Abstract
SulA halts cell division in Escherichia coli by binding to the major component of the division machinery FtsZ. We have solved the crystal structure of SulA alone and in complex with FtsZ from Pseudomonas aeruginosa. SulA is expressed when the SOS response is induced. This is a mechanism to inhibit cell division and repair DNA in the event of DNA damage. FtsZ is a tubulin-like protein that forms polymers, with the active-site GTPase split across two monomers. One monomer provides the GTP-binding site and the other, through its T7 loop nucleotide hydrolysis. Our structures show that SulA is a dimer, and that SulA inhibits cell division neither by binding the nucleotide-binding site nor by inducing conformational changes in FtsZ. Instead, SulA binds the T7 loop surface of FtsZ, opposite the nucleotide-binding site, blocking polymer formation. These findings explain why GTP hydrolysis and polymer turnover are required for SulA inhibition.
DNA damage or interference in DNA replication, either by chemicals or UV light in Escherichia coli, results in the induction of the SOS response, and cell division ceases. The SOS response induces the expression of >30 genes through RecA-induced LexA autoproteolysis (1). These genes are involved in a variety of processes, but the majority are involved in DNA repair (2). One of the SOS genes is sulA (3). The protein is synthesized in large amounts during SOS response, reaching ≈0.3% of total protein synthesis. SulA is sufficient to halt cell division (4). Turnover in the cell is rapid, with a half-life of 1.2 min. SulA is degraded by Lon, and in lon- cells, the half-life increases to 19 min (5).
SulA acts on the tubulin-like GTPase, FtsZ (6–8). During normal cell division, FtsZ polymerizes to form a ring at the middle of the bacterium (9, 10). FtsZ is the first and most numerous component of the septum. Other proteins, including FtsA and ZipA, are then recruited to the ring of FtsZ in a specific order to assemble the septal machinery (11, 12). In vitro, FtsZ can polymerize to form protofilaments (13), which can then associate through lateral interactions into higher-order structures such as sheets and rings. In vivo, ZipA (14) and ZapA (15) may be involved in regulating or controlling the polymerization reaction. Protofilaments consist of FtsZ monomers stacked one on top of the other, so that each monomer has two surfaces binding to two different FtsZ monomers. One surface contains the nucleotide-binding loops T1 to T6. The opposite surface contains the T7 loop, needed for the hydrolysis of GTP bound to the monomer next to it in the protofilament (16, 17). Hence the active-site GTPase is split across two monomers. The same protofilament arrangement is also seen with tubulin assembling into microtubules.
Induction of the SOS response causes the disappearance of FtsZ rings (18). SulA binds FtsZ in a 1:1 ratio, and GTP is needed for binding in vitro (19). Whether SulA affects the GTPase activity of FtsZ is less certain. Higashitani et al. (19), using a maltose-binding protein (MBP)-SulA fusion protein, found no inhibition of the GTPase activity of FtsZ, whereas Mukherjee et al. (20) and Trusca et al. (21), using a MBP-SulA fusion or a protein A-SulA fusion, respectively, found SulA does inhibit the GTPase activity of FtsZ. FtsZ mutants that are refractory to SulA map to either the nucleotide-binding surface of FtsZ or the opposite surface containing the T7 loop (22). SulA is likely to bind only one, not both, of these surfaces.
Because SulA is such a potent inhibitor of bacterial cell division, although only 18 kDa, it has been of pharmaceutical interest as a potential model for antibiotic action against FtsZ. A crystal structure of SulA:FtsZ allows investigation of the feasibility of mimicking SulA's action on FtsZ to inhibit cell division. Here we present the crystal structure of SulA from the opportunistic pathogen Pseudomonas aeruginosa alone and in complex with FtsZ from the same organism.
Materials and Methods
Protein Expression and Purification. SulA (PIR:A83269) from P. aeruginosa PAO1-LAC (American Type Culture Collection 47085) was cloned into pET28 by using the NcoI and XhoI sites. The resulting protein has a (His)6 tag, and the second residue is altered from Gln to Glu. The protein was expressed in C41 (DE3) cells and purified by using Ni2+-NTA chromatography. After gel filtration (Sephacryl S-200 column in 200 mM NaCl/20 mM Tris/5 mM DTT/1 mM EDTA/1 mM NaN3, pH 7.0), the protein was concentrated to 7 mg/ml and stored at -80°C. P. aeruginosa SulAΔ 35 (35–161) was cloned, expressed, and purified as above.
FtsZ from P. aeruginosa (1–318) was cloned and expressed as SulA with the addition of a N-terminal Strep-tag. The protein was purified by using Ni2+-NTA chromatography and gel filtrated (Sephacryl S-300 column) in 100 mM Tris/5 mM DTT/1 mM EDTA/1 mM NaN3, pH 8.0, for the protein mixed with full length SulA. FtsZ used for complex assembly with SulAΔ35 was gel filtrated in the same buffer but at pH 7.5. The protein was concentrated to 10 mg/ml and stored at -80°C. The selenomethionine-substituted protein was grown in the nonmethionine auxotrophic C41 (DE3) cells used for the native protein, by a previously described method (23, 24), and purified as for the native. Electrospray MS was used to check SeMet incorporation (native P. aeruginosa FtsZ: observed, 34,918.69 Da; calculated, 35,051.21 Da. SeMet: observed, 35,481.21 Da; calculated, 35,660.85 Da, indicating the N-terminal methionine is not present).
The P. aeruginosa SulA:P. aeruginosa FtsZ complex was assembled by mixing P. aeruginosa FtsZ and P. aeruginosa SulA in a ≈1:1.2 ratio. GTP and MgCl2 were added to 1 and 2 mM, respectively, and the mixture incubated for 15 min at 37°C. The mixture was gel filtrated (Sephacryl S-200 column in 20 mM Tris/5 mM DTT/5 mM MgCl2/0.1 mM GTP/1 mM NaN3, pH 8.0), and the complex was concentrated to 10 mg/ml and stored at -80°C. The complex of SulAΔ35:FtsZ was prepared as above, but the gel filtration buffer was 200 mM NaCl/20 mM Tris/5mM DTT/5 mM MgCl2/0.1 mM GTP/1 mM NaN3, pH 7.5.
Crystallization. SulA crystals were grown in 3–5 days by using the sitting drop vapor diffusion technique at 19°C. The reservoir was 3% 1,6-hexanediol, 50 mM CsCl, 5 mM Fe(III)Cl3,1% Jeffamine M-600, pH 5.5, and 0.1 M sodium citrate, pH 5.0. Drops were composed of 1 μl of protein and 1 μl of crystallization solution. Before flash cooling, the crystals were placed in mother liquor plus 30% Jeffamine M-600, pH 5.5. Crystals belong to space group P212121 and have cell dimensions of a = 79.7 Å, b = 91.7 Å, and c = 92.3 Å. Heavy metal derivatives were made by adding orthochloromercury phenol, KAu(CN)2, or K2OsO4 solutions directly to the drop to 3 mM. After 1 h, crystals were flash cooled as for the natives.
Selenomethionine-substituted SulA:FtsZ crystals were grown by the hanging drop vapor diffusion technique at 19°C. The reservoir was 8% polyethylene glycol 6000/2.4% 1,2,3-heptanetriol/80 mM LiCl/0.1 M Mes, pH 5.6, for the crystallization solution. Drops were composed of 1 μl of complex and 1 μl of crystallization solution. The crystals, flash cooled in mother liquor plus 6% 2-methyl-2,4-pentandiol, belong to space group C2, and have cell dimensions of a = 78.0 Å, b = 54.0 Å, c = 250.7 Å, and β = 101.1°.
SulAΔ35:FtsZ crystals were grown in 3–5 days by using the sitting drop vapor diffusion technique at 19°C. The reservoir was 7.5% polyethylene glycol (PEG) 20,000/7.5% PEG monomethyl ether 550/80 mM sodium formate/0.1 M Tris, pH 7.5. Drops were composed of 0.5 μl of protein and 0.5 μl of crystallization solution. Before flash cooling, the crystals were placed in mother liquor plus 15% glycerol. They belong to space group P212121 and have cell dimensions of a = 55.6 Å, b = 75.4 Å, and c = 241.0 Å.
Data Collection and Structure Determination. All crystals were indexed and integrated by using the MOSF LM package (25) and further processed by using the CCP4 package (26). For SulA, phases were provided by using MIRAS with a native and the three heavy metal derivative datasets. Sites were found by using SOLVE (27) and refined and phased to 3.5 Å by using SHARP (28). The resulting map was 4-fold averaged at 3.5 Å. Model building was done by using MAIN (29) and refinement with the CNS package (30).
For the SulA:SeMet-FtsZ crystals, a multiple anomalous dispersion (MAD) dataset was collected at ID14-4 European Synchrotron Radiation Facility, Grenoble. An initial 3.9-Å electron density map was generated by locating 24 selenium sites in the datasets PEAK, INFL, and HREM by using SnB (31) and phased by using SHARP. The resulting density was good enough to dock known FtsZ (32) and SulA (see above) structures by hand into the density, with both the density and position of the selenium sites in the FtsZ. The asymmetric unit contains one complex molecule that is a SulA dimer and two FtsZ molecules with GTP bound, one on either side of the SulA dimer. Refinement was done by using the CNS package, although refinement to acceptable final model parameters was not possible, probably due to the low quality of the diffraction data, which was very anisotropic.
For the SulAΔ35:FtsZ crystals, a native dataset was collected to 2.1 Å at ID14-4. Molecular replacement was done in CNS by using the complete SulA:FtsZ model that had been built into the MAD density. The FtsZ in this complex is clearly GDP bound. Model parameters are summarized in Tables 1 and 2.
Table 1. Crystallographic data.
| Crystal | λ, Å | Resolution, Å | I/σI* | Rm,† % | Multiplicity‡ | Completeness, %§ | PhP¶ | |
|---|---|---|---|---|---|---|---|---|
| P. aeruginosa SulA, space group P212121, a = 79.7 Å, b = 91.7 Å, c = 92.3 Å | ||||||||
| OCMP | 0.8638 | 3.5 | 12.9 (4.4) | 0.095 (0.274) | 3.9 | 99.8 (99.8) | 1.3 | 1.4 |
| K2OsO4 | 0.8638 | 4.0 | 13.3 (4.4) | 0.103 (0.217) | 3.8 | 94.8 (94.8) | 0.7 | 0.5 |
| KAu(CN)2 | 1.4880 | 3.5 | 12.7 (3.5) | 0.076 (0.335) | 4.0 | 99.9 (100.0) | 0.5 | 0.2 |
| NATI | 0.9330 | 2.9 | 14.0 (3.0) | 0.075 (0.336) | 3.1 | 95.9 (95.9) | ||
| P. aeruginosa SulA:P. aeruginosa FtsZ, space group C2, a = 78.0 Å, b = 54.0 Å, c = 250.7 Å, β = 101.1° | ||||||||
| PEAK | 0.9793 | 3.9 | 18.2 (6.6) | 0.064 (0.194) | 10.8 (5.4) | 96.4 (96.0) | ||
| INFL | 0.9797 | 3.9 | 17.2 (5.9) | 0.065 (0.216) | 10.4 (5.2) | 96.5 (96.5) | ||
| HREM | 0.9393 | 3.9 | 18.7 (9.0) | 0.068 (0.131) | 10.6 (5.3) | 96.4 (96.4) | ||
| P. aeruginosa SulAΔ35:P. aeruginosa FtsZ, space group P212121, a = 55.6 Å, b = 75.4 Å, c = 241.0 Å | ||||||||
| NATI | 0.9393 | 2.1 | 11.4 (3.7) | 0.096 (0.334) | 3.8 | 99.7 (99.7) | ||
Signal-to-noise ratio of merged intensities, in brackets for the highest-resolution shell
Rm: ΣhΣi|I(h, i) - I(h)|/ΣhΣiI (h, i), where I(h, i) are symmetry-related intensities, and I(h) is the mean intensity of the reflection with unique index h, in parentheses for the highest-resolution bin
Multiplicity for unique reflections, anomalous multiplicity in brackets
Completeness for unique reflections; anomalous completeness is identical, because inverse beam geometry was used, in parentheses for the highest-resolution bin. Correlation coefficients of anomalous differences at different wavelengths for the MAD experiment: PEAK vs. INFL, 0.47; PEAK vs. HREM, 0.57; INFL vs. HREM, 0.31
PhP, phasing power; acentric reflections for isomorphous and anomalous phasing, respectively. Figure of merit for all acentric reflections is 0.39
Table 2. Refinement statistics.
| Refinement statistics | |
|---|---|
| P. aeruginosa SulA | |
| Model | Four noncrystallographically related SulA molecules arranged in two different dimers: A and B, C and D SulA A, B, C, and D residues 43-161 |
| Diffraction data | NATI, 2.9 Å, all data |
| R factor, Rfree* | 0.247(0.358), 0.289(0.366) |
| B average/bonded† | 35.035 Å2, 3.113 Å2 |
| Geometry bonds/angles‡ | 0.009 Å, 1.497° |
| Ramachandran§ | 84.6%/0.0% |
| Protein Data Bank (PDB) ID | 1OFT |
| P. aeruginosa SulAΔ35: P. aeruginosa FtsZ | |
| Model | Four noncrystallographically related molecules |
| Two of FtsZ (A and B) and two of SulAΔ35 (X and Y) | |
| FtsZ A residues 11-317 | |
| FtsZ B residues 11-317 | |
| SulAΔ35 X residues 43-160 | |
| SulAΔ35 Y residues 43-161 | |
| Water molecules (442) | |
| Diffraction data | NATI, 2.1 Å, all data |
| R factor, Rfree* | 0.216(0.354), 0.255(0.405) |
| B average/bonded† | 43.68 Å2, 3.359 Å2 |
| Geometry bonds/angles‡ | 0.006 Å, 1.261° |
| Ramachandran§ | 93.8%/0.3% |
| Restrained NCS | FtsZ 0.7557 Å |
| SulAΔ35 0.4870 Å | |
| PDB ID | 1OFU |
Five percent of reflections were randomly selected for determination of the free R factor, prior to any refinement. High-resolution bin in brackets
Temperature factors averaged for all atoms and rms deviation of temperature factors between bonded atoms
rms deviations from ideal geometry for bond lengths and restraint angles
Percentage of residues in the “most favored region” of the Ramachandran plot and percentage of outliers (37)
Results and Discussion
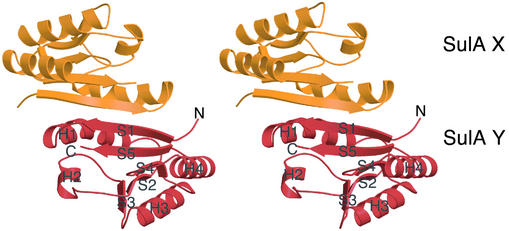
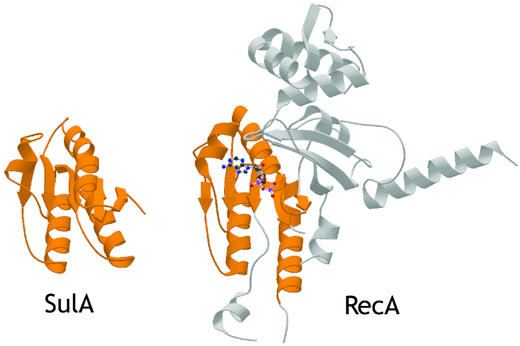
The crystal structure of SulA (Fig. 1) reveals it is a dimer with a fold unexpectedly similar to the RecA fold (33). Although the full length protein was crystallized, residues 1–42 are not visible in the electron density map. Residues 43–161 of SulA (32% sequence identity to E. coli SulA; Fig. 2) consist of a central region of parallel β-sheets surrounded by four α-helices; in the sequence order, they are S1-H1-S2-H2-S3-H3-S4-H4-S5 (Fig. 1). The dimer interface covers 22% of each monomer (1,495 Å2 of 6,580 Å2) and is hydrophobic in nature. It is composed of S1 (residues 45–52) and H1 (residues 55–69) from each monomer. The β strands are associated to form a continuous β-sheet across the dimer. The helices pair with each other from residues 64 to 68. Size exclusion chromatography of SulA confirms it is a dimer in solution (data not shown). The most similar structure to SulA is a region of RecA (residues 62–144 and 167–194), which includes the ATPase site (Fig. 3). There is an rms deviation of 2.4 Å over 112 Cα atoms of the 119 Cα atoms in the SulA structure. The sequence identity between the two regions is low at 16%. Unlike RecA, SulA is not an ATPase. SulA lacks a P-loop sequence between S1 and H1, and a tryptophan (Trp-91) side chain occupies the region where a nucleotide base would normally be. The reason for the strong structural similarity to RecA is unknown.
Fig. 1.
P. aeruginosa SulA has a fold similar to the nucleotide-binding core of RecA and forms a dimer. Stereo ribbon drawing of the SulA dimer with one monomer shown in orange and the other in red.
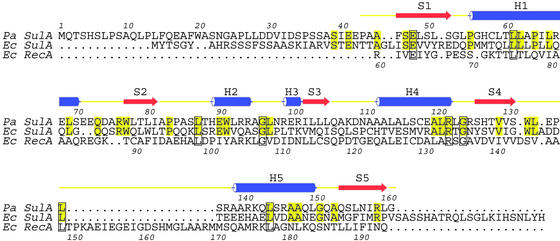
Fig. 2.
Structure-based sequence alignment [P. aeruginosa SulA homolog, Swiss-Prot Q9HZJ8, PDB ID code 1OFT, E. coli SulA, Swiss-Prot SULA_ECOLI, E. coli RecA, Swiss-Prot RECA_ECOLI, PDB ID code 2REB (33)]. Pa and Ec SulA have been aligned purely on the basis of their sequences. The overall identity between Pa and Ec SulA is 32%. The structures of Pa SulA and Ec RecA have been superimposed, and the resulting sequence alignment is shown. Only the N-terminal domain of RecA superimposes with SulA.
Fig. 3.
Structural similarity of the SulA monomer (PDB ID code 1OFT) and the N-terminal domain of E. coli RecA (PDB ID code 2REB; ref. 33). The structures have been aligned with an rms deviation of 2.4 Å over 112 Cα atoms (of 119 for SulA).
Crystallization of P. aeruginosa SulA:P. aeruginosa FtsZ was first achieved by using full length SulA and a C-terminal deletion of FtsZ (1–318). The C-terminal 76 residues of P. aeruginosa FtsZ are not predicted to have any secondary structure and were removed to enable crystallization of the complex. 90° light scattering, confirming that FtsZ still polymerized and that SulA could inhibit polymerization (data not shown). Yeast two-hybrid investigation of SulA:FtsZ interactions by using the E. coli proteins had found the equivalent region of FtsZ (residues 321–383) was not necessary for SulA:FtsZ interaction (34). Selenomethionine-substituted FtsZ was used in the crystallization of the complex to enable experimental phasing by using MAD. An electron density at 3.9 Å was produced and the known structures of FtsZ (32) and SulA (see above) placed by hand into the density. The structure contains a central dimer of SulA with one FtsZ molecule bound to each SulA. The result is an elongated complex of Z:A:A:Z. The interaction between FtsZ and SulA is via the T7 loop-containing face of FtsZ and not the nucleotide-binding face. For SulA, the interaction is via the surface opposite the dimer interface. The FtsZ is clearly bound to GTP. The N-terminal residues of SulA (1–42), which were not ordered in the previous structure, are not ordered in the complex structure either. When the complex structure did not refine to acceptable parameters, the decision was made to remove the disordered region of SulA.
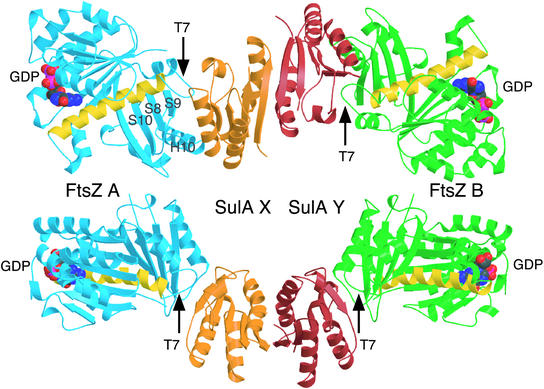
A higher-resolution structure of P. aeruginosa SulAΔ35:P. aeruginosa FtsZ at 2.1 Å was produced by using truncated SulA (SulAΔ35), enabling detailed examination of the complex (Fig. 4). This structure was solved by using the complex structure built with the electron density from the MAD experiment. It again contains a central SulA dimer with one FtsZ bound to each SulA. The FtsZ, though, is GDP bound. During model building, the extra density was noticed around Pro-43 of SulA, especially molecule X, not due to the ordering of N-terminal residues. The density appears to be a tryptophan residue from the N-terminal Strep-tag of FtsZ.
Fig. 4.
Crystal structure (2.1 Å) of the SulAΔ35:FtsZ complex (PDB ID code 1OFU). The SulA dimer is sandwiched between two FtsZ monomers via the T7 protofilament interface, leaving the GTP-binding regions exposed. Note how the FtsZ molecules are rotated exactly 180° relative to each other in the complex. The SulA dimer is in orange and red as in Fig. 1. The FtsZ molecules are shown in blue and green. The yellow helix is H7 and marks the transition from the N- to the C-terminal domain (8). GDP is shown in space-filling representation. The two views are rotated by 90° about x.
The binding of SulA to FtsZ is exclusively to the T7 loop-containing surface of FtsZ and covers it completely. Specifically, residues Pro-205 to Asn-208, which are part of the T7 loop, are covered by Arg-99 to Leu-104, which are part of the loop between H2 and S3 in SulA. Asp-270 to Leu-273 of FtsZ, which are part of the loop between S8 and H10, are covered by residues Val-120 to Gly-125 of SulA, which is H3 and part of the following loop. The region of FtsZ from Ile-299 to Ala-301, which is between S9 and S10, is occluded by SulA Gln-106 to Cys-108. Overall, the surface area of FtsZ involved in SulA binding is 12% (1,595 Å2 of 12,679 Å2) of its total and for SulA, 24% (1,595Å2 of 6,580Å2) of its surface area is involved in FtsZ binding. Comparison of the structure of FtsZ in complex with SulA with the known Methanococcus jannaschii FtsZ structure (32) indicates only minor differences between the two, even in the T7 loop region, despite the fact that they originate from two different organisms. There are no obvious changes to SulA on binding FtsZ, and any minor ones may be due to the higher resolution of the complex structure.
The structures of SulA and the SulAΔ35:FtsZ complex have some surprising features. The SulA dimer has not been detected before, possibly because of the problems of overexpressing E. coli SulA, which have been circumvented by fusions to MBP or protein A (20, 21). The N-terminal region of SulA (1–42) is flexible and absent from the structures. Sequence alignments of SulAs indicate this region is not conserved and varies in length (Fig. 2). Mutations of SulA that inhibit its interaction with FtsZ (34) are all found in the core of the molecule, possibly suggesting that they are mainly affecting the folding of SulA. The total region of SulA involved in covering FtsZ is 26 aa (residues 99–125), which are a β strand followed by an α-helix. This is probably too large a region for a peptide-mimicking drug. However, the T7 loop is bound by only five residues of SulA, and this may be sufficient for inhibition.
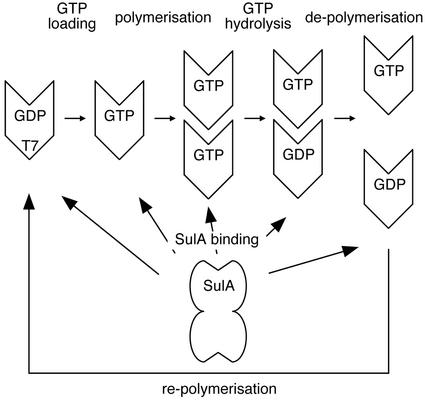
The basic mechanism of the FtsZ polymerization mechanism is known (Fig. 5; for a recent review see ref. 11). A pool of free FtsZ:GTP molecules can polymerize into protofilaments and other higher-order structures, such as sheets. Polymerization produces a complete active-site GTPase consisting of a T7 loop from one FtsZ molecule and the nucleotide-binding site from the adjoining FtsZ molecule in the protofilament. The T7 loop participates in nucleotide hydrolysis and, after polymerization, the nucleotide is hydrolyzed to GDP. Once this has occurred, the polymer can disassemble and the process is repeated if conditions are favorable.
Fig. 5.
Schematic drawing of the FtsZ polymerization/depolymerization cycle and SulA inhibition. FtsZ can polymerize only when in the GTP state. Once polymerized, FtsZ will hydrolyze GTP to GDP, because polymerization allows FtsZ to form complete active sites as they are split across two monomers. Depolymerization occurs some time after GTP hydrolysis. SulA, as shown in this study, will bind to FtsZ containing GTP and GDP. Because one of the two protofilament contacts is still available in the SulA:FtsZ complex structure, it is possible to bind the ends of protofilaments. The complex between SulA and FtsZ is very tight, and the FtsZ polymerization cycle means that each FtsZ molecule will at some point expose a free T7 interface. SulA will bind to this interface and remove it from the pool of available T7 interfaces for polymerization. Because SulA is produced in large amounts during SOS response, eventually all FtsZ will be monomeric and polymerization inhibited. Lon protease is needed to free FtsZ from SulA.
From our structure, we propose a mechanism of how SulA can act at all points in the polymerization cycle to inhibit cell division. Two independent surfaces on the FtsZ molecule are required for polymerization competence, the T7 loop surface and the nucleotide-binding surface. In theory, SulA could bind to either to block polymer formation. Our structure clearly shows SulA binds to the T7 loop-containing surface and by comparison with the M. jannaschii FtsZ (32) structure causes no major conformational changes in FtsZ. When this occurs in solution, there are two effects. First, the FtsZ:GTP “building blocks” for polymer growth are removed from the pool of available FtsZ for filament growth. This leads to disassembly of existing protofilaments via filament dynamics rather than active depolymerization (Fig. 5). Although SulA has no sequence homologues in eukaryotes, a similar mechanism is seen with the tubulin-binding protein stathmin, which sequesters tubulin dimers, although it disrupts lateral interactions between protofilaments rather than acting via the T7 loop to affect interactions within the protofilament (35). The second effect of SulA is binding to preformed FtsZ filaments. This “caps” the free T7 loop surface at one end of the protofilament; thus further growth is inhibited. Because the filament is a dynamic structure, it will disassemble as GTP is hydrolyzed within the protofilament.
For the sequestering mechanism to be effective in the cell, a high turnover of FtsZ between filaments and monomers in solution would be required. This is exactly what has recently been reported by using fluorescence recovery after photobleaching on a FtsZ-GFP fusion in vivo (36). The half-life of FtsZ in the Z ring is ≈30 sec. Using a mutant in the nucleotide-binding region of FtsZ (FtsZ84), which has ≈10% of the GTPase activity of the wild-type, it was found that the turnover rate in vivo between the Z-ring and the monomer depends on the GTPase activity. Several FtsZ mutants are refractory to SulA, and all have reduced GTPase activity varying from 50% to ≈0% (22). This finding strongly supports our model of SulA action as polymer turnover is related to GTPase activity. In these mutants, the polymers should be much less dynamic and, even when SulA sequesters the free T7-loop-containing surface, the rate of disassembly of the filament is reduced. It is currently impossible to separate a direct effect on the SulA affinity for FtsZ of these mutants from reduced turnover of FtsZ filaments and the apparent resulting reduction in “affinity” of SulA.
There has been some uncertainty as to whether SulA affects the GTPase activity of FtsZ. In our structures, one of the complexes is in the GTP state and one in the GDP state, probably due to residual nucleotide hydrolysis during the crystallization experiment. From the structure of the GTP-containing complex, it appears that hydrolysis is not necessary for interaction. If SulA drives depolymerization via the dynamic nature of FtsZ filaments, it would appear at the “bulk” level that SulA inhibits the GTPase activity of FtsZ. There would be less/no FtsZ able to polymerize and so no GTP hydrolysis through polymerization. Thus the effect on the GTPase is “indirect” by disrupting the active site formed between monomers.
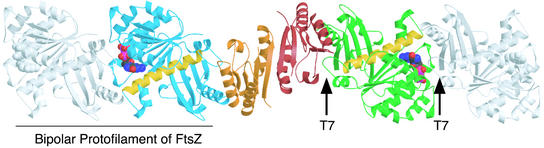
The mechanism we propose for SulA's action on FtsZ does not explain why SulA is dimeric. This feature of SulA was unexpected and has the interesting consequence that SulA can crosslink FtsZ protofilaments (Fig. 6). FtsZ binds exclusively to the other side of SulA from the dimer interface. This interface has 2-fold symmetry in one direction, which is then imposed on the FtsZ molecules. The result is that the FtsZ molecules bound to SulA are of exactly opposing polarity. It is then theoretically possible to add additional FtsZ monomers to the complex to form a bipolar protofilament arrangement (17) (Fig. 6). One possible idea is that this enables small substoichiometric amounts of SulA to rapidly halt cell division in the earliest stages of SulA synthesis during the SOS response. As further SulA is synthesized, the pool of free FtsZ is then “mopped up” by more of the newly synthesized SulA.
Fig. 6.
Model showing how SulA could crosslink the ends of FtsZ protofilaments. The complex is as in Fig. 4. FtsZ monomers, shown in gray, could theoretically bind to each and of the complex in a protofilament-like structure where the T7 loop from one monomer contacts the GTP-binding site from the previous monomer (17).
Acknowledgments
We thank Sew Peak-Chew (Medical Research Council–Laboratory of Molecular Biology, Cambridge) for performing MS and the synchrotron beamline scientists at 9.6 and 14.1 at the Synchrotron Radiation Source, Daresbury, and at 14-1 and 14-4 at the European Synchrotron Radiation Facility, Grenoble.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MAD, multiple anomalous dispersion; PDB, Protein Data Bank.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1OFT and 1OFU).
References
- 1.Fernandez de Henestrosa, A. R., Ogi, T., Aoyagi, S., Chafin, D., Hayes, J. J., Ohmori, H. & Woodgate, R. (2000) Mol. Microbiol. 35, 1560-1572. [DOI] [PubMed] [Google Scholar]
- 2.Walker, G. C. (1995) Trends Biochem. Sci. 20, 416-420. [DOI] [PubMed] [Google Scholar]
- 3.Husiman, O. & Dari, R. (1981) Nature 290, 797-799. [DOI] [PubMed] [Google Scholar]
- 4.Husiman, O., Dari, R. & Gottesman, S. (1984) Proc. Natl. Acad. Sci. USA 81, 4490-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizusawa, S. & Gottesman, S. (1983) Proc. Natl. Acad. Sci. USA 80, 358-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bi, E. & Lutkenhaus, J. (1990) J. Bacteriol. 172, 5602-5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erickson, H. P. (1995) Cell 80, 367-370. [DOI] [PubMed] [Google Scholar]
- 8.Nogales, E., Downing, K. H., Amos, L. A. & Löwe, J. (1998) Nat. Struct. Biol. 5, 451-458. [DOI] [PubMed] [Google Scholar]
- 9.de Boer, P., Crossley, R. & Rothfield, L. (1992) Nature 359, 254-256. [DOI] [PubMed] [Google Scholar]
- 10.Bi, E. F. & Lutkenhaus, J. (1991) Nature 354, 161-164. [DOI] [PubMed] [Google Scholar]
- 11.Addinall, S. G. & Holland, B. (2002) J. Mol. Biol. 318, 219-236. [DOI] [PubMed] [Google Scholar]
- 12.Bramhill, D. (1997) Annu. Rev. Cell Dev. Biol. 13, 395-424. [DOI] [PubMed] [Google Scholar]
- 13.Erickson, H. P., Taylor, D. W., Taylor, K. A. & Bramhill, D. (1996) Proc. Natl. Acad. Sci. USA 93, 519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hale, C. A., Rhee, A. C. & de Boer, P. A. J. (2000) J. Bacteriol. 182, 5153-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gueiros, F. J. & Losick, R. (2002) Genes Dev. 16, 2544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erickson, H. P. (1998) Trends Cell Biol. 8, 133-137. [DOI] [PubMed] [Google Scholar]
- 17.Löwe, J. & Amos, L. A. (1999) EMBO J. 18, 2364-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bi, E. & Lutkenhaus, J. (1993) J. Bacteriol. 175, 1118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higashitani, A., Higashitani, N. & Horiuchi, K. (1995) Biochem. Biophys. Res. Commun. 209, 198-204. [DOI] [PubMed] [Google Scholar]
- 20.Mukherjee, A., Cao, C. N. & Lutkenhaus, J. (1998) Proc. Natl. Acad. Sci. USA 95, 2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trusca, D., Scott, S., Thompson, C. & Bramhill, D. (1998) J. Bacteriol. 180, 3946-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai, K., Mukherjee, A., Xu, Y. & Lutkenhaus, J. (1994) J. Bacteriol. 176, 130-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van den Ent, F., Lockhart, A., Kendrick-Jones, J. & Lowe, J. (1999) Struct. Folding Des. 7, 1181-1187. [DOI] [PubMed] [Google Scholar]
- 24.Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. (1993) J. Mol. Biol. 229, 105-124. [DOI] [PubMed] [Google Scholar]
- 25.Leslie, A. G. W. (1991) Recent Changes to the MOSFLM Package for Processing Film and Image Plate Data (Science and Engineering Research Council Laboratory, Daresbury, Warrington, U.K.).
- 26.Collaborative Computational Project, no. 4 (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 27.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de la Fortelle, E. & Bricogne, G. (1997) Methods Enzymol. 276, 472-494. [DOI] [PubMed] [Google Scholar]
- 29.Turk, D. (1992) Ph.D. thesis (Technische Universität München, Munich).
- 30.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 31.Weeks, C. M. & Miller, R. (1999) J. Appl. Crystallogr. 32, 120-124. [Google Scholar]
- 32.Löwe, J. & Amos, L. A. (1998) Nature 391, 203-206. [DOI] [PubMed] [Google Scholar]
- 33.Story, R. M. & Steiz, T. A. (1992) Nature 355, 374-376. [DOI] [PubMed] [Google Scholar]
- 34.Huang, J., Cao, C. & Lutkenhaus, J. (1996) J. Bacteriol. 178, 5080-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gigant, B., Curmi, P. A., Martin-Barbey, C., Charbaut, E., Lachkar, S., Lebeau, L., Siavoshian, S., Sobel, A. & Knossow, M. (2000) Cell 102, 809-816. [DOI] [PubMed] [Google Scholar]
- 36.Stricker, J., Maddox, P., Salmon, E. D. & Erickson, H. P. (2002) Proc. Natl. Acad. Sci. USA 99, 3171-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laskowski, R. A., Macarthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283-291. [Google Scholar]