chromatin was immunoprecipitated for 16 h at 4°C by using anti-acetylated histone H3 or H4 antibodies (Upstate Biotechnology, Lake Placid, NY), after which PCR was performed by using ≈1/100 of the immunoprecipitated DNA. In addition, 1/100 of the solution before adding antibody was amplified as an internal control for the amount of DNA, and the 5′ region of GAPDH was amplified as a positive control. Primer sequences for the PCR are summarized in Table 3. Reaction products were electrophoresed in 2% Nusieve agarose gels.
Colony Formation Assay. Cells (1 × 105) were plated in 100-mm culture dishes for 24 h before transfection with expression vectors pcDNA3.1-CHFR, pcDNA3.1-ΔFHA, pcDNA-p53, or empty vector (5 μg each) by using Lipofectin (Invitrogen). After transfection, cells were selected for 14 days in medium containing 0.6 mg/ml G418 and stained with Giemsa. Colony counts were then performed with triplicate cultures by using National Institutes of Health IMAGE software.
Results
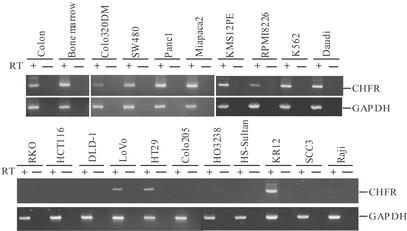
Loss of CHFR Expression. Earlier studies designed to characterize novel mitotic checkpoint genes led to the identification of CHFR (5). Although CHFR mRNA was found to be ubiquitously expressed in normal tissues, the mRNA was absent in a number of cancer cell lines (Fig. 1). Using RT-PCR, we found that 10 of the cell lines tested (RKO, HCT116, DLD-1, Colo205, Colo201, HO3238, Jurkat, HS-Sultan, SCC3, and Raji) did not express CHFR; 4 (LoVo, HT-29, BM314, and Raji) showed weak expression of the gene; and the remaining 29 showed high levels of expression (Fig. 1). When we sequenced the entire CHFR coding region of 23 cell lines, we found several nucleotide sequence changes, all of which were either silent or resulted in conservative amino acid substitutions outside of the FHA and RING finger domains. These sequence variants appear to be polymorphic changes because identical variations were detected in normal tissues (data not shown). We concluded, therefore, that pathways other than genetic inactivation were responsible for the silencing of CHFR in cancer cells.
Fig. 1.
RT-PCR analysis of CHFR expression in human tumor cell lines. Cell lines are indicated above the data. PCR was performed by using samples prepared with (RT+) or without (RT-) reverse transcriptase. GAPDH was amplified to show the integrity of the RNA.
Expression of the Human CHFR Depends on Its CpG Methylation Status. DNA methylation of 5′ regulatory regions harboring a higher than expected number of CpG dinucleotides is a key mechanism by which genes relevant to cancer initiation and progression are silenced. We therefore determined the location and density of CpG nucleotides within the CHFR locus. Database-assisted mapping of CHFR cDNA (GenBank accession no. AF170724) identified a genomic sequence on chromosome 12 (GenBank accession no. AC023047). Moreover, the 5′ region of CHFR fulfilled the strict criteria defining a CpG island (14): it spanned ≈1.5 kb and had a CpG frequency of 65% and a CpG/GpC ratio of 0.94 (Fig. 2A). By comparing the genomic DNA and cDNA sequences of CHFR (GenBank accession no. BI461698), the 5′-most end of the gene was determined to be 77 nt upstream of the known translation start site.
Fig. 2.
Methylation status of CHFR.(A) CpG island of CHFR: exons 1 and 2 are indicated by solid boxes, the transcription start site is indicated by an arrow, and the region analyzed by bisulfite PCR is indicated by a bar. (B) Re-expression of CHFR after treatment with the methyltransferase inhibitor 5-aza-dC. (C) Aberrant methylation of CHFR in human tumor cell lines. Cell lines are indicated above the data; the percentage of methylation is indicated below. (D) Representative results of analysis of CHFR methylation in human primary colorectal and head and neck cancers. N, normal tissue; T, primary cancers; OSCC, oral squamous cell cancer.
Having confirmed that CHFR contains a CpG island, we wondered whether expression of this gene could be perturbed experimentally with pharmacological agents that interfere with DNA methylation. To answer that question, we treated cell lines harboring a silenced CHFR with DNMT inhibitor 5-aza-dC, which restored CHFR expression, suggesting the silencing of CHFR was methylation-dependent (Fig. 2B).
Analysis of CHFR CpG Methylation in Human Cancers. To further investigate the relationship between CpG methylation and CHFR expression, we used bisulfite PCR to survey the methylation status of individual CpG dinucleotides within CHFR in human tissues and tumors (Figs. 2C and 8). Using a set of primers that encompass the region surrounding the putative transcription start site, we found six colorectal cancer cell lines (RKO, SW48, HCT116, DLD-1, Colo205, and Colo201) that harbored densely methylated CpG sites (>80%), four (Caco2, LoVo, HT29, and BM314) that showed partial methylation, and two (Colo320 and SW480) that did not present any detectable methylation. Thus, the 5′ CpG island of CHFR was aberrantly methylated in a majority of colorectal cancer cells, and the degree of methylation correlated precisely with levels of CHFR RNA.
To confirm that finding, we analyzed the 5′ regulatory region of CHFR by using bisulfite sequencing. After bisulfite treatment, individual DNA molecules were amplified, cloned, and sequenced, simultaneously revealing the methylation state at several CpG dinucleotides on a single allele. Consistent with the results obtained with bisulfite PCR, cell lines in which CHFR was silent exhibited virtually complete methylation of CpG dinucleotides (Fig. 9A, which is published as supporting information on the PNAS web site), whereas cell lines showing intermediate levels of CHFR expression were partially methylated at CpG dinucleotides. Thus, the density of CpG methylation detected by bisulfite sequencing correlated precisely with the expression pattern. In addition, bisulfite sequencing revealed that the density of methylation detected by bisulfite PCR was well correlated with that of methylation of each CpG site in primary colorectal tumors (Fig. 9B).
We then analyzed the methylation status in a panel of tumor specimens from patients with colorectal cancers to determine whether the aberrant methylation of the CHFR gene CpG island seen in cancer cell lines reflects an epigenetic processes ongoing in human cancers. Using bisulfite PCR, we detected aberrant methylation in 25 of 63 (40%) primary colorectal cancers and 27 of 51 (53%) colorectal adenomas (Fig. 2D). Notably, expression of CHFR mRNA in the absence of aberrant methylation was detected in specimens of normal colonic epithelium (Fig. 1). In addition, expression of CHFR in colorectal cancer xenografts showed that methylation significantly diminished CHFR expression, confirming that methylation-mediated gene silencing is not a cell line-specific event (Fig. 10, which is published as supporting information on the PNAS web site). Furthermore, these data imply that the methylation of CHFR and, presumably, its transcriptional silence are established at an early stage in the stepwise progression of colorectal carcinogenesis.
To determine the extent to which aberrant CHFR methylation is present in tumors derived from other tissue types, we first carried out bisulfite PCR in a panel of cancer cell lines derived from oral squamous cell carcinomas, hepatocellular carcinomas, and a number of hematological malignancies (Fig. 2C). Of the 43 cell lines examined, eight (HO3238, Jurkat, TALL1, HS-Sultan, SCC3, OSC20, KOSC3, and OSC40) showed a high degree of methylation (>80%), and five (KR12, Raji, Malt4, IM9, and OSC30) showed partial methylation (8–69%), frequencies similar to those observed in colorectal cancer cell lines. Analysis of corresponding primary tumors revealed aberrant methylation of CHFR in 16 of 54 (30%) primary head and neck cancers, a frequency similar to that observed in cell lines, but no methylation in samples of normal lymphocytes or tongue tissue obtained from the patients whose tumors were analyzed (Fig. 2D). Interestingly, no methylation was detected in hepatocellular carcinoma cell lines or primary hepatocellular cancers. The methylation frequencies for the various tumor types are summarized in Table 1.
Table 1. Frequency of CHFR methylation in human tumors.
| No. methylated (%) | |
|---|---|
| Cell lines | |
| Colorectal | 11/13 (85) |
| Head and neck | 4/10 (40) |
| ALL | 4/8 (50) |
| Myeloma | 4/7 (57) |
| Other hematopoietic | 2/5 (40) |
| Hepatocellular | 0/5 (0) |
| Other tumors | 0/8 (0) |
| Primary tumors | |
| Colorectal cancer | 25/63 (40) |
| Colorectal adenoma | 27/51 (53) |
| Head and neck | 16/54 (30) |
| Hepatocellular | 0/20 (0) |
CHFR CpG Methylation and Gene Expression Is Maintained by DNMT1 and DNMT3b. As mentioned above, expression of CHFR could be experimentally restored by treatment with the DNMT inhibitor 5-aza-dC (Fig. 2B). Therefore, to examine the pathway leading to methylation of CHFR in more detail, we used colorectal cancer cells (HCT116) genetically engineered for deletion of DNMT1 (MT1KO) (8), DNMT3b (3bKO), or both (DKO; genotype, DNMT1-/-DNMT3b-/-) (9). It was previously shown that MT1KO and 3bKO cells retain 80–90% of genomic 5-methylcytosine: a significant loss is observed within the centromeric repeat regions on specific chromosomes, but the methylation and expression status of individual hypermethylated genes (e.g., p16INK4A) is maintained in both cell types (7). However, the deletion of both DNMT1 and DNMT3b in DKO cells results in cooperative loss of methylation, virtually eliminating global 5-methylcytosine content, and a concomitant re-expression of single copy genes, including p16INK4A (9).
We wondered whether genetic ablation of DNMT1 and/or DNMT3b would result in demethylation and re-expression of CHFR in colorectal cancer cells. As shown in Fig. 3A, expression of CHFR mRNA was unaffected by genetic ablation of either DNMT1 or DNMT3b. In cells harboring simultaneous deletions of both methyltransferases, however, we detected significant levels of CHFR mRNA. Consistent with that finding, bisulfite PCR and sequencing revealed a complete loss of 5-methylcytosine from the CHFR CpG island, demonstrating the absolute requirement for these enzymes in the maintenance of CHFR methylation (Fig. 3 B and C) and establishing a strict causal relationship between DNA methylation and gene silencing in the regulation of CHFR expression in colorectal cancer.
Fig. 3.
Expression and methylation status of CHFR in DNMT knockout cells. (A) Re-expression of CHFR in methyltransferase knockout cells. The genotypes of the cell lines are indicated: MT1KO1 and MT1KO2, DNMT1-/-; 3bKO1 and 3bKO2, DNMT3b-/-; DKO2 and DKO3, DNMT1-/-DNMT3b-/-. (B) Bisulfite PCR analysis of the methylation status of the CHFR CpG island in HCT116 and methyltransferase knockout cells; M, methylated alleles. (C) Bisulfite sequencing of CHFR in WT and DNMT knockout cells. The PCR products were cloned into a vector, and at least nine clones were sequenced for each cell line. Open and closed areas represent unmethylated and methylated CpG dinucleotides, respectively.
Histone Deacetylation at the CHFR Locus in Human Cancer Cells. Methylation-dependent gene silencing is thought to be associated with a modified chromatin structure enriched in deacetylated histones (15). We therefore used the histone deacetylase inhibitor TSA to examine the potential role of histone acetylation in the regulation of CHFR expression. Quantitative realtime PCR was performed, and the results were normalized to the expression of GAPDH. When RKO colon cells, in which CHFR is densely methylated, were treated with TSA, we detected no changes in gene expression, even though parallel reactions with 5-aza-dC robustly elevated CHFR expression. On the other hand, treating RKO cells with both TSA and 5-aza-dC enhanced expression of CHFR to levels greater than were seen with either drug alone (Fig. 4A). This apparently synergistic interaction between TSA and 5-aza-dC is consistent with changes seen in a number of other genes in colorectal cancer cells (16) and is indicative of the integral relationship between altered chromatin structure and DNA methylation at the CHFR locus.
Fig. 4.
Expression and histone acetylation status of CHFR in a human tumor cell line. (A) Re-expression of CHFR in the RKO colorectal cancer cell line after treatment with 5-aza-dC and/or TSA. RKO cells were treated as follows: lane 1, mock; lane 2, 0.2 μM 5-aza-dC for 72 h; lane 3, 300 nM TSA for 24 h; and lane 4, 0.2 μM 5-aza-dC for 72 h plus 300 nM TSA for 24 h. The bar graph depicts the ratios of the intensities of the CHFR and GAPDH signals. (B) Chromatin immunoprecipitation analysis of the 5′ region of CHFR. Genomic DNA associated with acetylated histones H3 or H4 was prepared by immunoprecipitation using antibodies specific to acetylated histones H3 or H4. PCR was performed by using oligonucleotides specific for the 5′ region of CHFR. Input DNA was amplified to evaluate the total amount of DNA applied to the immunoprecipitation. As a control, the region around the transcription start site of GAPDH was amplified by using the DNA recovered from the chromatin immunoprecipitation assay.
The effect of TSA suggests that there is an accumulation of hyperacetylated H3 and H4 histones within transcriptionally active regulatory regions and an accumulation of the deacetylated forms of histones H3 and H4 in the regions of transcriptionally silent genes. To examine the status of the histones within the CHFR locus, we subjected various cell lines to chromatin immunoprecipitation using antibodies that specifically recognize the acetylated forms of histones H3 and H4. After amplification with primers specific for a portion of the CHFR CpG island, an enrichment of hyperacetylated histones was observed in cells expressing CHFR (Colo320, RPMI8226, and K562) (Fig. 4B), whereas cells harboring a methylation-dependently silenced CHFR gene (HS-Sultan, RKO, and DLD-1) showed a complete absence of hyperacetylated histones, and intermediate levels of histone acetylation were detected in cell lines showing partial methylation of the CHFR promoter (Raji and HT29). Thus, histone acetylation status correlated directly with CHFR expression and inversely with the methylation status of the CHFR CpG island.
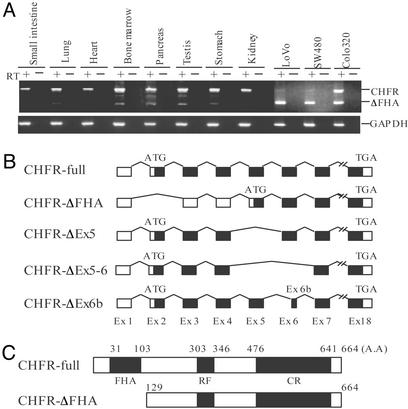
CHFR Protein Lacking the FHA Domain Is Expressed in Human Tumors. During the initial characterization of CHFR expression in cancer cells, we sequenced the entire cDNA in an attempt to identify mutations that would implicate genetic pathways in the functional inactivation of CHFR protein. Although we did not discover any mutations, we did find alternatively spliced CHFR transcripts that lacked exons 2, 5, or 6 (Fig. 5 A and B). Among these, the transcript lacking exon 2 was particularly interesting because the protein products would be predicted to lack the FHA domain, a peptide sequence that binds phosphorylated peptides (17, 18). We designated this isoform CHFR-ΔFHA (Fig. 5 B and C), and because expression of the CHFR-ΔFHA mRNA species had not been described previously, we asked whether these transcripts were expressed in normal or cancerous tissues. When small intestine, lung, heart, bone marrow, testis, kidney, stomach, and lymphocytes were surveyed for CHFR-ΔFHA RNA, we detected only low levels of expression. By contrast, relative expression levels of the ΔFHA isoform were much higher in human cancer cells, suggesting this isoform might play a functional role in cancer (Fig. 5A). In particular, CHFR-ΔFHA was the dominant isoform expressed in SW480 and LoVo cells.
Fig. 5.
(A) Expression of a truncated form of CHFR in tumor cell lines. The 396-bp products correspond to expected size of WT CHFR; the 241-bp products correspond to the FHA domain-deleted variant (CHFR-ΔFHA). LoVo, SW480, and Colo320 cells express CHFR-ΔFHA. GAPDH was amplified to show the integrity of the cDNA. (B) Schematic diagram showing various CHFR mRNA splicing variants. The names of the transcripts are shown on the left. Closed boxes indicate the coding regions. (C) Schematic diagram of intact CHFR and CHFR-ΔFHA proteins. FHA, fork-head associated domain; RF, RING finger domain; CR, cystein rich domain. Amino acid numbers (A.A) are shown on the top.
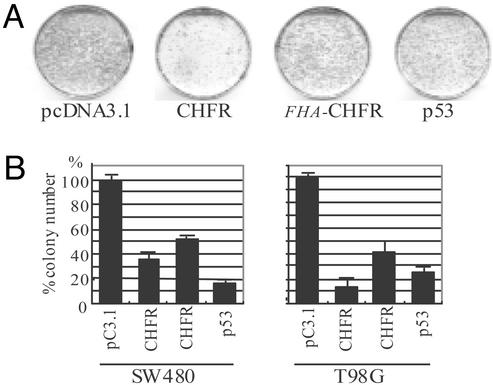
Ectopic Expression of CHFR Suppresses Cell Growth. The frequent inactivation of CHFR in human tumors suggests its product has tumor suppressor activity. To test this hypothesis, we transfected CHFR into various human cancer cell lines and then carried out G418 colony formation assays (Fig. 6). Introduction of fulllength CHFR markedly suppressed cell growth; expression of CHFR-ΔFHA was less effective, indicating a role for the FHA domain in the regulation of cell growth. Introduction of p53 also significantly suppressed cell growth, although the efficiency varied among the cell lines tested.
Fig. 6.
Suppression of growth by CHFR. (A) Colony formation assay using the T98G human tumor cell line. Cells were transfected with CHFR, CHFR-ΔFHA,or empty vector (pcDNA3.1) and plated. After 2 weeks, the cells were fixed with methanol and stained with Gimsa. (B) Quantitative analyses of colony numbers after transfection of CHFR, CHFR-ΔFHA, or p53 in several human tumor cell lines. Each experiment was repeated three times; the average numbers of colonies are shown.
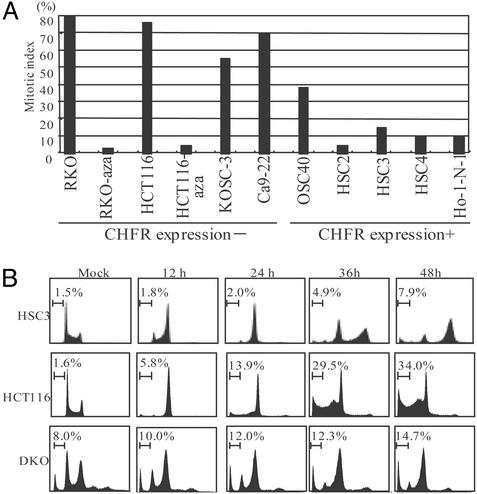
Loss of CHFR Is Associated with Impairment of a Checkpoint into Mitosis. To further analyze the functional consequences of CHFR methylation in human tumors, we determined the mitotic index in various tumor cell lines after treatment with docetaxel, a microtubule inhibitor. We found that the mitotic index was significantly higher in cell lines that showed CHFR methylation than in those that did not (Fig. 7A). Furthermore, the mitotic checkpoint was restored by treatment with 5-aza-dC, indicating its epigenetic inactivation. It is unlikely that this effect was due to the cellular stress caused by the chemical treatment, as DKO cells also showed a decreased mitotic index, and treatment of cell lines without CHFR methylation did not affect the mitotic index (Fig. 11 A and B, which is published as supporting information on the PNAS web site). The checkpoint was also restored by introduction of CHFR into cells using an adenoviral vector, which confirms the role of CHFR at the checkpoint into mitosis (Fig. 11 C and D). Furthermore, the importance of the FHA domain was demonstrated by the finding that introduction of Ad-ΔFHA was less able to restore the checkpoint than the intact protein.
Fig. 7.
Impairment of checkpoint into mitosis in CHFR-deficient cell lines. (A) Mitotic index of cancer cell lines after treatment with microtubule inhibitor. Cell lines were treated for 16 h with 1 μM docetaxel, after which the mitotic index was determined. (B) Cell cycle analysis. The indicated cell lines were treated for 0, 12, 24, 36, or 48 h and subjected to fluorescence-activated cell sorting analysis. The apoptotic cells were indicated as the sub-G1 fraction.
Finally, the effect of microtubule inhibition was further characterized by flow cytometry. Cell cycle analysis revealed that cell lines expressing CHFR arrested at G2/M within 48 h after treatment with docetaxel (Fig. 7B). On the other hand, cell lines lacking CHFR expression underwent apoptosis 48 h after docetaxel treatment. Restoration of the checkpoint was observed in DKO cells whose genotype other than DNMT1 and DNMT3b is thought to be almost identical to that of HCT116 cells, which is indicative of the role of epigenetic inactivation of CHFR in the altered checkpoint into mitosis in cancer cells.
Discussion
CHFR encodes a protein with FHA and RING finger domains that functions in the mitotic checkpoint pathway that governs transition from prophase to prometaphase (5, 19). Expression of this gene is apparently lost in several human cancer cell lines and primary lung and esophageal cancers (5–7). In the present article, we have shown that the silencing of CHFR is the result of DNA methylation of its 5′ CpG island and histone deacetylation. Although much evidence suggests that one or more mitotic checkpoints is impaired in cancer cells, so far few changes in the expression of molecules involved in the mitotic checkpoint pathway have been identified (3, 20, 21). Analysis of the methylation of BUB1 and MAD2L1, two other mitotic checkpoint genes, revealed no alterations in 13 colorectal cancer cell lines (data not shown). On the other hand, the importance of CHFR methylation in tumorigenesis is supported by our observations that CHFR is expressed in normal tissues and down-regulated in a wide variety of human tumors, that changes in the level of methylation are tumor-specific and closely associated with the loss of gene expression, and that introduction of ectopic CHFR reduces colony formation in tumor cells.
The fact that CHFR expression in methylated cell lines was restored by 5-aza-dC, a methyltransferase inhibitor, confirms that inactivation of the gene was caused by DNA methylation and not by the loss of transcription factors. Likewise, genetic ablation of DNMT1 and DNMT3b restored expression of CHFR, which is consistent with the previously observed, genomewide demethylation seen in the absence of these two enzymes (9). The mechanism by which DNA methylation represses gene expression is not fully understood, although recent studies indicate histone deacetylation plays a role (16, 22). Our chromatin immunoprecipitation assays showed that both histone H3 and H4 are deacetylated in cell lines where CHFR is silent, and that 5-aza-dC and TSA act synergistically to enhance CHFR expression. Histone deacetylation thus appears to be involved in the methylation-dependent silencing of CHFR, just as in the silencing of p16INK4A, p14ARF, and BRCA1 (13, 23). Nevertheless, our analysis showed that reducing DNA methylation can restore expression of CHFR despite histone deacetylation, indicating DNA methylation to be a more potent determinant of CHFR expression than histone acetylation.
The fact that cell lines with CHFR methylation showed higher mitotic indexes after treatment with microtubule inhibitor docetaxel than cell lines without methylation, and that treatment with 5-aza-dC reduced the mitotic index in methylated cell lines, suggests CHFR methylation is crucially involved in the mitotic checkpoint impairment seen in various cancers. Still, little is known about how CHFR contributes to the mitotic checkpoint. CyclinB/cdc2 activity remains high during the CHFR-dependent delay, so its activity is likely not a target of CHFR (5). On the other hand, CHFR has a RING finger domain and has been postulated to be involved in the activity of ubiquitin ligase. In fact, CHFR was recently shown to possess ubiquitin ligase activity and to be involved in the degradation of polo-like kinase 1 (PLK1) caused by mitotic stress (24, 25). Consequently, loss of CHFR may lead to accumulation of PLK1, in turn leading to mitotic checkpoint impairment.
CHFR also contains an FHA domain in its N terminus. This domain, which is involved in phosphopeptide binding, was first identified in a group of transcription factors, but is also present in a wide variety of other molecules, including cell cycle regulators Chk2 and Rad53 (17, 18). Disruption of the FHA domain would be expected to impair the ability of CHFR to interact with target molecules; indeed disruption of the FHA domains of Rad53 and CHK2 has been shown alter the cell cycle checkpoint (17). What's more, Scolnick and Halazonetis (5) reported that a CHFR mutant lacking the FHA domain exerted a dominant negative effect against WT CHFR. In similar fashion, we observed that expression of the CHFR-ΔFHA isoform appeared to alter the properties of the G2/M checkpoint in several tumor cell lines.
Much remains to be determined before the changes in the regulation of the cell cycle in human cancer cells are fully understood. Nevertheless, the present finding of aberrant CHFR methylation in large number of cancers, together with clarification of the functional significance of CHFR in the mitotic checkpoint pathway, has the potential to provide significant clinical benefits. For example, cancer cell lines that do not express CHFR were recently shown to be more susceptible to the microtubule inhibitor taxol (5, 24). This finding means that the silencing of CHFR expression by methylation can serve as a molecular marker useful for predicting the sensitivity to particular chemotherapeutic agents. In addition, because aberrant methylation of CHFR is tumor-specific, it can be used as a diagnostic marker to predict recurrence or metastasis of tumors.
Supplementary Material
Acknowledgments
We thank Dr. William F. Goldman for editing the manuscript and Setsuko Ishida and Mutsumi Ohe-Toyota for technical assistance. This study was supported by grants-in-aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology (to M.T., F.I., K.I., and T.T.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DNMT, DNA methyltransferase; 5-aza-dC, 5-aza-2′-deoxycytidine; TSA, trichostatin A.
References
- 1.Lengauer, C., Kinzler, K. W. & Vogelstein, B. (1998) Nature 396, 643-649. [DOI] [PubMed] [Google Scholar]
- 2.Michel, L. S., Liberal, V., Chatterjee, A., Kirchwegger, R., Pasche, B., Gerald, W., Dobles, M., Sorger, P. K., Murty, V. V. & Benezra, R. (2001) Nature 409, 355-359. [DOI] [PubMed] [Google Scholar]
- 3.Cahill, D. P., Lengauer, C., Yu, J., Riggins, G. J., Willson, J. K., Markowitz, S. D., Kinzler, K. W. & Vogelstein, B. (1998) Nature 392, 300-303. [DOI] [PubMed] [Google Scholar]
- 4.Jallepalli, P. V., Waizenegger, I. C., Bunz, F., Langer, S., Speicher, M. R., Peters, J. M., Kinzler, K. W., Vogelstein, B. & Lengauer, C. (2001) Cell 105, 445-457. [DOI] [PubMed] [Google Scholar]
- 5.Scolnick, D. M. & Halazonetis, T. D. (2000) Nature 406, 430-435. [DOI] [PubMed] [Google Scholar]
- 6.Shibata, Y., Haruki, N., Kuwabara, Y., Ishiguro, H., Shinoda, N., Sato, A., Kimura, M., Koyama, H., Toyama, T., Nishiwaki, T., et al. (2002) Carcinogenesis 23, 1695-1699. [DOI] [PubMed] [Google Scholar]
- 7.Mizuno, K., Osada, H., Konishi, H., Tatematsu, Y., Yatabe, Y., Mitsudomi, T., Fujii, Y. & Takahashi, T. (2002) Oncogene 21, 2328-2333. [DOI] [PubMed] [Google Scholar]
- 8.Rhee, I., Jair, K. W., Yen, R. W., Lengauer, C., Herman, J. G., Kinzler, K. W., Vogelstein, B., Baylin, S. B. & Schuebel, K. E. (2000) Nature 404, 1003-1007. [DOI] [PubMed] [Google Scholar]
- 9.Rhee, I., Bachman, K. E., Park, B. H., Jair, K. W., Yen, R. W., Schuebel, K. E., Cui, H., Feinberg, A. P., Lengauer, C., Kinzler, K. W., et al. (2002) Nature 416, 552-556. [DOI] [PubMed] [Google Scholar]
- 10.Toyota, M., Ahuja, N., Ohe-Toyota, M., Herman, J. G., Baylin, S. B. & Issa, J. P. (1999) Proc. Natl. Acad. Sci. USA 96, 8681-8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toyota, M., Ohe-Toyota, M., Ahuja, N. & Issa, J. P. (2000) Proc. Natl. Acad. Sci. USA 97, 710-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong, Z. & Laird, P. W. (1997) Nucleic Acids Res. 25, 2532-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magdinier, F. & Wolffe, A. P. (2001) Proc. Natl. Acad. Sci. USA 98, 4990-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takai, D. & Jones, P. A. (2002) Proc. Natl. Acad. Sci. USA 99, 3740-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bird, A. P. & Wolffe, A. P. (1999) Cell 99, 451-454. [DOI] [PubMed] [Google Scholar]
- 16.Cameron, E. E., Bachman, K. E., Myohanen, S., Herman, J. G. & Baylin, S. B. (1999) Nat. Genet. 21, 103-107. [DOI] [PubMed] [Google Scholar]
- 17.Sun, Z., Hsiao, J., Fay, D. S. & Stern, D. F. (1998) Science 281, 272-274. [DOI] [PubMed] [Google Scholar]
- 18.Li, J., Lee, G. I., Van Doren, S. R. & Walker, J. C. (2000) J. Cell. Sci. 113, 4143-4149. [DOI] [PubMed] [Google Scholar]
- 19.Cortez, D. & Elledge, S. J. (2000) Nature 406, 354-356. [DOI] [PubMed] [Google Scholar]
- 20.Yamaguchi, K., Okami, K., Hibi, K., Wehage, S. L., Jen, J. & Sidransky, D. (1999) Cancer Lett. 139, 183-187. [DOI] [PubMed] [Google Scholar]
- 21.Cahill, D. P., da Costa, L. T., Carson-Walter, E. B., Kinzler, K. W., Vogelstein, B. & Lengauer, C. (1999) Genomics 58, 181-187. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen, C. T., Gonzales, F. A. & Jones, P. A. (2001) Nucleic Acids Res. 29, 4598-4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rice, J. C. & Futscher, B. W. (2000) Nucleic Acids Res. 28, 3233-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaturvedi, P., Sudakin, V., Bobiak, M. L., Fisher, P. W., Mattern, M. R., Jablonski, S. A., Hurle, M. R., Zhu, Y., Yen, T. J. & Zhou, B. B. (2002) Cancer Res. 62, 1797-1801. [PubMed] [Google Scholar]
- 25.Kang, D., Chen, J., Wong, J. & Fang, G. (2002) J. Cell Biol. 156, 249-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.