Abstract
The MinCDE proteins of Escherichia coli are required for proper placement of the division septum at midcell. The site selection process requires the rapid oscillatory redistribution of the proteins from pole to pole. We report that the three Min proteins are organized into extended membrane-associated coiled structures that wind around the cell between the two poles. The pole-to-pole oscillation of the proteins reflects oscillatory changes in their distribution within the coiled structure. We also report that the E. coli MreB protein, which is required for maintaining the rod shape of the cell, also forms extended coiled structures, which are similar to the MreB structures that have previously been reported in Bacillus subtilis. The MreB and MinCDE coiled arrays do not appear identical. The results suggest that at least two functionally distinct cytoskeletal-like elements are present in E. coli and that structures of this type can undergo dynamic changes that play important roles in division site placement and possibly other aspects of the life of the cell.
Until recently, it was thought that prokaryotic cells did not contain internal cytoskeletal-like elements that play a role in cell shape determination and other cellular functions. This view was altered by the demonstration that the MreB and Mbl proteins of the Gram-positive bacterium Bacillus subtilis form coiled structures that extend along the length of the cell and play a role in cell shape maintenance (1). We report here that the Min proteins of Escherichia coli that are required for proper placement of the division septum, but play no apparent cytoskeletal role, are also organized into extended membrane-associated structures that coil around the cell between the two cell poles.
The MinC, MinD, and MinE proteins ensure that the division site is placed at the midpoint of the rod-shaped cell by preventing aberrant division events at positions other than the normal midcell site (reviewed in ref. 2). In this process, MinC acts as an inhibitor of septation that is given topological specificity by the action of MinE.
MinD is responsible for recruiting MinC and MinE to the membrane. When MinD is expressed in the absence of MinE and MinC, it is distributed around the periphery of the cell, indicating that MinD can associate with the cytoplasmic membrane independent of the other Min proteins (3–5). This view is supported by in vitro experiments showing that MinD can associate with phospholipid vesicles in the absence of MinC and MinE (6–8). In contrast, MinC and MinE appear to be located in the cytoplasm when expressed in the absence of MinD but become membrane-associated when either of the proteins is coexpressed with MinD (9–11).
MinE is a topological specificity factor that induces the redistribution of most of the cellular MinD and MinC into a membrane-associated polar zone at one end of the cell (3, 4, 10). This is followed by formation of the E-ring, a prominent ring-like MinE structure that is adjacent to the medial edge of the polar zone near midcell (11–13). Most of the remainder of the cellular MinE is located within the region of the MinD polar zone. The MinE ring and the MinC/MinD/MinE polar zone then undergo a process of rapid and repeated oscillation from pole to pole (Fig. 1H; refs. 3, 4, 10, 12, and 13). This oscillatory behavior is accomplished by the progressive shrinkage of the polar zone and E-ring to the end of the cell and their reformation at the opposite cell pole. The pole-to-pole oscillations occur many times within each division cycle. As a result, the time-averaged concentration of the MinC septation inhibitor is maintained at a lower level at midcell than elsewhere along the length of the cell. This process is believed to play an essential role in restricting division to its correct midcell site.
Fig. 1.
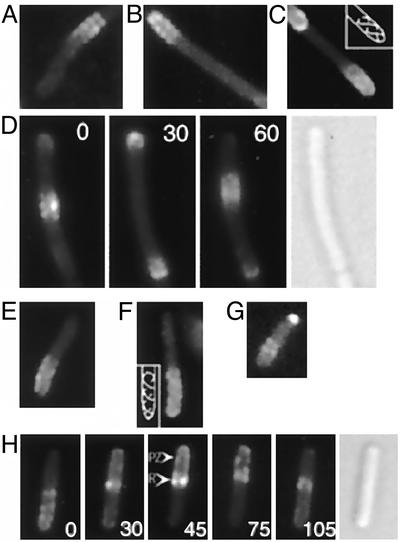
MinD and MinE structures in living cells. (A–D) Fluorescence images of YFP-MinD in unfixed cells of strain HL1/pFX40 [ΔminDE/Plac-yfp::minD minE]. (A–C) Random single images. (D) Time-lapse micrographs taken at 30-sec intervals, showing oscillation of a MinD zone between midcell and the cell poles. (E–H) Fluorescence images of MinE-YFP in unfixed cells of strain RC1/pFX55 [Δmin/Plac-minC minD minE::yfp]. (E–G) Random single images. (H) Time-lapse micrographs showing pole-to-pole oscillation of an E-ring and MinE polar zone. Time intervals, in sec, are indicated by numbers within the panels. In C and F, the images suggest the presence of two helical structures within the polar zone, as indicated in the Insets. Nomarski images are shown in D and H. PZ, polar zone; R, MinE ring.
In this article we show that the membrane-associated Min proteins are not randomly distributed within the polar zones, but instead are organized into extended coiled structures that wind around the cell cylinder. The previously described cycle of pole-to-pole oscillation of the polar zones and E-rings appears to occur by the dynamic redistribution of the Min proteins within this helical framework. We also show that the E. coli MreB protein forms extended coiled structures that extend along the length of the cell and resemble the structures that are formed by the homologous MreB protein of B. subtilis (1). The E. coli MreB coiled structures appear to differ from the MinCDE helical arrays that are described here. The results indicate that at least two structurally and functionally distinct cytoskeletal-like elements are present in E. coli and that structures of this type can undergo dynamic changes that play important roles in division site placement, and possibly other aspects of the life of the cell.
Materials and Methods
Strains and Growth Conditions. E. coli HL1 [ΔminDE zcf117::Tn10 recA::cat] (12) was kindly provided by Piet de Boer (Case Western Reserve University School of Medicine, Cleveland). Strains RC1 [Δlac Δara ΔminCDE] (4), PB103 and MC1000 (14), and PA340–129 [mre-129], kindly provided by M. Wachi (Tokyo Institute of Technology, Tokyo; ref. 15), have been described elsewhere. For examination of labeled MinC, MinD, or MinE, cells of strain RC1 containing the appropriate plasmid were grown in L-Broth in the presence of 10 μM isopropyl β-d-thiogalactoside (IPTG) for 2 h and images were acquired as described (16). For comparing the ability of MreB and YFP-MreB to complement the mre-129 mutation, exponentially growing PA340-129 cells containing pLE6 [Plac-mreB] or pLE7 [Plac-yfp::mreB] were grown in L-broth in the presence of 50 μM IPTG for ≈2 h at 37°C before microscopic examination.
Plasmids. The gfp moiety in gene fusions is the gfpmut2 gene (17). For full details of plasmid constructions see Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Microscopy. For optical sectioning, the Olympus BX60 microscope and UPlanApo, ×100, 1.35 oil objective lens was equipped with an Orbit piezo drive (Improvision, Lexington, MA) to obtain series of z-sections with a fixed spacing of 0.05 μm. Each stack of 40 optically sectioned fluorescent images was deconvoluted through 20 iterations by using the 3D restoration function in the OPENLAB version 3.1 program (Improvision) to obtain deconvolved images. Three-dimensional images were reconstituted from the stacks of the deconvolved images by using the OPENLAB or VOLOCITY program (Improvision).
Results
MinD Coiled Structures. Evidence that MinD polar zones might contain underlying higher-order structures came from fluorescence microscopy of cells that coexpressed fluorescently labeled MinD [yellow fluorescent protein (YFP)-MinD or GFP-MinD] and MinE. The microscopy revealed that inhomogeneities were present within many of the polar zones in a repetitive pattern that indicated the presence of organized cellular structures within the polar zones (Fig. 1 A–D). These structures were visible in raw images of 17% (18/104) of cells. The patterns appeared as a series of regularly spaced stripes or paired dots, suggesting that membrane-associated rings or coils were present within the polar zones. In some cells, the pattern suggested the presence of one or two extended coiled or helical structures (Fig. 1C). Structures similar to those described above were also present (Fig. 1D) within the MinD zones that are present along the length of long cells (3, 13).
Because a random micrograph represents an image obtained from a single focal plane, with out-of-focus information impinging from other levels within the cell, it is difficult to clearly define any extended cellular structures that are present in raw images. Therefore, we optically sectioned individual cells to obtain a series of images whose focal planes extended across the entire width of the cell cylinder. Most out-of-focus information was removed from each slice by an iterative deconvolution routine (see Materials and Methods). This routine sharpened the images of the periodic structures within the polar zone (Fig. 2 A2, B2, D2, and E2). The transcellular series of deconvolved images was then used to reconstruct the three-dimensional organization of MinD within the cell.
Fig. 2.
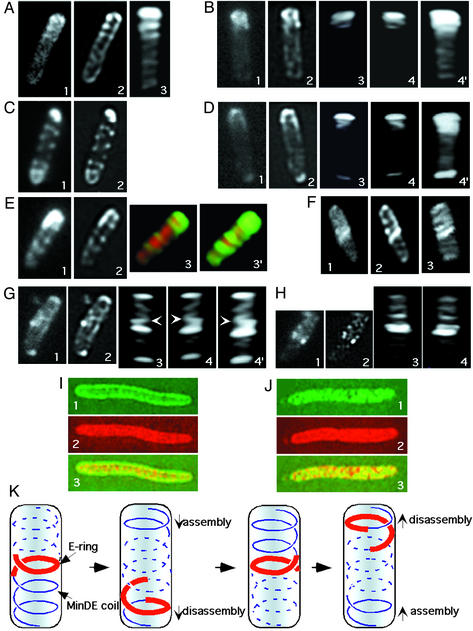
MinD and MinE structures in optically sectioned cells. Cells were fixed and subjected to optical sectioning and processing as described in Materials and Methods. The first image in each series is the initial raw image of one section without deconvolution; image 2 is the same section after deconvolution; and images 3, 4, and 4′ are three-dimensional reconstructions from the series of deconvolved images. (A and C) YFP-MinD fluorescence, strain RC1/pFX40 [Δmin/Plac-yfp::minD minE]. (B and D) GFP-MinD fluorescence, strain RC1/pFX9 [Δmin/Plac-gfp::minD minE]. In image 4, the three-dimensional image shown in image 3 was rotated ≈180° around the long axis of the cell. Image 4′ is identical to image 4 except that contrast has been increased to permit visualization of the less bright structures in the lower part of the cell. (E) YFP-MinD and CFP fluorescence, strain RC1/pFX40/pLE18 [Δmin/Plac-yfp::minD minE/Para-cfp]. Images 1 and 2 show YFP-MinD. Images 3 and 3′ show overlays of YFP-MinD (green) and CFP (red) fluorescence. CFP acts as a cytoplasmic marker. Image 3′ is identical to image 3, except that contrast has been increased to permit better visualization of the coils in the lower portion of the cell. (F–H) MinE-GFP fluorescence, RC1/pSY1083G [Δmin/Plac-minC minD minE::gfp]. In image 4, the three-dimensional representation shown in image 3 was rotated 180° around the long axis of the cell. Image 4′ is identical to image 4 except that contrast has been increased to permit visualization of the less bright structures in the lower part of the cell. The arrows in G3, G4, and G4′ indicate the junction between the coiled E-ring and the coiled array in the polar zone. (I) YFP-MinD (green, images 1 and 3) and CFP (red, images 2 and 3) in strain RC1/pYLS97/pLE18 [Δmin/Plac-yfp::minD/Para-cfp]. CFP is used as a cytoplasmic marker. In image 3, the representations in images 1 and 2 are overlaid. (J) MinE-YFP (green, images 1 and 3) and CFP (red, images 2 and 3) in strain RC1/pLE11/pLE18 [Δmin/Plac-minE::yfp/Para-cfp]. In image 3, the representations in images 1 and 2 are overlaid. (K) Graphic model illustrating coiled arrays at different stages of the oscillation cycle. Red lines represent the MinE ring. Blue lines represent MinDE coiled structures within the polar zones (solid lines) and in the opposite end of the cell (interrupted lines). For simplicity, a single coiled array is shown in each end of the cell, although the results suggest that some cells may contain more than one coiled array, and the helical structure may, in some cases, extend over the entire length of the cell.
The deconvoluted images and three-dimensional reconstructions indicated that MinD within the polar zone was organized in a coiled structure that extended from the cell pole toward midcell (Fig. 2 A, B, and D). The coiled structures were present in polar zones that extended only a short distance from the end of the cell (Fig. 2 B and D) as well as in longer zones that extended farther from the pole toward midcell (Fig. 2 A). The loop closest to the end of the cell was often brighter than the other coils within the polar zone. This appearance could be explained by a higher concentration of MinD in the basal loop or could reflect the presence of two or more tightly packed basal coils that could not be visually resolved.
The structures that were visible in the deconvolved images and three-dimensional reconstructions corresponded in number and position to the structures that were visible in unprocessed raw images (Fig. 2 A–C and E). This result establishes that the coiled MinD structures were not artifacts of the deconvolution and three-dimensional rendering procedures. Similar structures were present in deconvolved images and three-dimensional reconstructions of most cells that contained visible polar zones, even when the unprocessed raw images did not show obvious structural organization. This finding suggests that the periodic coiled structures represent the basic structure of the MinD polar zone, although this can only be stated with certainty for cells showing evidence of the structures in both raw and processed images. The periodic structures were seen in both fixed (Fig. 2) and living (Fig. 1) cells, confirming that they were not fixation artifacts. Time-lapse studies of living cells showed that MinD zones that contained the coiled structures oscillated within the cell (Fig. 1D), confirming that the coils were not postmortem artifacts.
Interestingly, the coiled structures were not limited to the polar zones. Thus, in many cells, a periodic MinD structure was visible in the opposite end of the cell that resembled the coiled array of the polar zone, but was much less intense. In raw images the structures appeared as a series of poorly resolved diagonal stripes (Fig. 2 A1, C1, and E1). After deconvolution they appeared as a series of parallel curved diagonal bands whose appearance suggested that they represented one face of a helical array that coiled around the cell (Fig. 2 A2, C2, and E2). In three-dimensional reconstructions the YFP-MinD structures in the nonpolar zone region of the cell appeared to coil around the cell (Fig. 2 E3 and E3′). The resolution of the micrographs was insufficient to indicate whether the coiled arrays in the two ends of the cell were continuous or discontinuous. As previously observed (13), some cells contained polar zones at both poles (Fig. 2C), one presumably representing a polar zone during the disassembly stage where the zone shrinks toward the pole, and the other presumably representing a polar zone during the stage of assembly (2). In these cells a YFP-MinD helical array extended between the two polar zones (Fig. 2C).
Quantitative Western immunoblots showed that the concentration of Yfp::MinD in RC1/pFX40 [Δmin/Plac-yfp::minD minE] cells grown under the present conditions was ≈60% of the MinD concentration in MC1000, the minCDE+ parent of RC1, confirming that the coiled structures were not because of overproduction of Yfp::MinD.
Free cyan fluorescent protein (CFP) was used as a marker for the cytoplasmic compartment to determine the spatial relation of the coiled structures to other cellular compartments. Doublelabel analysis of cells that coexpressed CFP, YFP-MinD, and MinE showed that the coils of YFP-MinD were located on the outer surface of the cytoplasmic compartment (Fig. 2 E3 and E3′), indicating that the coiled structure was located just beneath or associated with the inner surface of the cytoplasmic membrane.
When YFP-MinD was expressed in the absence of MinE, the fluorescence was located in the periphery of the cell, on the surface of the cytoplasmic compartment that was marked by free CFP (Fig. 2I). This was consistent with previous reports that MinD can associate with the cell membrane (3–5) or with phospholipid vesicles (6–8) in the absence of the other Min proteins. There was no reproducible ordered structure visible within the peripheral MinD compartment.
MinE Coiled Structures. Study of cells that coexpressed MinD with MinE-YFP or MinE-GFP showed that MinE was also organized into extended coiled structures within the cell. Random micrographs (Fig. 1 E, F, and H) and images of individual optical sections (Fig. 2 F1, G1, and H1) often showed a periodic series of crosswise or diagonal stripes that extended from one end of the cell to the E-ring near midcell, corresponding in position to the MinE polar zones (12, 16). The stripes within the MinE polar zones were less bright than the corresponding structures in the MinD polar zones, consistent with the difference in intensity of MinD and MinE polar zones, and were therefore usually less well defined in raw images. Deconvolved images and three-dimensional reconstructions suggested that the stripes reflected the presence of a series of coils that extended from the end of the cell to the E-ring (Fig. 2 F–H).
The MinE rings, identified by their high fluorescence intensity, appeared not to be closed rings. Instead, the high intensity MinE bands at the medial edge of the polar zone appeared to be composed of 1–2 loops of a coiled structure (Figs. 1G and 2F and 2 G3, G4, and G4′). In some images the coils that comprised the E-ring appeared to be continuous with the MinE coiled structure within the polar zone (Fig. 2G). The relative brightness of the E-ring made it difficult in most cells to resolve the detailed relationship of the E-ring to the coiled arrays of MinE in the two ends of the cell. A similar series of less intense MinE bands or coils was also present in the opposite end of the cell, extending from the region of the E-ring to the distal cell pole (Fig. 2F). The bands were generally oriented at an angle to the long axis of the cell. The MinE structures in the two ends of the cell resembled the MinD coiled structures that were located in the same regions (see above) in their packing density and general appearance.
When MinE-YFP was expressed in the absence of MinD, the labeled MinE was located in the interior of the cell where it approximated the general location of free CFP, which was used as a cytoplasmic marker in the same cells (Fig. 2 J). This finding confirmed previous reports that the membrane localization of MinE requires MinD (11).
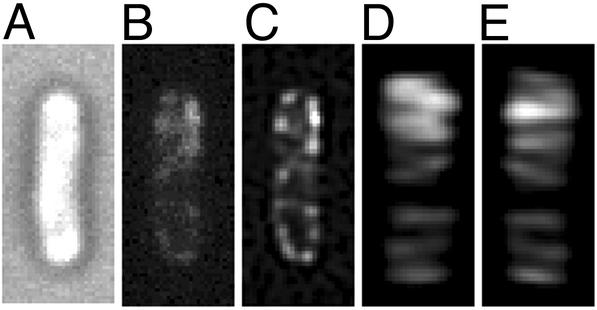
Cellular Organization of MinC. Study of GFP-MinC revealed that MinC also was organized into long-range coiled structures when coexpressed with MinD and MinE in strain RC1/pYLS49–2 [Δmin/Plac-gfp::minC minD minE]. The structures were visible in raw images (Fig. 3B) and in deconvolved images and threedimensional reconstructions (Fig. 3 C–E). The distribution of GFP-MinC was similar to that of MinD (see above), with brightly fluorescent coils within polar zones and less intense coils or loops extending to the opposite end of the cell (Fig. 3 D and E).
Fig. 3.
GFP-MinC in the presence of MinD and MinE. A cell of strain RC1/pYLS49–2 [Δmin/Plac-gfp::minC minD minE] is shown. (A) Nomarski image. (B) Raw image without deconvolution. (C) Deconvolved image of the same optical section shown in B.(D and E). Three-dimensional reconstruction from the set of optically sectioned images, viewed at 0° (D) and 180° (E) rotation around the long axis of the cell.
Cellular Distribution of MreB. It has previously been shown that the MreB protein of B. subtilis is organized into an extended structure that coils around the cell along its length. We therefore examined the cellular distribution of the E. coli homolog of the B. subtilis MreB protein to determine whether it also formed a coiled structure that might be related to the MinD and MinE structures described here.
The mreB gene was cloned from the chromosome of E. coli PB103 and an in-frame yfp::mreB fusion was constructed. The yfp::mreB gene product expressed from pLE7 [Plac-yfp::mreB] by growth in the presence of IPTG was ≈40% as effective as wild-type mreB expressed from pLE6 [Plac-mreB] in complementing an mreB mutant strain (PA340–129; ref. 15), as indicated by the number of cells of normal shape remaining in the induced cultures.
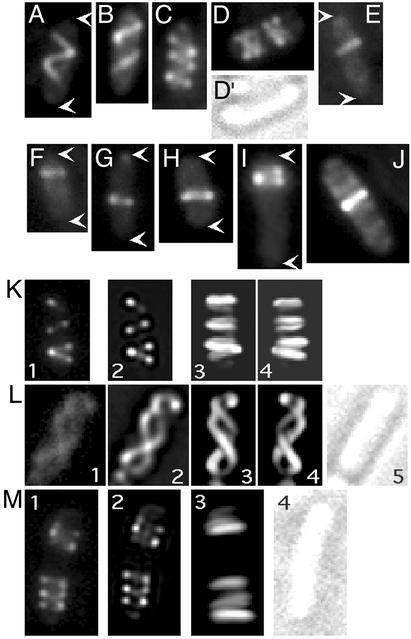
When yfp::mreB was induced by growth of strain MC1000/pLE7 [wt/Plac-yfp::mreB] in the presence of IPTG, fluorescence microscopy revealed that many cells contained extended coiled structures that wound around the cell along its length (Fig. 4). The structures were visible in raw images (Fig. 4 A–J), deconvolved and reconstructed images (Fig. 4 K–M), and in both living and fixed cells, indicating that they were not processing or fixation artifacts. The packing density of the coils and their distribution along the cell varied considerably. In ≈82% of cells (90/109) the YFP-MreB coiled structure was quite extended. In these cells, one or two turns of the coiled array extended along most of the length of the cell (Fig. 4 A, B, and K), with a packing density approximately half of that of the MinD coiled structures described above. In ≈13% of cells (14/109) the loops were more tightly packed, with a packing density similar to that of the MinD coiled structures (Fig. 4 C, D, and M). The more tightly packed structures were sometimes present in two clusters located in opposite ends of the cell, with a space between them (Fig. 4 D and M), suggesting that the cells might be predivisional or might have begun septal ingrowth. Approximately 5% of cells (5/109) contained a single or double coil or loop at one location, with no apparent preference for any specific position along the length of the cell (Fig. 4 E–J). The B. subtilis MreB protein has also been reported to form single loops, especially in younger cells (1).
Fig. 4.
YFP-MreB structures. Fluorescence images of selected cells of strain MC1000/pLE7 [wt/Plac-yfp::mreB] after growth at 30° for 3–4 h in the presence of 10 μM IPTG, containing extended helical arrays (A, B, K, and L), compressed helical arrays (C, D, and M), and single coils or loops (E–J). White arrows indicate the ends of cells. (A–J) Raw images. In K–M, cells were subjected to optical sectioning. Image 1 is a raw image of a single section before deconvolution and image 2 is the same section after deconvolution. In K, L, and M, images 3 and 4 are three-dimensional reconstructions of the stacks of deconvolved images, and in K and L, image 4, the three-dimensional representation in image 3 was rotated ≈180° around the long axis of the cell.
Discussion
This article establishes that E. coli normally contains at least two long-range coiled structures that wind around the cytoplasmic compartment: the MreB cytoskeletal element and the MinCDE helical arrays. These cytoskeletal-like elements appear to be normal components of the E. coli cell.
The MreB and MinCDE proteins carry out quite different functions. The mreB gene product has been implicated in maintenance of the rod shape of E. coli and B. subtilis, based on the observation that cells of mreB mutants grow as irregular spheres instead of rods (1, 15, 18–20). In contrast, the E. coli Min proteins are not required to maintain the rod shape of the cell, but play an essential role in determining placement of the division septum (14). This function requires formation of E-rings and MinC/MinD/MinE polar zones and their pole-to-pole oscillation. These events result from the dynamic redistribution of the protein components of the MinC/MinD/MinE coiled structures.
Are the MreB and MinCDE coiled arrays part of the same structure? The MreB coiled structures generally differed from the MinCDE structures in location and appearance, with a packing density in most cells that was only about half that of the MinCDE coiled arrays. Additionally, some cells contained only a single MreB loop or ring, with no apparent preferred location along the length of the cell, whereas single MinD or MinC loops were not observed, except at the extreme end of the cell (data not shown). The variety of arrangements of the MreB coiled arrays may be because of changes in MreB organization as cells progress through the division cycle or may reflect varying levels of Yfp-MreB expression in different cells in the population. Some cells contained more densely packed MreB coiled arrays, accounting for <15% of the population. In contrast, MinD polar zones with their more densely packed coiled structures were present in most cells. These observations suggest that MinCDE and MreB are not part of the same structure in most cells, but further work will be needed to firmly establish whether any relationship exists between the two coiled structures or whether they might be associated with a common structural element at any stage of their history.
One of the interesting observations in this article was the finding that the coiled arrays of MinC, MinD, and MinE were not restricted to the polar zones and E-rings, as might have been expected. Instead, less intense MinC/MinD/MinE coiled structures were also present in the other half of the cell, extending from the edge of the polar zone to the opposite cell pole. This finding implies that the MinCDE coiled structures that comprise the polar zones are not completely disassembled when MinC, MinD, and MinE leave the zones during the disassembly phase of the pole-to-pole oscillation cycle. As a result, the basic MinCDE helical array remains in place, possibly as part of a permanent helical framework that is located along the length of the cell. This outcome raises several questions.
First, what is the relationship between the coiled array in the polar zone and the coiled array in the other half of the cell? The coiled structures in the two halves of the cell may be continuous, composed of a single structure that extends from one pole to the other, with the polar zone being explained by the presence of a higher concentration of Min proteins in one portion of the array. This possibility is perhaps made less likely by the indication that the helices in the two parts of the cell were wound in opposite directions (Fig. 2 A2 and F2). Alternatively, the coiled arrays in the two halves of the cell may be separate helical structures that extend from a cell pole to the central edge of the polar zone or perhaps from one pole to the other. This finding would explain the presence of images suggesting the presence of two overlapping coiled structures within the polar or internal zones (Fig. 1 C and F).
Second, does the residual coiled array in the opposite end of the cell play a role in the dynamic behavior of the Min proteins? We speculate that the coiled array that is already in place at the opposite end of the cell may act as a template or scaffold for the incorporation of MinD into the polar domains during the oscillation sequence. We consider two possibilities. In the first scenario, MinD molecules that are released during disassembly of the old polar zone directly associate with the helical framework that is already in place at the opposite end of the cell to form the new polar zone. In the second scenario, MinD molecules that are released from the disassembling polar zone initiate new helical strands by nucleation at the opposite pole, by using the helical framework that is already in place at the end of the cell as a scaffold to guide the growth of the new structure (Fig. 2K). MinE and MinC would either move together with MinD or secondarily associate with the MinD helical array. It is presently thought that the movement of the Min proteins from one pole to the other occurs by release of the proteins from the polar zone and E-ring followed by their movement within the cytoplasm to the opposite end of the cell where they reassociate with the membrane to form a new polar zone (3, 21). This possibility would be compatible with either of the two models, although the pole-to-pole movement of one or more of the proteins could involve another mechanism, such as movement of the proteins along the coiled track.
It is likely that MinD and MinE are part of a common structure within the polar array, although this has not been unequivocally shown, and it has not been established whether MinC is part of such a MinD/MinE extended structure, although this also seems likely. MinD can associate with phospholipid vesicles in the absence of the other Min proteins, requiring only the presence of ATP or suitable ATP analogs (6, 7). Under these in vitro conditions, MinD forms long linear structures. It is likely that the coiled structures that are present in cells that coexpress MinD and MinE are based on a similar extended MinD structure, with each coiled structure probably consisting of an array of protofilaments (7). In this article, when YFP-MinD was expressed in the absence of MinE, coiled or filamentous structures were not observed in vivo, whereas filamentous structures were seen in the in vitro studies. It cannot be excluded that MinD does form these structures in vivo when expressed in the absence of the other Min proteins but that they are relatively unstable in the absence of MinE.
Instead of being a free-standing ring, the E-ring appears to be a part of the MinE or MinD/MinE coiled array that comprises the polar zone, representing the terminal loop of the polar coiled array. Polar zones are thought to assemble progressively from the pole and to terminate growth when they approach midcell. The terminal MinE loop is well positioned to act as a stop-growth signal to terminate growth of the polar coiled array when it approaches midcell, an essential step in regulating division site placement. The idea that the E-ring acts as a stop-growth signal was previously suggested by the observation that MinD polar zones do not terminate near the normal midcell site in mutants that are blocked in E-ring formation (16).
The geometry of the coiled structures may be determined solely by the structure of MinD or a MinD–MinE complex, possibly in combination with other proteins. In this view, the growing polar structures would spontaneously assume a radius of curvature that leads to the observed helical organization when associated with the inner side of the cytoplasmic membrane. It is also possible that the basic MinDE helical array assembles on a track that is formed by another cytoskeletal-like element, probably not MreB. Although there are no obvious candidates for another cytoskeletal protein in E. coli aside from MreB, it is clear that bacterial cells can contain more than one long-range structural element. This finding has been demonstrated in B. subtilis, where, in addition to MreB, the Mbl protein, which has no apparent E. coli homolog, forms a coiled structure that extends around the cell (1). The FtsZ protein of B. subtilis also forms extended coiled structures that extend from midcell toward the cell poles at an intermediate stage in formation of the polar septum during sporogenesis (22). Overexpression of the E. coli ftsZ gene also gives rise to coiled FtsZ structures (23). In addition, although not a normal cellular component, the plasmid-encoded ParM protein forms long filamentous structures that extend along the length of the E. coli cell in plasmidcontaining cells (24).
Finally, do the MinC/MinD/MinE helical arrays function only to localize the Min proteins as part of the division site placement process, or may the structures also carry out other cellular functions? It is clear that the coiled arrays do not carry out any essential cellular function because ΔminCDE strains can grow and divide. The coiled array also does not appear to function in cell shape determination because the basic rod shape of the cells is maintained in ΔminCDE strains. It is of interest, however, that several observations have suggested a possible role for the Min proteins in chromosome segregation. Thus, MinD shows significant homology to Par proteins that play essential roles in the equipartition of unit copy number plasmids into daughter cells (25, 26). It also has been reported that chromosome segregation is impaired in ΔminCDE and min mutant strains (27–29). These observations raise the possibility that the MinD helical track might play a role in chromosome segregation, possibly by providing a track for the transfer of oriC from midcell to the cell poles during the process of chromosome partition (30, 31). It also has been reported that the MinC and MinD proteins of B. subtilis regulate the cellular localization of the SpoIIIE protein, which is localized to the polar forespore septum, an important step in facilitating the transfer of chromosomal DNA from mother cell to forespore (32). It will, therefore, be of considerable interest to determine whether the MinCDE coiled structure plays a role in chromosome segregation or other functions in E. coli that are not directly related to division site placement.
Supplementary Material
Acknowledgments
We thank Dr. M. J. Osborn, Dr. Ann Cowan, and Dr. Glenn King for advice throughout this project, and Dr. X. Fu for kindly providing the pFX series of plasmids. This work was supported by National Institutes of Health Grants GM-60632 and GM-53276 (to L.R.).
Abbreviations: YFP, yellow fluorescent protein; CFP, cyan fluorescent protein.
See commentary on page 7423.
References
- 1.Jones, L., Carballido-Lopez, R. & Errington, J. (2001) Cell 104, 913-922. [DOI] [PubMed] [Google Scholar]
- 2.Rothfield, L. I., Shih, Y.-L. & King, G. F. (2001) Cell 106, 13-16. [DOI] [PubMed] [Google Scholar]
- 3.Raskin, D. & de Boer, P. (1999) Proc. Natl. Acad. Sci. USA 96, 4971-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rowland, S. L., Fu, X., Sayed, M. A., Zhang, Y., Cook, W. R. & Rothfield, L. I. (2000) J. Bacteriol. 182, 613-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szeto, T., Rowland, S., Rothfield, L. & King, G. F. (2002) Proc. Natl. Acad. Sci. USA 99, 15693-15698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu, Z., Gogol, E. & Lutkenhaus, J. (2002) Proc. Natl. Acad. Sci. USA 99, 6761-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suefuji, K., Valluzzi, R. & RayCahudhuri, D. (2002) Proc. Natl. Acad. Sci. USA 99, 16776-16781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lachner, L., Raskin, D. & de Boer, P. (2003) J. Bacteriol. 185, 735-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raskin, D. & de Boer, P. (1999) J. Bacteriol. 181, 6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu, Z. & Lutkenhaus, J. (1999) Mol. Microbiol. 34, 82-90. [DOI] [PubMed] [Google Scholar]
- 11.Raskin, D. & de Boer, P. (1997) Cell 91, 685-694. [DOI] [PubMed] [Google Scholar]
- 12.Hale, C., Meinhardt, H. & de Boer, P. (2001) EMBO J. 20, 1563-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu, X., Shih, Y.-L., Zhang, Y. & Rothfield, L. I. (2001) Proc. Natl. Acad. Sci. USA 98, 980-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Boer, P. A., Crossley, R. E. & Rothfield, L. I. (1989) Cell 56, 641-649. [DOI] [PubMed] [Google Scholar]
- 15.Wachi, M., Doi, M., Tamaki, S., Park, W., Nakajima-lijima, S. & Matsuhashi, M. (1987) J. Bacteriol. 169, 4935-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shih, Y.-L., Fu, X., King, G. F., Le, T. & Rothfield, L. I. (2002) EMBO J. 21, 3347-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cormack, B. P., Valdivia, R. H. & Falkow, S. (1996) Gene 173, 33-38. [DOI] [PubMed] [Google Scholar]
- 18.Wachi, M. & Matsuhashi, M. (1989) J. Bacteriol. 171, 3123-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doi, M., Wachi, M., Ishno, F., Tomioka, S., Ho, M. & Matsuhashi, M. (1988) J. Bacteriol. 170, 4619-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamaki, S., Matsuzawa, H. & Matsuhashi, M. (1980) J. Bacteriol. 141, 52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, Z. & Lutkenhaus, J. (2001) Mol. Cell 7, 1337-1343. [DOI] [PubMed] [Google Scholar]
- 22.Ben-Yehuda, S. & Losick, R. (2002) Cell 109, 257-266. [DOI] [PubMed] [Google Scholar]
- 23.Ma, X., Ehrhardt, D. & Margolin, W. (1996) Proc. Natl. Acad. Sci. USA 93, 12998-13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Møller-Jensen, J., Jensen, R. B., Lachner, L., Löwe, J. & Gerdes, K. (2002) EMBO J. 21, 3119-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Motallebi-Veshareh, M., Rouch, D. A. & Thomas, C. M. (1990) Mol. Microbiol. 4, 1455-1463. [DOI] [PubMed] [Google Scholar]
- 26.de Boer, P. A., Crossley, R. E., Hand, A. R. & Rothfield, L. I. (1991) EMBO J. 10, 4371-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaffé, A., D'Ari, R. & Hiraga, S. (1988) J. Bacteriol. 170, 3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulder, E., El'Bouhali, M., Pas, E. & Woldringh, C. L. (1990) Mol. Gen. Genet. 221, 87-93. [DOI] [PubMed] [Google Scholar]
- 29.Akerlund, T., Bernander, R. & Nordström, K. (1992) Mol. Microbiol. 6, 2073-2083. [DOI] [PubMed] [Google Scholar]
- 30.Webb, C. D., Teleman, A., Gordon, S., Straight, A., Belmont, A., Lin, D. C., Grossman, A. D., Wright, A. & Losick, R. (1997) Cell 88, 667-674. [DOI] [PubMed] [Google Scholar]
- 31.Niki, H. & Hiraga, S. (1998) Genes Dev. 12, 1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharp, M. & Pogliano, K. (2002) EMBO J. 21, 6267-6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsien, R. Y. (1998) Annu. Rev. Biochem. 67, 509-544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.