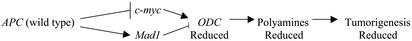Abstract
Most sporadic colon adenomas acquire mutations in the adenomatous polyposis coli gene (APC) and show defects in APC-dependent signaling. APC influences the expression of several genes, including the c-myc oncogene and its antagonist Mad1. Ornithine decarboxylase (ODC), the first enzyme in polyamine synthesis, is a transcriptional target of c-myc and a modifier of APC-dependent tumorigenesis. A single-nucleotide polymorphism exists in intron 1 of the human ODC gene, which lies between two myc-binding domains. This region is known to affect ODC transcription, but no data exist on the relationship of this polymorphism to risk of colorectal neoplasia in humans. We show that individuals homozygous for the minor ODC A-allele who reported using aspirin are ≈0.10 times as likely to have an adenoma recurrence as non-aspirin users homozygous for the major G-allele. Mad1 selectively suppressed the activity of the ODC promoter containing the A-allele, but not the G-allele, in a human colon cancer-derived cell line (HT29). Aspirin (≥10 μM) did not affect ODC allele-specific promoter activity but did activate polyamine catabolism and lower polyamine content in HT29 cells. We propose that the ODC polymorphism and aspirin act independently to reduce the risk of adenoma recurrence by suppressing synthesis and activating catabolism, respectively, of colonic mucosal polyamines. These findings confirm the hypothesis that the ODC polymorphism is a genetic marker for colon cancer risk, and support the use of ODC inhibitors and aspirin, or other nonsteroidal antiinflammatory drugs (NSAIDs), in combination as a strategy for colon cancer prevention.
Clinical, epidemiological, and molecular studies provide compelling evidence that most colorectal cancers arise from adenomas (1–4). Stop codon mutations in, or deletions of, the adenomatous polyposis coli gene (APC) are the cause of familial adenomatous polyposis, a hereditary form of colon cancer in humans (5, 6). Mice that do not express wild-type APC develop intestinal neoplasia similar to humans with familial adenomatous polyposis (7). Recent evidence indicates that most (>90%) sporadic colorectal adenomas acquire somatic mutations in the APC gene and show defects in APC-dependent signaling (8).
Most colon adenomas do not develop into invasive cancers. Candidate genes that suppress APC-dependent tumorigenesis have been identified (9, 10). Differential expression of these “modifier” genes (genes that modify the phenotype of a specific genotype) might account for differences in adenoma incidence, and might be useful as markers of cancer risk in humans.
APC influences the expression of a number of genes. One of these genes is the c-myc oncogene (11). Myc is a member of a family of proteins that regulate transcription by binding to specific mycbinding elements (CACGTG), termed E-boxes, of genes affecting both proliferation and apoptosis (12). One transcriptional target of c-myc is ornithine decarboxylase (ODC), the first enzyme in polyamine synthesis (13) whose transcription is activated by myc. ODC expression is increased in the normal mucosa of individuals with genotype-positive familial adenomatous polyposis, compared with genotype-negative family controls (14), and in colonic neoplasia of individuals without genetic risk of colon cancer (15). This finding suggests that the net effect of producing the normal APC protein on ODC expression is negative.
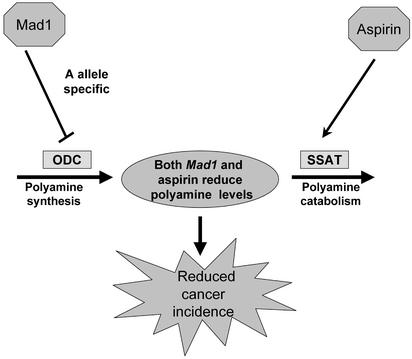
Recent studies from our group show that wild-type APC suppresses c-myc, activates the c-myc antagonist Mad1, and reduces the expression of ODC RNA via a mechanism involving changes in both c-myc and an E-box in the ODC promoter (16). Furthermore, both ODC RNA and polyamine contents are up-regulated in the intestinal mucosa of mice expressing a mutant APC, compared with normal littermates (17). Intestinal tumorigenesis in these mice is elevated and can be suppressed by treatment with a specific inhibitor of ODC (17). Together, these data suggest that elevated levels of polyamines are involved in APC-dependent intestinal cancer, as depicted in Scheme 1. Wild-type APC coordinately regulates the expression of c-myc and its antagonist, Mad1, causing a suppression of ODC expression, a decrease in polyamine synthesis, and reduced tumorigenesis. Mutation in APC leads to increased ODC expression, increased polyamines, and tumorigenesis.
Scheme 1.
ODC promoter activity is influenced by cooperative interactions involving neighboring E-boxes (18). A polymorphic site, situated between two E-boxes, has been identified in the human ODC promoter and shown to affect c-myc-dependent ODC promoter activity in rodent fibroblasts (19). No information is available on the effect of this polymorphic site on ODC promoter activity in human intestinal cells or tissues.
NSAIDs, including aspirin, are the most widely investigated chemopreventive agents in colorectal neoplasia. Experimental, clinical, and epidemiological evidence indicates that aspirin and other NSAIDs prevent or inhibit the development of colorectal neoplasia (14, 20–24). Regular use of these drugs is also associated with a lower risk of colorectal adenomas (24); furthermore, recent clinical trial data also support the chemopreventive effect of aspirin on adenoma recurrence (25, 26). However, mechanisms responsible for the protective effect of NSAIDs on colorectal neoplasia remain unresolved.
Possible associations between ODC genotypes and the risk of colorectal neoplasia have not yet been evaluated. Given the suggested role of polyamine levels in APC-dependent intestinal carcinogenesis, as suggested by our studies in mouse and human models (16, 17), and because APC has been implicated in the majority of sporadic human colon adenomas and cancers (8), we assessed the relationship between the ODC polymorphism and the risk of adenoma recurrence in participants in a colon cancer prevention trial. We further investigated whether this association was modified by aspirin use. Our epidemiological analyses reveal a substantial and statistically significant effect of the ODC polymorphism on risk of adenoma recurrence in aspirin users. We also present experimental studies in cell culture models that can explain the epidemiological results, suggesting that one likely mechanism for both the ODC polymorphism and aspirin is their effect on intracellular polyamine pools.
Methods
Epidemiological Studies. Study population. Analyses were conducted among participants in an adenoma recurrence trial, the design and results of which have been published (27, 28). Because the wheat bran fiber intervention had no significant effect on adenoma recurrence, we evaluated data from all participants in the trial who provided a blood sample. Men and women 40–80 years of age who had removal of one or more colorectal adenomas 3 mm or larger at colonoscopy within 3 months of study entry were recruited. Self-administered questionnaires were used to obtain risk factor data, including aspirin use. Use of aspirin was collected in two separate instruments. At baseline, participants were asked whether they used aspirin in the previous month. A questionnaire was later developed that ascertained the use of aspirin in the previous 10 years; however, because this was not uniformly administered at baseline and was not collected on all participants, we did not use this variable for our primary analysis. Comparisons of these two data sources indicate that ≈70% of individuals who reported using aspirin the previous month also reported its use in the previous 10 years. Recurrence was defined as recurrence of one or more colorectal adenomas or a colon cancer occurrence anytime after randomization. The study was approved by the University of Arizona Human Subjects Committee.
ODC genotyping. Genomic DNA was extracted from whole blood by using a phenol-chloroform method. Information on the genotyping assay has been published (19). Briefly, an allelic discrimination assay was developed based on the sequence difference at the polymorphic PstI site. Allele-specific TaqMan probes were synthesized with different 5′ fluorescent labels (6-carboxyflourescein or VIC) and the same 3′ quencher dye (6-carboxytetramethylrhodamine). Each PCR reaction included 20 ng of genomic DNA, 30 pmol of each primer, 12.5 pmol of each TaqMan probe, and 1× TaqMan Universal PCR Master Mix (Applied Biosystems) in a volume of 50 μl. Of the 1,304 study participants who underwent one or more follow-up colonoscopies, 690 provided a blood specimen for genotyping; in two of these samples, there was insufficient DNA available for genotyping. Based on 27 duplicate samples, the genotyping assay was shown to be 100% reliable.
Statistical analysis. We used logistic regression analysis (29) to calculate the odds ratio (OR) and 95% confidence interval to test the association between the ODC genotype, aspirin use, and adenoma recurrence, where recurrence was classified as present or absent. We first computed crude ORs by multiplying the number of non-aspirin users who did not recur by the number of aspirin users who recurred and dividing this by the product of the number of aspirin users who did not recur and the number of non-aspirin users who recurred. We conducted similar analyses for ODC and recurrence. We also constructed statistical multivariate models that included age, gender, and number of study colonoscopies conducted after baseline. Established or suspected risk factors for colorectal neoplasia were also considered for inclusion in additional multivariate models. We assessed whether the relation between ODC genotype and adenoma recurrence differed among aspirin and non-aspirin users by conducting separate logistic regression models for each group; we further assessed the joint effect of aspirin and ODC by using individuals with the combination of homozygosity for the major G-allele and no reported aspirin use as the reference group and comparing this to users of aspirin who were also homozygous for the minor A-allele. We assessed the interaction between ODC and aspirin use by including an interaction term for these variables in the logistic regression model and comparing it to the model without the interaction term.
Cell Line Experiments. Cell culture. The human colon cancer cell line HT29 and stable clones transfected with wild-type APC (HT29-APC) or β-galactosidase (HT29-β-gal) downstream of the metallothionein promoter [the latter two lines provided by Bert Vogelstein of The Johns Hopkins Medical Institute (11)], were maintained in McCoy's 5A medium supplemented with 10% FBS and 1% penicillin/streptomycin (Invitrogen) solution at 37°C in a humidified atmosphere of 5% CO2.
Plasmids. Full-length Mad1 cDNA was isolated from pLTR-Mad (obtained from Robert Eisenman, Fred Hutchinson Cancer Research Center, Seattle) and inserted into the pcDNA3.1 (Invitrogen). The pGL3-ODC/A or pGL3-ODC/G plasmids contain nucleotides -462 to +3070 from the ODC gene, cloned into promoterless pGL3-Enhancer vector (Promega; ref. 19). The spermidine/spermine N1-acetyltransferase reporter plasmid (SSAT-Luc), containing 3.43 kb of SSAT 5′ flanking sequence, was obtained from Robert Casero (The Johns Hopkins University, Baltimore).
Transfections. To examine the effect of Mad1 overexpression on ODC promoter activity and the effect of aspirin on SSAT promoter activity, transient transfections were performed with LipofectAMINE reagent (Invitrogen). For the Mad1 overexpression studies, either pGL3-ODC/A or pGL3-ODC/G was cotransfected with either pcDNA3.1 or pcDNA-Mad1 in a molar ratio of 3:1. For the SSAT studies, 1 μg of SSAT-Luc was cotransfected with 0.3 μg of pCMV-β-gal expression plasmid, which acted as the transfection efficiency control.After 5 h of incubation, cells were supplemented with complete medium containing 20% FBS and left to grow overnight. On the next day, the media was changed. For SSAT studies, varying concentrations of aspirin or its vehicle were added along with the fresh medium. After 48 h, transfected cells were washed with PBS and lysed, and luciferase activities were measured by using 10 μl of cell extract and 50 μl of luciferase reagent (Promega). Relative luciferase units (RLU) were measured by using a Turner Designs TD-20/20 luminometer, and measurements were normalized to protein concentration. β-Gal activity was measured by using the β-gal Assay kit (Invitrogen) according to the manufacturer's protocol. Experiments were performed in triplicate and repeated at least three times. Differences in ODC allele-specific promoter activities were assessed by ANOVA among groups.
RNA isolation and analysis. Total RNA was obtained from cells by extraction using TRIzol reagent as described by others (30) and our group (16). GAPDH was used to control for variation in RNA loading. Northern blot autoradiograms were quantitated by densitometric analysis (IMAGEQUANT, Molecular Dynamics). Data are expressed as the ratio of the integrated densities of 32P-labeled hybridization bands for the SSAT and GAPDH genes.
Protein expression. Cell extracts were obtained by lysing cells in a radioimmunoprecipitation buffer (50 nM Tris·HCl, pH 7.4/150 mM NaCl/1 mM phenylmethylsulfonyl fluoride/1 mM EDTA/5 mg/ml aprotinin/5 mg/ml lenpeptin/1% Triton X-110/1% sodium deoxycholate/0.1% SDS) on ice. Specific protein content was measured by Western blotting as described (16). All antibodies were obtained from Santa Cruz Biotechnology. Polyamine analysis. One million cells were grown overnight, and the medium was changed the following day and supplemented with either ethanol or aspirin (20 or 100 μM, respectively). After 7 days in culture, the cells were trypsinized, washed, and sonicated in 0.1 M HCl. After sonication, the preparation was adjusted to 0.2 M HClO4, and the supernatant was analyzed by reverse-phase high-performance liquid chromatography with 1,7-diaminoheptane as an internal standard (31). Protein was determined by bicinchoninic acid (BCA) assay (32).
Enzyme activity. SSAT enzyme activity was measured by estimation of labeled N1-acetylspermidine synthesized from [14C]acetyl-CoA and unlabeled spermidine, as described (33). The ODC enzyme activity was measured by evaluating the release of 14CO2 from l-[14C]ornithine, as described (16).
Results
ODC Genotype, Aspirin Use, and Adenoma Recurrence. No significant difference in regard to risk factors of interest was observed between the subset of 688 individuals used for the present analyses and that of the total population of 1,304 (Table 2 and Supporting Methods, which are published as supporting information on the PNAS web site, www.pnas.org). Furthermore, no significant risk factor differences were found among different ODC genotypes (Table 3, which is published as supporting information on the PNAS web site). Among the 688 individuals genotyped for ODC, 56% were homozygous for the G-allele and 6% were homozygous for A-allele; the remaining 38% were heterozygotes at this locus. Use of aspirin was reported by 209 (30%) of the study participants.
In statistical analyses that controlled for age, gender, and number of colonoscopies (Table 1), individuals who reported using aspirin were 0.32 less likely to have an adenoma recurrence. In analyses of ODC genotype effects, no difference in the odds of recurrence was found for heterozygous individuals as compared with those homozygous for the G-allele. However, as shown in Table 1, participants homozygous for the A-allele had an estimated 0.48 odds of recurrence as compared with homozygous G individuals.
Table 1. Association between adenoma recurrence and either ODC genotype or aspirin use.
| Recurrence/total (%) | Crude OR* (95% confidence interval) | Adjusted OR† (95% confidence interval) | P value | |
|---|---|---|---|---|
| Aspirin use | ||||
| No | 250/479 (52.2) | 1.00 (Referent) | 1.00 (Referent) | |
| Yes | 91/209 (43.5) | 0.71 (0.51-0.98) | 0.68 (0.48-0.98) | 0.04 |
| ODC genotype | ||||
| GG | 192/382 (50.3) | 1.00 (Referent) | 1.00 (Referent) | |
| GA | 134/264 (50.8) | 1.02 (0.75-1.40) | 0.96 (0.68-1.34) | 0.80 |
| AA | 15/42 (35.7) | 0.55 (0.28-1.07) | 0.48 (0.24-0.99) | 0.05 |
OR from logistic regression models. Non-aspirin users are the referent group for the model assessing the effect of aspirin and recurrence; individuals homozygous for the G-allele are the referent group for the model assessing the effect of ODC and recurrence
Multivariate models include age, gender, number of colonoscopies conducted after baseline, and both aspirin use and genotype
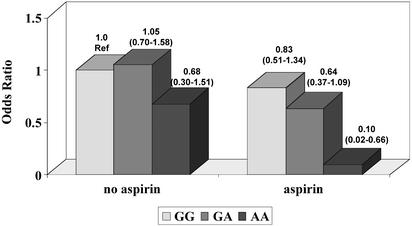
To assess whether the association between ODC genotype and adenoma recurrence varied by aspirin use, we used individuals who reported no aspirin use and were homozygous for the G-allele as the reference group (Fig. 1). Among non-aspirin users, the effect of ODC on adenoma recurrence was weaker than that among the total population; however, among aspirin users, the combined effect of aspirin use and homozygosity for the A-allele resulted in a significant reduction in odds of recurrence (OR = 0.10). The confidence interval is wide, suggesting that the point estimate could be as low as 0.02 or as high as 0.66. Furthermore, when an interaction term for ODC and aspirin use was included in the model, the P value for this term was 0.13; the P value for the difference between the model with and that without the interaction term was 0.10.
Fig. 1.
Odds ratios for adenoma recurrence as a function of ODC genotype and aspirin use. Results of logistic regression models that control for age, gender, and number of colonoscopies are shown (95% confidence intervals). Individuals homozygous for the G-allele and who reported no aspirin use are the reference (Ref) group. The number of individuals who recurred over the total in each group among non-aspirin users are 137/266 (51.5%), 100/182 (55.0%), and 13/31 (41.9%) for GG, AG, and AA variants, respectively; the corresponding figures among aspirin users are 55/116 (47.4%), 34/82 (41.5%), and 2/11 (18.2%).
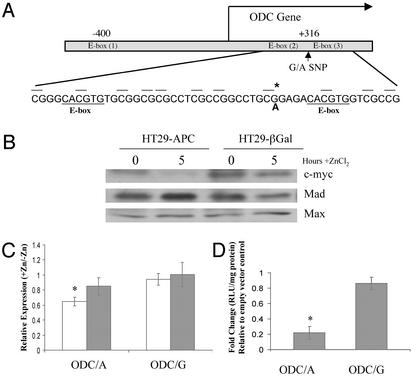
Functional Significance of ODC Promoter Activity in Human Colon Cells. The locations of three consensus E-boxes in the human ODC promoter are depicted in Fig. 2A. One of these E-boxes is found in the region 5′ of the transcriptional start site. Two closely spaced E-boxes are found 3′ of the transcription start site, within intron 1. The A316G SNP (A/G SNP located 316 nucleotides 3′ of the transcription start site) is located between these two E-boxes and may regulate which transcription factors can bind or how strongly they bind (18).
Fig. 2.
A single-nucleotide polymorphism (SNP) in the ODC promoter and its significance for APC-dependent ODC promoter activity. (A) ODC gene structure. There are three consensus-binding sites for the c-myc:Max and Mad1:Max heterodimers within the ODC gene. These sites, called E-boxes, have the consensus sequence CACGTG. E-box (1) is located in the 5′ flanking region of the gene, and E-box (2) and E-box (3) are separated by 28 nt in the first intron. The G/A SNP (*) is positioned between the two E-boxes in intron 1 at +316. Also marked in this diagram are the nearby CpG sequences. (B) Effect of wild-type APC expression on the level of three E-box transcription factors. HT29-APC and control HT29-β-gal cells were treated with 300 μM ZnCl2 for 5 h. Whole-cell lysates from untreated and treated cells were run on an SDS/PAGE gel and Western blots were performed for c-myc, Mad, and Max, as described in Methods. (C) Effect of wild-type APC expression on allelespecific ODC promoter activity. HT29-APC (open bars) and HT29-β-gal (filled bars) cells were transfected with either pGL3-ODC/A or pGL3-ODC/G, as described in Methods. Transfected cultures were then treated with or without 300 μM ZnCl2 for an additional 24 h and harvested, and lysates were analyzed for luciferase activity. Results are presented as the ratios of mean values ± standard deviations of triplicate measurements from cultures treated with ZnCl2 divided by those from similarly transfected cells not treated with ZnCl2. The result is an average of three experiments. Bars indicate standard deviations. *, Statistically significant effects (P < 0.05). (D) Effect of Mad1 expression on allele-specific ODC promoter activity. By using HT29 cells, either pGL3-ODC/A or pGL3-ODC/G was cotransfected with pcDNA3.1 or pcDNA-Mad1 in a molar ratio of 3:1. The cells were harvested after 48 h, and lysates were analyzed for luciferase activity. Experiments were performed in triplicate. Bars indicate standard deviation. *, Statistically significant effects (P < 0.05).
To determine whether the A316G SNP has any functional consequence for ODC expression in colon-derived cells, we evaluated allele-specific ODC promoter activity in two cell culture models. First, we used HT29 colon tumor cells conditionally expressing wild-type APC under control of the zinc-inducible metallothionein promoter (11), because wild-type APC is expressed in the normal colonic mucosa of humans (8). Fig. 2B demonstrates that protein levels of c-myc are suppressed, whereas those of Mad1 are induced, within 5 h after addition of zinc chloride in this cell model. Protein levels of Max, the binding partner for both c-myc and Mad1, are unchanged. A previous cDNA microarray analysis from our laboratory revealed wild-type APC-dependent changes in Mad1 RNA and protein, and presented evidence for APC-dependent regulation of ODC transcription, in the HT29 cell model (16). ODC allele-specific promoter activity was assessed in this model 24 h after addition of zinc chloride (Fig. 2C). As judged by a luciferase assay, zinc chloride treatment of HT29-APC cells transfected with pGL3-ODC/A caused a statistically significant decrease (≈40%) in the activity of the ODC promoter containing the A-allele (P = 0.016), whereas this treatment had no statistically significant effect (P > 0.1) on luciferase expression in HT29-APC cells transfected with pGL3-ODC/G or on HT29-β-gal cells transfected with either ODC promoter reporter (data not shown).
Consequently, we hypothesized that the increase in Mad1 expression caused by wild-type APC may result in an allele-specific reduction of ODC transcription. To test this possibility, we constructed a plasmid containing full-length Mad1 cDNA under the control of the CMV promoter (pcDNA-Mad1). Cotransfections were performed in HT29 cells with pGL3-ODC/A or pGL3-ODC/G and either pcDNA3.1 (empty vector control) or pcDNA-Mad1. As shown in Fig. 2D, constitutive expression of Mad1 led to a significant (78%) A-allele-specific inhibition of promoter activity (P = 0.007) and a nonsignificant (16%) G-allele-specific reduction (P = 0.307). The reason(s) for the quantitative differences in suppression of ODC promoter activities reported in the experiments shown in Fig. 2 C and D is unknown, but they may be due to possible differences in absolute levels of Mad1 expression in the two experimental conditions and/or the effects of zinc treatment used for induction of wild-type APC (Fig. 2C).
The direct relevance of these transient transfection experiments to the function of the endogenous ODC gene remains to be established. We were concerned that our transfection experiments might be compromised by differences in promoter methylation, because the region around the ODC polymorphism is a CpG island and the polymorphism converts a CpG to CpA. We therefore conducted bisulfate sequencing of DNA in this region isolated from normal human colonic mucosa and normal human prostatic tissue. We found that CpGs in a 100-nt region around the polymorphic site are either not methylated or are methylated at a very low frequency (<10%; data not shown). Thus, the results of our transient transfection promoter–reporter experiments are not confounded by potential methylation of the endogenous ODC promoter in human tissues, including normal colonic mucosa.
Aspirin Does Not Influence ODC Allele-Specific Promoter, RNA, or Enzyme Activity. The ODC SNP and aspirin could be acting independently to predict risk of adenoma recurrence. Alternatively, the ODC SNP could be a factor predicting responsiveness to the protective effects of aspirin. To address these hypotheses, cells were transiently transfected with ODC allele-specific promoters and then treated with various concentrations of aspirin to determine whether the two ODC alleles are differentially affected by aspirin. Results of this experiment showed that aspirin, in the therapeutic range (10–100 μM), had no statistically significant effect on ODC promoter activity of either allele (Fig. 6, which is published as supporting information on the PNAS web site). Furthermore, aspirin had no effect on ODC RNA or enzyme activity in HT29 cells (Fig. 7, which is published as supporting information on the PNAS web site).
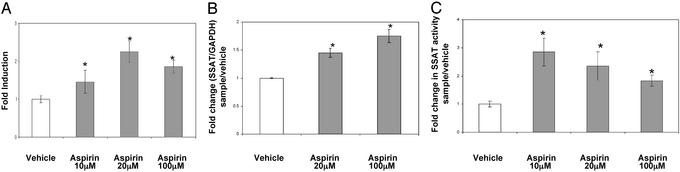
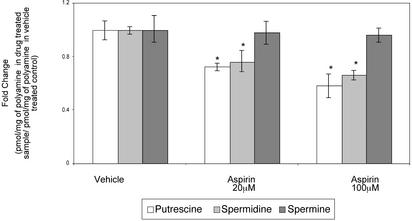
Aspirin and the ODC SNP Affect a Common Metabolic Pathway. Others have recently shown that the NSAID indomethacin induces the expression of an enzyme, SSAT, that is involved in polyamine catabolism and export (34). To determine whether aspirin might be affecting polyamine metabolism, both ODC and SSAT gene expression and enzyme activities were measured. Aspirin (10–100 μM) induces the activity of the SSAT promoter and enzyme activity in HT29 human colon cells (Fig. 3). Furthermore, as shown in Fig. 4, this increase in promoter activity is reflected by a decrease in intracellular levels of putrescine and spermidine, but not spermine, at doses of 20 and 100 μM in HT29 cells. Both spermidine and spermine are substrates for SSAT. Reduced putrescine levels most likely result from export of monoacetylspermidine, thus limiting putrescine production via the flavindependent polyamine oxidase (15). The mechanism responsible for the lack of effect on spermine pools is unknown. In colon tumor cells and tissues, spermidine and spermine levels are similar, whereas putrescine levels are lower (15). Thus, a 20–50% reduction in putrescine and spermidine pools represents a significant decrease in total cellular polyamine contents.
Fig. 3.
Aspirin induces SSAT promoter, RNA, and enzyme activity in HT29 cells. (A) SSAT promoter activity. HT29 cells were transfected with reporter SSAT-Luc along with pCMV-β-gal plasmid. Relative luciferase units (RLU) were calculated after normalizing to the protein and β-gal activities in the cell lysates. Fold induction was calculated after dividing the RLU of sample by the RLU of vehicle. Bars indicate standard deviation. *, Statistically significant difference (P < 0.05) compared with vehicle. (B) SSAT RNA. HT29 cells were seeded and grown for 24 h. The cells were then treated with indicated amounts of aspirin for 48 h and harvested, and total RNA was extracted. Total RNA was analyzed by probing for SSAT and GAPDH. Fold induction was calculated by dividing normalized sample values to the GAPDH control. Bars indicate standard deviation. *, Statistically significant difference (P < 0.05) compared with vehicle. (C) SSAT enzyme activity. HT29 cells were grown overnight and then treated with either various concentrations of aspirin or its vehicle for 24 h. Cells were harvested and SSAT enzyme activity was measured as described in Methods. The results are an average from three different experiments. Bars indicate standard deviation. *, Statistically significant difference (P < 0.05) compared with vehicle.
Fig. 4.
Decrease in polyamine levels in HT29 cells caused by aspirin. Cells were treated for 48 h with either 20 or 100μM aspirin or its vehicle (ethanol). Polyamine levels were normalized to the protein in the samples. Fold changes were then calculated after dividing the polyamine level in the sample by the polyamine level in the vehicle control and plotted. Bars indicate standard deviations. *, Statistically significant difference (P < 0.05) compared with vehicle.
Discussion
In our well characterized study population, we found evidence that, when homozygous, the variant A-allele of the ODC gene is associated with lower estimated odds of adenoma recurrence. No effect on recurrence was shown for heterozygous (GA) individuals. Our findings further indicate that aspirin use modifies the association of the ODC polymorphism and adenoma recurrence, as shown by a statistically significant decrease in the likelihood of recurrence among individuals homozygous for the ODC A-allele who also reported aspirin use.
To test the plausibility of these findings, we examined the effect of the polymorphism on ODC promoter activity in cell culture models of colon epithelial cells, including HT29 and Caco-2 cells. Only results for HT29 are shown here, but corroborative results were obtained with Caco-2 cells. Normal colonic epithelial cells express wild-type APC, whereas HT29 cells express mutant APC (11). To model normal colonic mucosa, we conditionally expressed wild-type APC in HT29 cells. We found that APC expression in these cells induced the transcriptional repressor Mad1 and repressed the expression of the transcriptional activator c-myc (Fig. 2; ref. 16). Microarray, and validation by other techniques including Western blotting, detected wildtype APC-dependent changes in c-myc and Mad1, but not Max, or two other E-box-binding proteins, upstream stimulating factor 1 or 2 expression (ref. 16; K.E.F. and E.W.G., unpublished work). We are currently working to determine the effects of wild-type APC on other Max-binding proteins. Moreover, the activity of the ODC promoter containing the minor A-allele was selectively suppressed in both an APC- and Mad1-dependent manner. These findings are consistent with those reported by another group (18), who showed that the two E-boxes flanking the ODC polymorphic site act cooperatively to influence ODC promoter activity, and that regions flanking these E-boxes are involved in ODC promoter activity.
Both genetically altered animals overexpressing ODC and the use of ODC inhibitors suggest that this gene is involved in the development of a number of epithelial cancers (35–38). However, no human data exist on the role of the ODC polymorphism and carcinogenesis. The SNP in intron 1 of the ODC gene is functional in that it conveys allele-specific responses to E-box transcription factors, including repressors (this work) and activators (19). Our results indicate that the ODC polymorphism and aspirin work via unique mechanisms to influence a common metabolic pathway. APC influences ODC expression (16), and the ODC genotype appears to influence APC-dependent ODC transcription, thereby affecting the level of polyamine synthesis. Aspirin does not affect ODC allele-specific promoter activity, and we have no evidence that the ODC genotype affects cellular responses to aspirin. Aspirin does not affect ODC RNA levels or enzyme activity in HT29 cells, but it activates polyamine catabolism, specifically increasing the expression and activity of SSAT and causing polyamine contents to drop. Thus, both the ODC A-allele and aspirin act to suppress tissue polyamine contents. This reduction, in turn, might reduce the risk of colorectal neoplasia, a suggestion supported by experimental models (15, 17) and human studies (see ref. 15 for review). These studies form the rationale for ongoing colon cancer prevention studies aimed at reducing intestinal mucosal polyamine contents (39).
Our findings support the model depicted in Fig. 5, which can explain our observation that only the homozygous minor A-allele is associated with reduced cancer risk. Because either one or two G-alleles would be sufficient to block Mad1 binding and facilitate increased ODC promoter activity and polyamine synthesis, one would expect significantly depressed polyamine levels to be present only in the intestinal mucosa of those normal individuals carrying two A-alleles. We are currently testing this prediction.
Fig. 5.
Model depicting actions of ODC polymorphism and aspirin on polyamine metabolism, and the consequences for adenoma recurrence. Before acquisition of somatic APC mutations, colonic cells in individuals with the minor A-allele would have reduced polyamine synthesis due to selective suppression of ODC promoter activity by Mad1. Aspirin would act, in an ODC allele-independent manner, to reduce further polyamine levels by activating the transcription of SSAT, thereby increasing polyamine catabolism and export. Cancer risk would be directly related to colonic mucosal polyamine contents, being decreased by decreased polyamine levels.
Our data do not address whether aspirin acts early or late in colon carcinogenesis. The effect of the ODC SNP would appear to be an early event. Before an APC mutation, this SNP would influence the level of polyamine synthesis, while aspirin would act independently on polyamine catabolism. Experimental studies, reviewed elsewhere (15), indicate that ODC inhibitors are potent inhibitors of colon carcinogenesis. These data suggest that colon cancer risk, in individuals with ODC GG and GA genotypes, could be markedly reduced by combination chemoprevention with NSAIDs and ODC inhibitors. Several studies are currently underway to test this hypothesis in humans with elevated risk factors for colon cancer (15, 39).
Based on data from numerous sources, use of aspirin and other NSAIDs has been proposed as a possible chemopreventive strategy for reducing the risk of colorectal neoplasia. Results from genetic and pharmacological studies suggest that the antitumor effects of NSAIDs are mediated, at least in part, through inhibition of cyclooxygenase. Additional chemopreventive effects independent of cyclooxygenase have also been proposed (40). Given the recent clinical trial data, where a significant reductions in adenoma recurrence rates were observed among 1,084 participants with previous adenomatous polyps (25) as well as among 517 patients with previous colorectal cancer (26), research related to the understanding of mechanisms potentially responsible for this protection is warranted. Aspirin and other NSAIDs clearly influence the expression of enzymes involved in polyamine metabolism (this article and ref. 33). Whether polyamines are involved in the mechanism by which aspirin reduces colon cancer risk remains to be determined.
Limitations of our human data include the limited statistical power, given the low prevalence of the ODC SNP, which is reflected in the wide confidence interval for the joint effect of aspirin use and the ODC A-allele. Clearly, these findings need to be confirmed by larger studies. Furthermore, the lack of detailed information regarding dose and duration of aspirin use limits the analysis of this variable. Lastly, studies of adenoma recurrence have limitations (41), including a short length of follow-up and assessment of only early events in neoplastic progression.
Given the recent multicenter trial data indicating a chemopreventive effect of aspirin on colorectal adenoma recurrence (25, 26), it is important to understand the mechanism(s) responsible for this preventive effect. Although important research is currently being devoted to selective cyclooxygenase-2 inhibitors, unresolved issues regarding mechanisms by which aspirin exerts its anticancer activity continue to surface. If our results are confirmed by larger studies, particularly those applied to trial data and/or colorectal cancer end-points, they indicate that the ODC A-allele is an important genetic variant that acts in concert with aspirin to markedly reduce the risk of colorectal neoplasia.
Supplementary Material
Acknowledgments
We thank Cheryl Kramer, Nancy Hart, and the clinic staff; and Bert Vogelstein for providing the HT29-APC and HT29-β-gal cells. This work was supported in part by Public Health Service Grants CA-41108, CA-23074, CA-72008, and CA-95060 from the National Cancer Institute, and by the Lankenau Hospital Foundation. M.E.M. is supported by Career Development Award KO1 CA79069-10 from the National Cancer Institute.
Abbreviations: NSAID, nonsteroidal antiinflammatory drug; OR, odds ratio; SNP, single-nucleotide polymorphism; SSAT, spermidine/spermine N1-acetyltransferase.
References
- 1.Morson, B. C. (1974) Cancer 34, Suppl., 845-849. [DOI] [PubMed] [Google Scholar]
- 2.Muto, T., Bussey, H. J. & Morson, B. C. (1975) Cancer 36, 2251-2270. [DOI] [PubMed] [Google Scholar]
- 3.Fearon, E. R. & Vogelstein, B. (1990) Cell 61, 759-767. [DOI] [PubMed] [Google Scholar]
- 4.Leslie, A., Carey, F. A., Pratt, N. R. & Steele, R. J. C. (2002) Br. J. Surg. 89, 845-865. [DOI] [PubMed] [Google Scholar]
- 5.Nishisho, I., Nakamura, Y., Miyoshi, Y., Ando, H., Horii, A., Koyama, K., Utsunomiya, J., Baba, S. & Hedge, P. (1991) Science 253, 665-659. [DOI] [PubMed] [Google Scholar]
- 6.Groden, J., Thliveris, A., Samowitz, W., Carlson, M., Gelbert, L., Albertsen, H., Joslyn, G., Stevens, J., Spirio, L., Robertson, M., et al. (1991) Cell 66, 589-600. [DOI] [PubMed] [Google Scholar]
- 7.Su, L. K., Kinzler, K. W., Vogelstein, B., Preisinger, A. C., Moser, A. R., Luongo, C., Gould, K. A. & Dove, W. F. (1992) Science 256, 668-670. [DOI] [PubMed] [Google Scholar]
- 8.Iwamoto, M., Ahnen, D. J., Franklin, W. A. & Maltzman, T. H. (2000) Carcinogenesis 21, 1935-1940. [DOI] [PubMed] [Google Scholar]
- 9.Keshav, S., McKnight, A. J., Arora, R. & Gordon, S. (1997) Cell Proliferation 30, 369-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobbie, Z., Heinimann, K., Bishop, D. T., Muller, H. & Scott, R. J. (1997) Hum. Genet. 99, 653-657. [DOI] [PubMed] [Google Scholar]
- 11.He, T.-C., Sparks, A. B., Rago, C., Hermeking, H., Zawel, L., da Costa, L. T., Morin, P. J., Vogelstein, B. & Kinzler, K. W. (1998) Science 281, 1509-1512. [DOI] [PubMed] [Google Scholar]
- 12.Jamerson, M. H., Johnson, M. D. & Dickson, R. B. (2000) Oncogene 19, 1065-1071. [DOI] [PubMed] [Google Scholar]
- 13.Bello-Fernandez, C., Packham, G. & Cleveland, J. L. (1993) Proc. Natl. Acad. Sci. USA 90, 7804-7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giardiello, F. M., Hamilton, S. R., Krush, A. J., Piantadosi, S., Hylind, L. M., Celano, P., Booker, S. V., Robinson, C. R. & Offerhaus, J. A. (1993) N. Engl. J. Med. 328, 1313-1316. [DOI] [PubMed] [Google Scholar]
- 15.Meyskens, F. L. & Gerner, E. W. (1999) Clin. Cancer Res. 5, 945-951. [PubMed] [Google Scholar]
- 16.Fultz, K. E. & Gerner, E. W. (2002) Mol. Carcinog. 34, 10-18. [DOI] [PubMed] [Google Scholar]
- 17.Erdman, S. H., Ignatenko, N. A., Powell, M. B., Flohm-Mangone, K. A., Holubec, H., Guillen-Rodriguez, J. M. & Gerner, E. W. (1999) Carcinogenesis 20, 1709-1713. [DOI] [PubMed] [Google Scholar]
- 18.Walhout, A. J., Gubbels, J. M., Bernards, R., van der Vliet, P. C. & Timmers, H. T. (1997) Nucleic Acids Res. 25, 1493-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo, Y., Harris, R. B., Rosson, D., Boorman, D. & O'Brien, T. G. (2000) Cancer Res. 60, 6314-6317. [PubMed] [Google Scholar]
- 20.Giovannucci, E., Egan, K. M., Hunter, D. J., Stampfer, M. J., Colditz, G. A., Ascherio, A. & Willett, W. C. (1995) N. Engl. J. Med. 333, 609-614. [DOI] [PubMed] [Google Scholar]
- 21.Reddy, B. S., Maruyama, H. & Kelloff, G. (1987) Cancer Res. 47, 534-536. [PubMed] [Google Scholar]
- 22.Moorghen, M., Ince, P., Finney, K. J., Sunter, J. P., Appleton, D. R. & Watson, A. J. (1988) J. Pathol. 156, 341-347. [DOI] [PubMed] [Google Scholar]
- 23.Steinbach, G., Lynch, P. M., Phillips, R., Wallace, M., Hawk, E., Gordon, G., Sherman, J., Wakabayashi, N., Saunders, B., Shen, Y., et al. (1999) Am. J. Gastroenterol. 94, 2687. [Google Scholar]
- 24.Thun, M. J., Henley, S. J. & Patrono, C. (2002) J. Natl. Cancer Inst. 94, 252-266. [DOI] [PubMed] [Google Scholar]
- 25.Baron, J. A., Cole, B., Sander, R. S., Haile, R. W., Ahnen, D. J., Bresalier, R., McKeown-Eyssen, G., Summers, R. W., Rothstein, R., Burke, C. A., et al. (2003) N. Engl. J. Med. 348, 891-899. [DOI] [PubMed] [Google Scholar]
- 26.Sandler, R. S., Halabi, S., Baron, J. A., Budinger, S., Paskett, E., Keresztes, R., Petrelli, N., Pipas, J. M., Karp, D. D., Loprinzi, C. L., et al. (2003) N. Engl. J. Med. 348, 883-890. [DOI] [PubMed] [Google Scholar]
- 27.Martínez, M. E., Reid, M. E., Guillén-Rodríguez, J., Marshall, J. R., Sampliner, R., Aickin, M., Ritenbaugh, C., van Leeuwen, B., Mason-Liddil, N., Giuliano, A., et al. (1998) Cancer Epidemiol. Biomarkers Prev. 7, 813-816. [PubMed] [Google Scholar]
- 28.Alberts, D. S., Martinez, M. E., Roe, D. J., Guillen-Rodriguez, J. M., Marshall, J. R., van Leeuwen, B., Reid, M. E., Ritenbaugh, C., Vargas, P. A., Bhattacharyya, A. B., et al. (2000) N. Engl. J. Med. 342, 1156-1162. [DOI] [PubMed] [Google Scholar]
- 29.Hosmer, D. W. & Lemeshow, S. (2000) Applied Logistic Regression (Wiley, New York).
- 30.Chomczynski, P. & Sacchi, N. (1987) Anal. Biochem. 162, 156-159. [DOI] [PubMed] [Google Scholar]
- 31.Seiler, N. & Knodgen, B. (1980) J. Chromatogr. 221, 227-235. [DOI] [PubMed] [Google Scholar]
- 32.Smith, P. K., Krohn, R. I., Hermanson, G. T., Mallia, A. K., Gartner, F. H., Provenzano, M. D., Fujimoto, E. K., Goeke, N. M., Olson, B. J. & Klenk, D. C. (1985) Anal. Biochem. 150, 76-85. [DOI] [PubMed] [Google Scholar]
- 33.Ignatenko, N. A. & Gerner, E. W. (1996) Cell Growth Differ. 7, 481-486. [PubMed] [Google Scholar]
- 34.Turchanowa, L., Dauletbaev, N., Milovic, V. & Stein, J. (2001) Eur. J. Clin. Invest. 31, 887-893. [DOI] [PubMed] [Google Scholar]
- 35.Weeks, C. E., Herrman, A. L., Nelson, F. R. & Slaga, T. J. (1982) Proc. Natl. Acad. Sci. USA 79, 6028-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson, H. J., Meeker, L. D., Herbst, E. J., Ronan, A. M. & Minocha, R. (1985) Cancer Res. 45, 1170-1173. [PubMed] [Google Scholar]
- 37.Nowels, K., Homma, Y., Seidenfeld, J. & Oyasu, R. (1986) Cancer Biochem. Biophys. 8, 257-263. [PubMed] [Google Scholar]
- 38.O'Brien, T. G., Megosh, L. C., Gilliard, G. & Soler, A. P. (1997) Cancer Res. 57, 2630-2637. [PubMed] [Google Scholar]
- 39.Meyskens, F. L. (1998) Hematol. Oncol. Clin. North Am. 12, 935-941. [DOI] [PubMed] [Google Scholar]
- 40.Hwang, D. H., Fung, V. & Dannenberg, A. J. (2002) J. Natl. Cancer Inst. 4, 91-97. [Google Scholar]
- 41.Martínez, M. E. (2001) J. Natl. Cancer Inst. 93, 1764-1765. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.