Abstract
The ALL-1 gene is directly involved in 5–10% of acute lymphoblastic leukemias (ALLs) and acute myeloid leukemias (AMLs) by fusion to other genes or through internal rearrangements. DNA microarrays were used to determine expression profiles of ALLs and AMLs with ALL-1 rearrangements. These profiles distinguish those tumors from other ALLs and AMLs. The expression patterns of ALL-1-associated tumors, in particular ALLs, involve oncogenes, tumor suppressors, antiapoptotic genes, drug-resistance genes, etc., and correlate with the aggressive nature of the tumors. The genes whose expression differentiates between ALLs with and without ALL-1 rearrangement were further divided into several groups, enabling separation of ALL-1-associated ALLs into two subclasses. One of the groups included 43 genes that exhibited expression profiles closely linked to ALLs with ALL-1 rearrangements. Further, there were evident differences between the expression profiles of AMLs in which ALL-1 had undergone fusion to other genes and AMLs with partial duplication of ALL-1. The extensive analysis described here pinpointed genes that might have a direct role in pathogenesis.
Chromosome band 11q23 is a region of recurrent rearrangements in human acute leukemias. These rearrangements, usually in the form of reciprocal chromosome translocations, affect 5–10% of children and adults with acute lymphoblastic leukemia (ALL) and acute myeloblastic leukemia (AML). The most common translocations are t(4;11) and t(9;11), accounting for 40% and 27%, respectively, of all 11q23 rearrangements. There is a strong association between leukemia phenotype and particular rearrangements. Thus, t(4;11) occurs nearly exclusively in ALL, and 85% of cases with t(9;11) are AMLs (1, 2). Essentially all 11q23 abnormalities involve the ALL-1 gene (also termed MLL, HRX, or HTRX), which rearranges with >30 partner genes to produce fusion proteins composed of ALL-1 N terminus and the C terminus of the partner protein (3, 4). A second and less frequent type of ALL-1 rearrangement does not involve partner genes but rather partial duplications of ALL-1 N-terminal segments (5). ALL-1-associated leukemias show some unusual and intriguing features (reviewed in refs. 6 and 7). First, they predominate infant acute leukemias, amounting to 80% of infants with ALL and 65% of those with AML. Second, they account for the majority of therapy-related (secondary) leukemias, developing in 5–15% of primary cancer patients treated with drugs, such as etoposide (VP16), that inhibit DNA topoisomerase II. Third, in infant leukemia and in therapy-related leukemia the disease arises after a brief latency. In fact, studies of monozygotic twins and newborns with leukemia and analysis of neonatal blood spots from children who were diagnosed with leukemia indicate that in most or all infant leukemias ALL-1 rearrangements occur in utero. The short latency suggests that ALL-1 fusion proteins induce leukemia with few, if any, additional mutations. Fourth, prognosis of patients with 11q23 abnormalities is dismal. Recent large studies indicated that <25% of infants and adults >40 years old with ALL and t(4;11), or with AML and t(9;11), were curable (1, 2, 8).
The unique biological and clinical features of 11q23-associated leukemias, in conjunction with their induction by altered versions of ALL-1, a highly intricate chromatin modifier (9), prompted us to look for molecular clues for those features by examining the expression profiles of these leukemias.
Materials and Methods
Patients, Specimens, and DNA Microarrays. Apart from two individuals, all patients with 11q23 abnormalities were adults. The samples were provided by the GIMEMA Italian Multicenter Study Group. Informed consent was obtained from the patients. Also included in the analysis were four AML cell lines with t(9;11) (MONO-MAC-1, MONO-MAC-6, THP-1, and MOLM-13) and one with t(6;11) (ML-2), and two ALL cell lines with t(4;11), RS-(4;11) and B-1. Genes picked up in the supervised analysis, as well as most of those pointed out as separating ALLs with and without t(4;11) in nonsupervised analysis, had similar expression profiles in cell lines and primary tumors. The primary tumors included 12 ALLs with t(4;11) obtained from 10 adults, one child, and one infant, and 10 AMLs of adults, including 5 with t(9;11), 3 with ALL-1 partial duplication, and single cases of t(10;11) and t(11;19). Controls comprised 10 AMLs of adults, 11 ALLs of adults, and 2 ALLs of children. Details regarding the patients the sample identifies may be found in Table 2 and Supporting Text, which are published as supporting information on the PNAS web site, www.pnas.org. Bone marrow samples were obtained from newly diagnosed patients. The samples were composed of at least 80% blasts and were subjected to Ficoll gradient centrifugation before extraction of RNA. Details of microarray analysis may be found in Supporting Text.
Preprocessing and Filtering of Data. The expression data were organized in a matrix of ns = 52 columns (hybridizations) and 12,600 rows (genes on the chip). Denote by Ags the “average difference” of gene g in sample s. First, we thresholded the data; we set Tgs = Ags for sizeable values, Ags ≥ 10, and replaced low values, Ags < 10, by Tgs = 10. Next, log was taken, Egs = log2Tgs, and the genes were filtered on the basis of their variation across the samples. Denote by E Ēgs the average of the Egs values obtained for gene g over all ns tumor samples and by σg their standard deviation. Only those genes that satisfied σg > 1.1 were studied; 3,090 genes passed this filtering procedure. After removal of non-human Affymetrix controls and genes appearing on only one of the versions of the U95A chip, we were left with 3,064 genes (of 12,600). All further analysis was done on these genes.
Supervised Analysis. We used supervised analysis (hypothesis testing) to identify genes, one at a time, whose expression levels can be used to separate tumors into two known classes A and B of nA and nB samples, respectively. We used the Wilcoxon rank sum test to find genes differentially expressed between the two groups of samples (e.g., AML samples with chromosome translocations versus those without; other comparisons are listed in Results). We used the rank sum test because it is nonparametric, i.e., it does not assume normal distribution of the data. For each gene g we make the null hypothesis, according to which all nA + nB expression levels were drawn from the same distribution. The test produces a statistic Wg and a P value for each gene g.A P value of Pg = 0.05 means that the probability of erroneously concluding that a gene does separate the two groups is 5%, which is the standard value used in the literature. However, here we deal with multiple comparisons; at this level of Pg it is expected that 150 of 3,000 random, identically distributed genes will be falsely identified as separating the two groups of samples. To control the number of false positives, we used the false discovery rate (FDR) method (10). The N tested genes are ordered according to their increasing Pg values, and a parameter q, which controls the fraction of false positives, is set. We then identify the minimal index j such that for all i > j we have Pi ≥ 1 × q/N. The null hypothesis is rejected for all genes with index i ≤ j. This procedure yields a list of genes for which the expected fraction of false positives is q.
Unsupervised Analysis: Clustering. We used the coupled two-way clustering method (11). We assume that each group of genes is important for one particular process of interest. Thereby, the noise generated by the large majority of genes that are not relevant for that process is eliminated; furthermore, by using a group of correlated genes, the noise of the individual measurements is averaged out and reduced. The relevant subsets of genes and samples are identified by means of an iterative process, which uses, at each iteration level, stable gene and sample clusters that were generated at the previous step. Before each clustering operation the rows of the data matrix (genes) are centered (mean = 0) and normalized (SD = 1). The ability to focus on stable clusters that were generated by any clustering operation is essential for the coupled two-way clustering method; otherwise, there would be a computationally unfeasible number of gene/sample cluster pairs to test (11). Because most clustering methods do not have a reliable inherent stability measure for clusters, we used Superparamagnetic Clustering (SPC), a physics-based algorithm (12) that does provide a stability index, ΔT(C), to each cluster C. SPC was tested on data from a large number of problem areas including image analysis, speech recognition, computer vision, and gene expression (refs. 11 and 12 and references therein). A parameter T controls the resolution at which the data are viewed; as T increases, clusters break up and the outcome is a dendrogram. A cluster C is “born” at T = T1(C), the value of T at which its “parent” cluster breaks up into two or more subclusters, one of which is C. As T increases further, to T2(C) > T1(C), C itself breaks up and “dies”; ΔT(C) = T2(C) - T1(C) is the stability index provided by SPC. The larger ΔT(C) is, the more statistically significant and stable (against noise in the data and fluctuations) is the cluster C (13).
Results
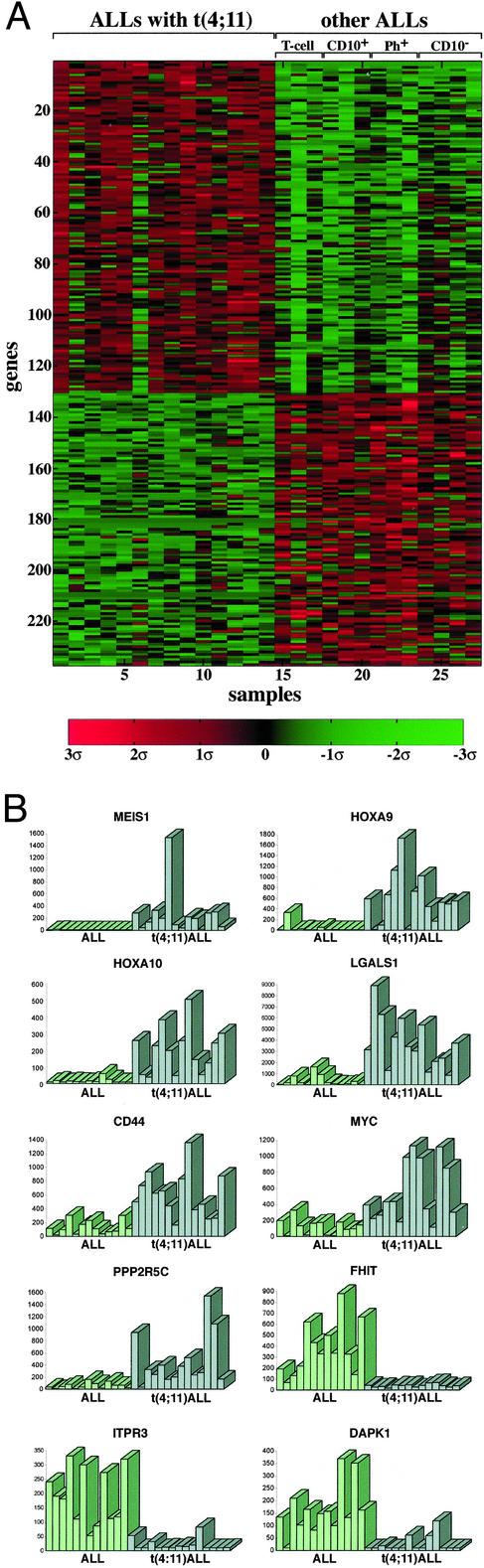
Expression Profiles of ALLs with t(4;11). Leukemic cells of ALLs with t(4;11) display features of precursor B cells with IgH rearrangements, negative for CD10 and positive for CD19, but also show some characteristics of myeloid cells (1). This and their capability to differentiate in vitro into monocyte-like cells had suggested that the leukemic clones originate from an early precursor cell. Hence, this leukemia is classified as pro-B cell ALL. To determine whether the expression repertoire of ALLs with t(4;11) is unique, we compared it to the transcription profiles of a set of ALL samples lacking t(4;11). These consisted of CD10- pro-B cell ALLs, Ph chromosome-positive early pre-B cell ALLs, CD10+ early pre-B cell ALLs, and T cell ALLs. Supervised analysis indicated that at a FDR of 0.05 there were 130 overexpressed and 107 underexpressed genes in ALLs with t(4;11), in comparison to ALLs lacking the abnormality (Fig. 1A). To evaluate the consistency of the pattern, the relative expression of each gene in all of the samples was displayed in the form of bars (see examples in Fig. 1B). The top genes on the lists of overexpressed or underexpressed genes in ALLs with t(4;11) are shown in Table 1. The complete lists may be found in Table 3, which is published as supporting information on the PNAS web site. The clear difference in expression profiles between ALLs with the t(4;11) abnormality and other types of ALLs establishes that the former belong to a unique and distinguishable class of ALL.
Fig. 1.
Supervised analysis of genes distinguishing ALLs with ALL-1 rearrangement [t(4;11)] from other ALLs (A), and relative levels of expression of selected genes (B). Expression levels greater and smaller than the mean 0 are shown in red and green, respectively.
Table 1. Genes most correlated with ALLs carrying the t(4;11) aberration, compared to other ALLs.
| No.* | Also scored as t(4;11)-specific† | GenBank accession no. | P value | Fold change | Confidence interval | |
|---|---|---|---|---|---|---|
| Overexpressed in t(4;11) ALLs | ||||||
| 1 | √ | D16532 | VLDLR, very low density lipoprotein receptor | 0.000004 | 17.51 | (10.67-28.74) |
| 2 | √ | U85707 | MEIS1, myeloid ecotropic viral integration site 1 homolog (mouse) | 0.000004 | 14.50 | (7.64-27.51) |
| 3 | √ | AC004080 | HOXA10, homeo box A10 | 0.000022 | 10.80 | (6.17-18.90) |
| 4 | √ | AI535946 | LGALS1, lectin, galactoside-binding, soluble 1 (galectin 1) | 0.000024 | 23.00 | (8.82-59.98) |
| 5 | √ | M54992 | CD72 antigen | 0.000037 | 4.35 | (2.80-6.74) |
| 6 | √ | U41813 | HOXA9, homeo box A9 | 0.000041 | 20.12 | (7.22-56.09) |
| 7 | AF098641 | CD44, CD44 isoform (Indian blood group system) | 0.000056 | 4.43 | (2.65-7.41) | |
| 9 | √ | AA099265 | RECK, reversion-inducing-cystein-rich protein with kazal motifs | 0.000063 | 3.58 | (2.03-6.31) |
| 10 | √ | M14087 | HL14, β-galactoside-binding lectin | 0.000068 | 6.94 | (3.50-13.76) |
| 11 | √ | Z69030 | PPP2R5C, protein phosphatase 2, regulatory subunit B (B56), γ isoform | 0.000069 | 7.55 | (3.60-15.83) |
| 13 | D83767 | D8S2298E (reproduction 8) | 0.000086 | 3.16 | (2.01-4.97) | |
| 14 | AF016004 | GPM6B, glycoprotein M6B | 0.000095 | 13.06 | (5.98-28.53) | |
| 15 | X96753 | CSPG4, chondroitin sulfate proteoglycan 4 (melanoma-associated) | 0.000097 | 7.97 | (3.56-17.85) | |
| 16 | √ | D78177 | QPRT, quinolinate phosphoribosyltransferase | 0.000104 | 7.31 | (3.86-13.86) |
| 17 | √ | V00568 | MYC, v-myc myelocytomatosis viral oncogene homolog (avian) | 0.000126 | 5.93 | (2.68-13.12) |
| 18 | X61118 | LMO2, LIM domain only 2 (rhombotin-like 1) | 0.000126 | 3.85 | (2.05-7.22) | |
| 20 | √ | M58597 | FUT4, fucosyltransferase 4 [α-(1,3) fucosyltransferase, myeloid-specific] | 0.000187 | 3.52 | (2.17-5.71) |
| Underexpressed | ||||||
| 131 | √ | U46922 | FHIT, fragile histidine triad | 0.000010 | -8.18 | (-5.16)-(-12.97) |
| 132 | √ | U70321 | TNFRSF14, tumor necrosis factor receptor superfamily, member 14 | 0.000012 | -24.73 | (-12.05)-(-50.72) |
| 133 | √ | U01062 | ITPR3, inositol 1,4,5-triphosphate receptor type 3 | 0.000013 | -10.69 | (-6.49)-(-17.63) |
| 134 | √ | M16594 | GSTA2, glutathione S-transferase A2 | 0.000017 | -3.48 | (-2.27)-(-5.34) |
| 135 | √ | U03858 | FLT3LG, fms-related tyrosine kinase 3 ligand | 0.000024 | -2.24 | (-1.56)-(-3.20) |
| 136 | √ | AB007895 | KIAA0435 | 0.000037 | -4.38 | (-2.47)-(-7.77) |
| 137 | J05257 | DPEP1, dipeptidase 1 (renal); renal metabolism of glutathione | 0.000046 | -3.24 | (-2.07)-(-5.07) | |
| 138 | √ | X53586 | ITGA6, integrin α6 | 0.000056 | -15.57 | (-6.59)-(-36.79) |
| 139 | √ | J03600 | ALOX5, arachidonate 5-lipoxygenase | 0.000056 | -4.57 | (-2.63)-(-7.94) |
| 141 | √ | L34059 | CDH4, cadherin 4, type 1, R-cadherin (retinal) | 0.000069 | -6.89 | (-3.48)-(-13.64) |
| 142 | AF041434 | PTP4A3, protein tyrosine phosphatase type IVA, member 3 | 0.000085 | -4.11 | (-2.36)-(-7.18) | |
| 143 | √ | X76104 | DAPK1, death-associated protein kinase 1 | 0.000093 | -7.90 | (-3.94)-(-15.85) |
Gene numbers at the left match numbers in Table 3 and appear in the same order as in Fig. 1. Missing numbers (8, 12, 19, and 140) correspond to genes that were present more than once on the array and already appear in the table
Also included within the group of 43 (three genes appear twice) genes, associated with specific features of t(4;11) ALLs, in Fig. 2
Examination of the genes whose expression pattern distinguishes t(4;11) ALLs from other ALLs reveals a substantial number of genes associated with growth control, cell transformation, or malignancy. Those genes may be classified into several functional categories.
Overexpressed oncogenes: (i) HOX A9 and MEIS1, which form a sequence-specific DNA-binding complex (14) and are frequently coactivated in spontaneous AML of BXH-2 mice (15) [forced coexpression of the two genes in murine bone marrow cells rapidly induces AML (16)]; (ii) HOX A10, which induces AML in mice (17); (iii) LMO2 (RMBT2), whose overexpression, resulting from chromosome translocations, is associated with T cell ALL (18); (iv) MYC, which has a critical role in cell proliferation and is deregulated in human lymphomas and other tumors (19); (v) LGALS1 (galectin1), which cooperates with RAS in cell transformation (20) and inhibits T cell proliferation and survival (21); and (vi) PDGFRβ (platelet-derived growth factor receptor β), which is a tyrosine kinase and is deregulated through chromosome translocations and gene fusions in chronic myeloproliferative diseases (22).
Overexpressed genes involved in drug resistance: (i) CD44, which is associated with aggressive B-CLL (23) and conferring resistance to several widely used anticancer drugs (24); (ii) DHFR (dihydrofolate reductase), which confers resistance to methotrexate; (iii) BLMH (bleomycine hydrolase); and (iv) CAT (catalase), which protects from oxidative stress.
Overexpressed genes involved in protection from apoptosis and in survival: (i) CDC2 (cell division cycle 2; p34; CDK1), which preserves the viability of cancer cells in response to microtubule poisons and anticancer drugs like vincristine and taxol, by increasing expression of the apoptosis inhibitor survivin (25); (ii) PPP2R5C (phosphatase 2A), which is implicated in regulation of growth, transcription, and signal transduction, and is required for survival and protects from apoptosis in Drosophila (26); (iii) MAP3K5 (mitogen-activated protein kinase kinase kinase 5), which is involved in activation of the p38 MAP kinase required for initiation of the G2/M checkpoint (27) and is selectively activated in non-small cell lung cancer (28).
Underexpressed proapoptotic genes: (i) ITPR3 (inositol 1,4,5-triphosphate receptor type 3), which mediates the release of intracellular calcium and consequently actively promotes apoptosis (29); (ii) IGFBP3 (IGF-binding protein 3), which has proapoptotic activity both dependent and independent of p53 (30); and (iii) JUN, which is implicated as positive modulator of apoptosis induced in hematopoietic progenitor cells of the myeloid linkage (31) [down-regulation of JUN might account for the failure of glucocorticoid therapy (32)].
Underexpressed tumor suppressors and growth inhibitors: (i) FHIT (fragile histidine triad), which is a target of chromosome aberrations and inactivated in many cancers, including lung, esophagus, stomach, breast, kidney, and leukemias (33); (ii) DAPK1 (death-associated protein kinase 1), mediating IFN′s activity and countering oncogene-induced transformation by activation of a p19ARF/p53 apoptotic checkpoint (34); and (iii) MADH1 (mothers against decapentaplegic homologue 1; SMAD1), which is a transcription modulator mutated in various forms of cancer (35).
Overexpressed genes acting in cell cycle progression and cell proliferation: (i) CCNA1 (cyclin A1), which functions in S phase and mitosis and the expression of which is elevated in a variety of tumors including AMLs (36); (ii) BMYB (myb-like 2), which is required for proliferation of hematopoietic cells (37) and directly activates the antiapoptotic gene ApoJ/clusterin (38); and (iii) CDKN3 (cyclin-dependent kinase inhibitor 3), which interacts with cyclin-dependent kinases and is overexpressed in breast and prostate cancer (39).
Our battery of ALLs lacking t(4;11) consisted of tumors at various stages of differentiation, including pre-B, pro-B, and T cell ALLs. Therefore, the differences in expression found should be due in part to the differences in differentiation stage between t(4;11) to the other ALLs. Hence, we now tried to (i) identify those genes whose expression pattern is directly correlated with the t(4;11) abnormality, either resulting from the abnormality or specifically associated with the cell type in which the chromosome translocation occurred; (ii) separate the genes above from genes whose expression reflects (sensitive to) the differences between the early and late differentiation stages (pro-B vs. pre-B and T cell tumors); (iii) identify genes associated with unique features of CD10- ALLs. To this end we defined three groups of ALL samples: (i) t(4;11) tumors (pro-B cells), (ii) CD10- tumors (pro-B cells), and (iii) the rest of the ALLs (pre-B and T cells). Three distinct supervised analyses were performed, which separate (i) t(4;11) ALLs from the rest of ALLs, (ii) t(4;11) ALLs from CD10- ALLs, and (iii) CD10- ALLs from the rest of the ALLs.
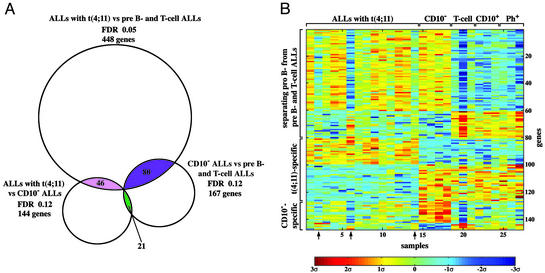
The genes that participate in one or more separations were identified (see the Venn diagram of Fig. 2A). Three overlapping groups were found, containing 77 (three genes appear twice), 43 (three genes appear twice), and 20 (one gene appears twice) genes. Lists of the genes are in Tables 4–6, which are published as supporting information on the PNAS web site. Each of these groups contained genes that are overexpressed or underexpressed; the expression matrix of the three groups is shown in Fig. 2B. Seventyseven genes separate both pro-B cell t(4;11) ALLs from pre-B and T cell ALLs, as well as the latter from pro-B cell CD10- ALLs. Having been picked in both separations, this group of 77 genes distinguishes pro-B cell ALLs [both with and without the t(4;11) chromosome translocation] from pre-B and T cell ALLs. The 43 genes of the second intersection simultaneously separate t(4;11) ALLs from CD10- ALLs and from pre-B and T cell ALLs. Being singled out in both separations, this group of genes is neither associated with the differences between pro-B vs. pre-B and T cells and tumors, nor does it involve specific features of CD10- tumors. Rather, the expression of these 43 genes is affected directly by the t(4;11) abnormality and probably by other unique features of the pro-B cells in which the t(4;11) aberration occurred. The majority of these 43 genes also appear in Table 1. The last group of 20 genes separates CD10- from t(4;11) ALLs, as well as from pre-B and T cell ALLs. Being selected in both separations, these 20 genes are likely to be associated with unique features of CD10- tumors.
Fig. 2.
Intersections of genes separating three types of ALLs (see text). (A) Three groups of genes, encompassing 77 (three appear twice), 43 (three appear twice), and 20 (one appears twice) genes, were found to participate each in two separations. (B) The expression matrix of these three groups. Levels of expression higher or lower than the mean 0 are shown in red/yellow and blue, respectively. Arrows point to samples with variant expression profile (see text).
Inspection of Fig. 2B points to three t(4;11) tumors, samples 2, 6, and 14, that show a variant transcription profile. Although the expression pattern of the 43 genes, specifically correlated with t(4;11) ALLs, is similar in these three tumors and in the rest of t(4;11) ALLs (see genes 81–126 of Fig. 2B), the transcription profile of the three tumors with regard to genes 1–80 (which distinguish pro-B from pre-B and T cell tumors) is closer to pre-B and T cell ALLs, unlike the profile of the other 11 t(4;11) samples. The three tumors also show some quantitative variation from the other t(4;11) ALLs in transcription of the genes whose expression is associated with CD10- ALLs (genes 127–147; Fig. 2B). These results suggest the existence of two subfamilies of ALLs with the t(4;11) chromosome translocation, distinguished by their expression patterns.
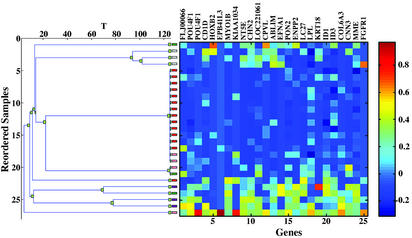
Finally, we applied the coupled two-way clustering method (11, 12) in an unsupervised analysis. A group of 25 genes was found to be consistently underexpressed in ALLs with t(4;11) compared with the other ALLs (Fig. 3; Table 7, which is published as supporting information on the PNAS web site). The cluster of samples with low expression of these genes includes 13/14 of t(4;11) and 3/4 of CD10- ALLs. This is consistent with the close similarity in biological and clinical features between these two types of tumors. A second group of 132 genes separated the seven cell lines included in the analysis from the 45 primary tumors. All these genes were underexpressed in the cell lines (Fig. 5 and Table 8, which are published as supporting information on the PNAS web site).
Fig. 3.
Clustering the ALL samples on the basis of their expression levels over a cluster of 25 genes, G7 (which was obtained by the coupled two-way clustering). (Left) The resulting dendrogram; each leaf corresponds to an ALL sample, with t(4;11) ALLs colored red and CD10- ALLs rose. (Right) The expression matrix, with rows corresponding to samples and columns to genes. 13/14 of t(4;11) samples and 3/4 of CD10- ALLs are in the central cluster of samples with low expression levels.
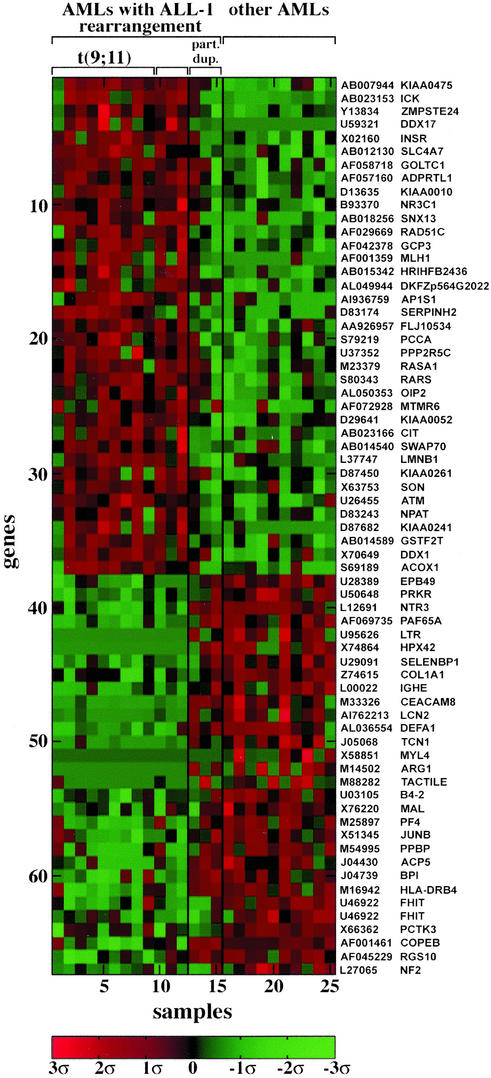
Transcription Profile of AMLs with ALL-1 Rearrangements. AMLs with 11q23 translocations and ALL-1 rearrangements were compared in their expression profiles to AMLs with normal karyotypes. At a FDR of 0.15 (85% confidence) we identified 66 genes overexpressed or underexpressed in AMLs with 11q23 abnormalities (Fig. 4; Table 9, which is published as supporting information on the PNAS web site). Three primary AMLs with ALL-1 partial duplication (5) were compared with the other AMLs with regard to expression of the 66 genes. Two of the three tumors resembled AMLs without 11q23 abnormalities, whereas the third appeared closer to the tumors with chromosome translocations (Fig. 4). The similarity between AMLs without 11q23 aberrations and AMLs with ALL-1 partial duplications was further evidenced in the failure to separate the two groups at an acceptable FDR. (In parallel, AMLs with 11q23 abnormalities were separated from AMLs with ALL-1 partial duplications at a FDR of 0.3; some of this analysis is shown in Fig. 6 and Table 10, which are published as supporting information on the PNAS web site.) These results, if confirmed with additional samples, suggest molecular differences between AMLs triggered by recombination of the ALL-1 gene to partner genes and AMLs triggered by ALL-1 partial duplications. The variations might be reflected in biological and clinical features.
Fig. 4.
Genes distinguishing AMLs with 11q23 chromosome translocations and ALL-1 rearrangements (samples 1–12) from other AMLs (samples 16–25). Samples 13–15 of AMLs with ALL-1 partial duplication were not included in the supervised analysis but were added later for the purpose of comparison.
Examination of the list of genes most correlated with AMLs carrying 11q23 abnormalities (Table 9) shows that some are involved in cancer, proliferation, or apoptosis. These include the overexpressed insulin receptor, which enhances DNA synthesis and inhibits apoptosis (40), the overexpressed repair gene RAD 51, which is up-regulated in breast and pancreatic cancers (41) and probably increases drug resistance, the overexpressed PPP2R5C phosphatase, the underexpressed JUNB, which up-regulates the tumor suppressor gene p16 and represses cyclin D1 (42) and whose knockout in mice induces myeloproliferative disease (43), the underexpressed tumor suppressor FHIT, the underexpressed double stranded RNA-activated protein kinase proapoptotic PRKR, which acts in the context of IFN′s pathway and up-regulates FAS and BAX (44), and the underexpressed DEFA1 (defensin), which is involved in immune response.
Having identified genes differentially expressed in ALLs with t(4;11) compared with ALLs without t(4:11) and in AMLs with 11q23 abnormalities compared with AMLs without such abnormalities, we intersected the results of these two tests (we used a FDR level of 0.15 for both) to find the genes in common. We identified 50 (two appear twice) such genes that were overexpressed or down-regulated in the relevant tumors (Fig. 7 and Table 11, which are published as supporting information on the PNAS web site). For all these genes the difference was high for one type of tumors (e.g., ALLs), but modest for the second type (e.g., AMLs). The genes that were overexpressed in the samples with ALL-1 rearrangements included the phosphatase PPP2R5C and the MCM4 gene, whose product is an essential component of the prereplicative complex (45). The underexpressed genes included FHIT and JUNB.
Discussion
Our results indicate distinct transcription profiles of ALL-1-associated tumors. This is likely to be reflected in the unusual clinical and biological characteristics of these tumors, such as short latency, poor prognosis, expression of myeloid genes in ALL, etc. Some of the genes pinpointed in our study of ALLs with t(4;11), which were mostly in adults, were also indicated (Table 3) in our previous preliminary analysis (46) and in recent investigations that dealt with ALLs from infants and children (47, 48).
Examining the genes overexpressed or underexpressed in tumors with ALL-1 rearrangements (in particular in ALLs) indicates a constellation of expression patterns previously associated with and/or highly favorable for malignant transformation and cancer. This includes activation of oncogenes (MYC, HOX A9 and MEIS1, LMO2, etc.), inactivation of tumor suppressor genes such as FHIT and DAPK1, suppression of apoptosis by down-regulation of proapoptotic genes and up-regulation of survival genes, suppression of host immune response (up- and down-regulation of galectin 1 and defensin, respectively), up-regulation of genes conferring drug resistance, such as CD44, DHFR (dihydrofolate reductase), and BLMH (bleomycine hydrolase), and overexpression of genes involved in cell proliferation (e.g., cyclin A1 and myb-like 2). Some of the overexpressed genes we identified, like VLDL, PDGFRβ (platelet-derived growth factor receptor β), HOX A9, MEIS1, and insulin receptor, are also to be found expressed in normal hematopoietic stem cells (49) but the majority of genes are not. We suggest that at least some of the genes alluded to by our study contribute directly to the aggressive nature of the disease and to its known resistance to therapy.
In an attempt to identify genes whose expression correlates more strictly with the t(4;11) genotype, we removed genes that distinguish pro-B from pre-B and T cell tumors, as well as genes associated with the CD10- phenotype. The 43 genes left (Table 5) are closely linked to the t(4;11) genotype and would be good candidates for future biological experiments. Examination (Fig. 8, which is published as supporting information on the PNAS web site) of the expression level of the 43 genes in two other studies (47, 48) demonstrates (in particular the investigation of Armstrong et al.) that nearly all of these genes separate ALLs with and without ALL-1 rearrangements. Therefore, the expression profile of the 43 genes distinguishes both adults (most patients of ours) and children (the other two studies) with ALLs and ALL-1 rearrangements. Another approach taken to identify genes more likely to be associated with the pathogenesis was based on the (unproven) speculation that ALL-1 fusion proteins trigger malignancy by a similar mechanism in both ALLs and AMLs. Thus, we looked for genes that behave in similar fashion (up- or down-regulated) in ALLs and AMLs with ALL-1 rearrangements (Fig. 7). At the top of the list we find PPP2R5C, FHIT, and JUNB.
Compartmentalization of the genes into two groups whose expression distinguishes t(4;11) from other ALLs resulted in the unexpected identification of two subclasses of t(4;11) tumors (Fig. 2B). The subclasses are discerned by the expression profile of the 77 genes separating pro-B from pre-B and T cell tumors. Because t(4;11) tumors are generally considered to be pro-B cell ALLs, it is perplexing that, with regard to genes separating pro-B from pre-B and T cell ALLs, the smaller subclass of t(4;11) appears close to pre-B and T cell tumors. Comparison of the clinical records of the corresponding two subclasses of patients (Table 2; samples ht17, ht21, and ht27 in this table show the variant profile) indicates that in the first group there are 2/3 long-term survivors, but in the second group the outcome is worse (2/9). How widespread the distribution of t(4;11) patients into two groups is and whether there is a significant correlation with survival remain to be determined.
The supervised analysis of AMLs with ALL-1 rearrangements vs. control AMLs showed a less uniform pattern, as well as a lower number of separating genes. This suggests that the two groups of tumors are more heterogeneous. Unexpectedly, two of the three AMLs with ALL-1 partial duplications showed expression profiles resembling AML controls. The generality of this observation should be decided by analyzing additional tumors.
Supplementary Material
Acknowledgments
O.R.-A. thanks Amnon Amir for helpful thoughts and ideas. These studies were supported by National Cancer Institute Grant CA 50507 and by grants from the Israel Academy of Science, the Binational Science Foundation (U.S. and Israel), the Israel Cancer Research Fund, the Ridgefield Foundation, the Minerva Foundation, and the Germany–Israel Science Foundation.
Abbreviations: ALL, acute lymphoblastic leukemia; AML, acute myeloblastic leukemia; FDR, false discovery rate.
References
- 1.Johansson, B., Moorman, A. V., Haas, O. A., Watmore, A. E., Cheung, K. L., Swanton, S. & Secker-Walker, L. M. (1998) Leukemia 12, 779-787. [DOI] [PubMed] [Google Scholar]
- 2.Swansbury, G. J., Slater, R., Bain, B. J., Moorman, A. V. & Secker-Walker, L. M. (1998) Leukemia 12, 792-800. [DOI] [PubMed] [Google Scholar]
- 3.Gu, Y., Nakamura, T., Alder, H., Prasad, R., Canaani, O., Cimino, G., Croce, C. M. & Canaani, E. (1992) Cell 71, 701-708. [DOI] [PubMed] [Google Scholar]
- 4.Tkachuk, D. C., Kohler, S. & Cleary, M. L. (1992) Cell 71, 691-700. [DOI] [PubMed] [Google Scholar]
- 5.Schichman, S. A., Canaani, E. & Croce, C. M. (1995) J. Am. Med. Assoc. 273, 571-576. [PubMed] [Google Scholar]
- 6.DiMartino, J. F. & Cleary, M. L. (1999) Br. J. Haematol. 106, 614-626. [DOI] [PubMed] [Google Scholar]
- 7.Biondi, A., Cimino, G., Pieters, R. & Pui, C. H. (2000) Blood 96, 24-33. [PubMed] [Google Scholar]
- 8.Pui, C. H., Gaynon, P. S., Boyett, J. M., Chessells, J. M., Baruchel, A., Kamps, W., Silverman, L. B., Biondi, A., Harms, D. O., Vilmer, E., et al. (2002) Lancet 359, 1909-1915. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura, T., Mori, T., Tada, S., Krajewski, W., Rozovskaia, T., Wassell, R., Dubois, G., Mazo, A., Croce, C. M. & Canaani, E. (2002) Mol. Cell 10, 1119-1128. [DOI] [PubMed] [Google Scholar]
- 10.Benjamini, Y. & Hochberg, Y. (1995) J. R. Stat. Soc. B 57, 289-300. [Google Scholar]
- 11.Getz, G., Levine, E. & Domany, E. (2000) Proc. Natl. Acad. Sci. USA 97, 12079-12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blatt, M., Wiseman, S. & Domany, E. (1996) Phys. Rev. Lett. 76, 3251-3254. [DOI] [PubMed] [Google Scholar]
- 13.Levine, E. & Domany, E. (2001) Neural Comput. 13, 2573-2593. [DOI] [PubMed] [Google Scholar]
- 14.Shen, W. F., Rozenfeld, S., Kwong, A., Komuves, L. G., Lawrence, H. J. & Largman, C. (1999) Mol. Cell. Biol. 19, 3051-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura, T., Largaespada, D. A., Shaughnessy, J. D., Jr., Jenkins, N. A. & Copeland, N. G. (1996) Nat. Genet. 12, 149-153. [DOI] [PubMed] [Google Scholar]
- 16.Kroon, E., Krosl, J., Thorsteinsdottir, U., Baban, S., Buchberg, A. M. & Sauvageau, G. (1998) EMBO J. 17, 3714-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thorsteinsdottir, U., Sauvageau, G., Hough, M. R., Dragowska, W., Lansdorp, P. M., Lawrence, H. J., Largman, C. & Humphries, R. K. (1997) Mol. Cell. Biol. 17, 495-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boehm, T., Foroni, L., Kaneko, Y., Perutz, M. F. & Rabbitts, T. H. (1991) Proc. Natl. Acad. Sci. USA 88, 4367-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nesbit, C. E., Tersak, J. M. & Prochownik, E. V. (1999) Oncogene 18, 3004-3016. [DOI] [PubMed] [Google Scholar]
- 20.Paz, A., Haklai, R., Elad-Sfadia, G., Ballan, E. & Kloog, Y. (2001) Oncogene 20, 7486-7493. [DOI] [PubMed] [Google Scholar]
- 21.Rabinovich, G. A., Baum, L. G., Tinari, N., Paganelli, R., Natoli, C., Liu, F. T. & Iacobelli, S. (2002) Trends Immunol. 23, 313-320. [DOI] [PubMed] [Google Scholar]
- 22.Cross, N. C. & Reiter, A. (2002) Leukemia 16, 1207-1212. [DOI] [PubMed] [Google Scholar]
- 23.Eistere, W., Hilbe, W., Stauder, R., Bechter, O., Fend, F. & Thaler, J. (1996) Br. J. Haematol. 93, 661-669. [DOI] [PubMed] [Google Scholar]
- 24.Fujita, Y., Kitagawa, M., Nakamura, S., Azuma, K., Ishi, G., Higashi, M., Kishi, H., Hiwasa, T., Koda, K., Nakajima, N. & Harigaya, K. (2002) FEBS Lett. 528, 101-108. [DOI] [PubMed] [Google Scholar]
- 25.O'Connor, D. S., Wall, N. R., Porter, A. C. & Altieri, D. C. (2002) Cancer Cell 2, 43-54. [DOI] [PubMed] [Google Scholar]
- 26.Li, X., Scuderi, A., Letsou, A. & Virshup, D. M. (2002) Mol. Cell. Biol. 22, 3674-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bulavin, D. V., Higashimoto, Y., Popoff, I. J., Gaarde, W. A., Basrur, V., Potapova, O., Appella, E. & Fornace, A. J. (2001) Nature 411, 102-107. [DOI] [PubMed] [Google Scholar]
- 28.Greenberg, A. K., Basu, S., Hu, J., Yie, T. A., Tchou-Wong, K. M., Rom, W. N. & Lee, T. C. (2002) Am. J. Respir. Cell Mol. Biol. 26, 558-564. [DOI] [PubMed] [Google Scholar]
- 29.Blackshaw, S., Sawa, A., Sharp, A. H., Ross, C. A., Snyder, S. H. & Khan, A. A. (2000) FASEB J. 14, 1375-1379. [DOI] [PubMed] [Google Scholar]
- 30.Furstenberger, G. & Senn, H. J. (2002) Lancet Oncol. 3, 298-302. [DOI] [PubMed] [Google Scholar]
- 31.Liebermann, D. A., Gregory, B. & Hoffman, B. (1998) Int. J. Oncol. 12, 685-700. [DOI] [PubMed] [Google Scholar]
- 32.Pallardy, M. & Biola, A. (1998) C. R. Seances Soc. Biol. Ses Fil. 192, 1051-1063. [PubMed] [Google Scholar]
- 33.Pekarsky, Y., Zanesi, N., Palamarchuk, A., Huebner, K. & Croce, C. M. (2002) Lancet Oncol. 3, 748-754. [DOI] [PubMed] [Google Scholar]
- 34.Raveh, T., Droguett, G., Horwitz, M. S., DePinho, R. A. & Kimchi, A. (2001) Nat. Cell Biol. 3, 1-7. [DOI] [PubMed] [Google Scholar]
- 35.Hata, A., Shi, Y. & Massague, J. (1998) Mol. Med. Today 4, 257-262. [DOI] [PubMed] [Google Scholar]
- 36.Yam, C. H., Fung, T. K. & Poon, R. Y. (2002) Cell Mol. Life Sci. 59, 1317-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arsura, M., Introna, M., Passerini, F., Mantovani, A. & Golay, J. (1992) Blood 79, 2708-2716. [PubMed] [Google Scholar]
- 38.Cervellera, M., Raschella, G., Santilli, G., Tanno, B., Ventura, A., Mancini, C., Sevignani, C., Calabretta, B. & Sala, A. (2000) J. Biol. Chem. 275, 21055-21060. [DOI] [PubMed] [Google Scholar]
- 39.Lee, S. W., Reimer, C. L., Fang, L., Iruela-Arispe, M. & Aaronson, S. A. (2000) Mol. Cell. Biol. 20, 1723-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tseng, Y. H., Ueki, K., Kriauciunas, K. M. & Kahn, C. R. (2002) J. Biol. Chem. 277, 31601-31611. [DOI] [PubMed] [Google Scholar]
- 41.Maacke, H., Opitz, S., Jost, K., Hamdorf, W., Henning, W., Kruger, S., Feller, A. C., Lopens, A., Diedrich, K., Schwinger, E.& Sturzbecher, H. W. (2000) Int. J. Cancer 88, 907-913. [DOI] [PubMed] [Google Scholar]
- 42.Shaulian, E. & Karin, M. (2001) Oncogene 20, 2390-2400. [DOI] [PubMed] [Google Scholar]
- 43.Passegue, E., Jochum, W., Schorpp-Kistner, M., Mohle-Steinlein, U. & Wagner, E. F. (2001) Cell 104, 21-32. [DOI] [PubMed] [Google Scholar]
- 44.Gil, J. & Esteban, M. (2000) Apoptosis 5, 107-114. [DOI] [PubMed] [Google Scholar]
- 45.You, Z., Ishimi, Y., Masai, H. & Hanaoka, F. (2002) J. Biol. Chem. 277, 42471-42479. [DOI] [PubMed] [Google Scholar]
- 46.Rozovskaia, T., Feinstein, E., Mor, O., Foa, R., Blechman, J., Nakamura, T., Croce, C. M., Cimino, G. & Canaani, E. (2001) Oncogene 20, 874-878. [DOI] [PubMed] [Google Scholar]
- 47.Yeoh, E. J., Ross, M. E., Shurtleff, S. A., Williams, W. K., Patel, D., Mahfouz, R., Behm, F. G., Raimondi, S. C., Relling, M. V., Patel, A., et al. (2002) Cancer Cell 1, 133-143. [DOI] [PubMed] [Google Scholar]
- 48.Armstrong, S. A., Staunton, J. E., Silverman, L. B., Pieters, R., den Boer, M. L., Minden, M. D., Sallan, S. E., Lander, E. S., Golub, T. R. & Korsmeyer, S. J. (2002) Nat. Genet. 30, 41-47. [DOI] [PubMed] [Google Scholar]
- 49.Ivanova, N. B., Dimos, J. T., Schaniel, C., Hackney, J. A., Moore, K. A. & Lemischka, I. R. (2002) Science 298, 601-604. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.