Abstract
RRS1-R confers broad-spectrum resistance to several strains of the causal agent of bacterial wilt, Ralstonia solanacearum. Although genetically defined as recessive, this R gene encodes a protein whose structure combines the TIR-NBS-LRR domains found in several R proteins and a WRKY motif characteristic of some plant transcriptional factors and behaves as a dominant gene in transgenic susceptible plants. Here we show that PopP2, a R. solanacearum type III effector, which belongs to the YopJ/AvrRxv protein family, is the avirulence protein recognized by RRS1-R. Furthermore, an interaction between PopP2 and both RRS1-R and RRS1-S, present in the resistant Nd-1 and susceptible Col-5 Arabidopsis thaliana ecotypes, respectively, was detected by using the yeast split-ubiquitin two-hybrid system. This interaction, which required the full-length R protein, was not observed between the RRS1 proteins and PopP1, another member of the YopJ/AvrRxv family present in strain GMI1000 and that confers avirulence in Petunia. We further demonstrate that both the Avr protein and the RRS1 proteins colocalize in the nucleus and that the nuclear localization of the RRS1 proteins are dependent on the presence of PopP2.
Plants rely on an innate immune response for their survival after pathogen attack. Specific recognitions between pathogen Avr and plant R proteins are crucial for the onset of the resistance response and determine the issue of many plant–pathogen interactions by triggering plant defense. Disease results from the inactivation or absence of one or both partners (1). It has been postulated that R gene products are receptors for pathogen-encoded Avr components (2). Despite the cloning of numerous R and Avr genes, only two plant R proteins, Pto and Pi-ta, were shown to interact physically with their pathogen Avr counterparts, Avr-Pto and Avr-Pita, respectively, by using the yeast two-hybrid system (3–5). The hypothesis that R proteins are part of protein complexes was recently substantiated by the identification of multiprotein complexes including R and Avr proteins (6–9). Additionally, whereas Avr bacterial proteins expressed in plants carrying the cognate R protein generally induce a cell death program (10), termed the hypersensitive response, closely linked to resistance, they can also cause disease-like symptoms when expressed in plants lacking the appropriate R protein. Bacterial pathogenicity effectors such as Avr proteins are injected into the host cell via a type III secretion system (TTSS) (11). According to the guard model (9, 12), such effectors can associate and induce modifications of plant targets functioning as negative regulators of basal defense responses leading to disease development in plants lacking the corresponding R protein. In a resistant host, the plant target that interacts with both R and Avr proteins is guarded by the R protein, preventing its manipulation by pathogen effectors. The recent characterization of RIN4, a negative regulator of plant defense, strengthens this model (13).
Most resistance proteins belong to the nucleotide-binding–leucine-rich repeat (NB-LRR) class of R proteins and contain a LRR domain that mediates protein–protein, peptide–ligand binding, and protein–carbohydrate interactions (14, 15) and acts as a specificity determinant for pathogen recognition (16). LRR may also contribute to signaling (17). A conserved NB domain critical for ATP or GTP binding (18) is also found in NB-LRR R proteins and shares similarities with NBS regions of effectors of programmed cell death such as Apaf-1 and Ced4 (19, 20). NB-LRR proteins fall into two subclasses based on their N-terminal motifs. One group possesses an N-terminal coiled-coil domain, whereas the second subclass shares similarities to the cytoplasmic Toll IL-1 receptor (TIR) domains of human and Drosophila Toll-like receptors (9). The conserved TIR, NBS, and LRR domains play crucial roles in pathogen perception and in subsequent downstream signaling.
The Arabidopsis thaliana RRS1-R resistance gene confers broad-spectrum resistance to several strains of Ralstonia solanacearum, the causal agent of bacterial wilt (21). It encodes the first member of the TIR-NBS-LRR subclass of R proteins, which possesses a C-terminal WRKY motif characteristic of some plant transcriptional factors (22, 23). Although genetically defined as recessive, this R gene behaves as a dominant gene in transgenic plants. Several lines of evidence are in favor of RRS1-R as a dominant R gene because: (i) it has sequence similarities with other R genes; (ii) its requirement for salicylic acid and the signaling component NDR1, and (iii) the observation that in transgenic resistant Nd-1 plants carrying RRS1-S the susceptible allele failed to induce wilt disease.
In this study, we report on the identification of PopP2 as the corresponding Avr protein to RRS1-R and demonstrate that the two proteins interact in yeast. We also provide evidence of the nuclear colocalization of PopP2 and of the RRS1 proteins. Detection of the RRS1 proteins in this organelle is observed only in the presence of the avirulence protein.
Materials and Methods
Plant Materials and Bacterial Strains. Arabidopsis accessions used in this study were Col-5 (a glabrous derivative of Col-0) and Nd-1. Disease resistance phenotypes were determined by root inoculation of 4-wk-old plants with R. solanacearum strains as reported (21). Bacterial internal growth curves were measured as described (21).
Split-Ubiquitin Analysis. All constructs were expressed in the Saccharomyces cerevisiae strain JD53. The single-copy Cub-URA3 fusion vector (24) was used as a Gateway destination vector (Invitrogen). The RRS1-R, RRS1-S, popP1, and popP2 coding sequences were amplified by PCR and recombined into the bait vector by using the Gateway system. The following Gateway primers were used: for RRS1-R, (GWF) TG ATG ACC AAT TGT GAA AAG GAT GAG GA (forward primer) and (GWR) GAA AGT AAA AAT TAT AAT CAT CGA AGA ATG TTG A (reverse primer) led to the amplification of a 4,204-bp fragment; for RRS1-S, (GWF) TG ATG ACC AAT TGT GAA AAG GAT GAG GA (forward primer) and (GWR) CGC AGA TGG AGG AGG AAG TGG AAC GAG T (reverse primer) amplified a 3,934-bp fragment; for the TIR-NBS-coding sequence (TIR-NBS domains of RRS1-S and RRS1-R), (GWF) TG ATG ACC AAT TGT GAA AAG GAT GAG GA (forward primer) and (GWR) CCC CAG GAT CCT GTA CTG TTT CTC CAT (reverse primer) led to the amplification of a 1,450-bp fragment; for the TIR-NBS-LRR coding sequence (TIR-NBS-LRR domains of RRS1-S and RRS1-R), (GWF) TA ATG GTC GAC ATG ACC AAT TGT GA (forward primer) and (GWR) CAC ATA GCA CTA GTT TTG TCT TTG GAT CC (reverse primer) led to the amplification of 3,304- and 3,310-bp fragments, respectively; for popP1 and popP2, (GWF) TA ATG AAA AGA CTA TTC AGA GCA TTG GGC GT (forward primer) and (GWR) CCG ACT CCA GGG CAT GTC GAA TTT TTC (reverse primer), and (GWF) TA ATG AAG GTC AGT AGC GCA AAC GCA GGC G T(forward primer) and (GWR) CGT TGG TAT CCA ATA GGG AAT CCT GCA GCA GT (reverse primer) led to the amplification of a 1,165-bp and a 1,525-bp fragment, respectively. All PCRs were done by using the Pfx polymerase (Invitrogen), and DNA fragments obtained were recombined into the pDONR201 vector by using the Gateway system.
Yeast Transformation. Standard procedures for yeast growth and yeast transformation were used (25). The 5-fluoroorotic acid (5-FOA) selective plates contained minimal medium containing yeast nitrogen base without amino acids (Difco) and glucose, supplemented with lysine, leucine, uracil, and 1 mg/ml 5-FOA.
Recombinant DNA Methods. Molecular biology techniques were performed by using standard protocols unless otherwise noted (26, 27).
Construction of a popP2 Mutant Strain. A HindIII–XbaI 0.74-kb fragment in the 5′-internal part of the popP2 gene was amplified with the following primers: 5′-AAG CTT AGC TAC ATT GCT TGC TTG GCT C-3′ and 5′-TCT AGA ATA TCC ATA TGC AGG GGC-3′. A KpnI–EcoRI 0.9-kb fragment encompassing the 3′-terminal end of the popP2 gene and its downstream sequences was PCR-cloned with the following primers: 5′-TGG TAC CAC ACG ACT GAT CGT GCT T-3′ and 5′-TGA ATT CTG GTC AAG AAG TCC TTC C-3′. These 0.74- and 0.9-kb DNA fragments were cloned upstream and downstream, respectively, of a promotorless lacZ gene followed by a gentamycin-resistance cassette in the plasmid vector pCZ367 (S. Cunnac and S.G., unpublished data). The resulting plasmid was linearized and used to transform R. solanacearum strain GMI1000 to select a marker-exchange event leading to the deletion of the 3′-terminal half of the popP2 gene. The structure of the disrupted popP2 locus in the resulting strain GRS100 was checked by both PCR amplification and Southern hybridization.
Complementation of Virulent Strains by a Functional popP2 Gene. The full-length popP2 gene and its 5′-promoter region were amplified from genomic DNA by using the primers 5′-AAG CTT GAC TAC TGC GCG AAA TTG GCG-3′ and 5′-AGA TCT ATC GCC GAC CGA CCA GCG TG-3′ and then cloned as a 1.87-kb HindIII–BglII fragment in the pLAFR6 vector (B. Staskawicz, University of California, Berkeley) digested by HindIII and BamHI. This pLAFR6::popP2 construct was introduced into the popP2 mutant strain GRS100 and in the virulent strain Rd15 by electroporation as described (28).
Generation of the GFP and Red Fluorescent Protein (RFP) Fusion Proteins. For expression in plant cells, the popP2 coding region was PCR-amplified from genomic DNA by using the following primers: 5′-TCT AGA TGA AGG TCA GTA GCG CAA AC-3′ and 5′-AGA TCT GGT TGG TAT CCA ATA GGG AA-3′. The resulting 1.47-kb fragment was cloned as a XbaI–BglII insert downstream from the cauliflower mosaic virus 35S promoter in the plasmid pGR2935 (A. Robert-Seilaniantz and J. Cullimore, personal communication), a derivative of the binary vector pGreen (29). The popP295-488 coding sequence was amplified from genomic DNA by using the following primers: (GWF) TA ATG GGT GTG GAT CAT CCT TTG CCG GGG CGC ACG T (forward primer) and (GWR) CGT TGG TAT CCA ATA GGG AAT CCT GCA GCA GT (reverse primer) and led to the amplification of a 1,246-bp fragment. The N-terminal fusions with RFP were generated by using the Gateway-compatible pGR0029RFP2 plasmid. This vector was generated by the insertion of the HindIII fragment of the RedGate plasmid (www.evry.inra.fr/public/projects/ppr/redgate.html) into the unique HindIII site of pGR0029 (29). The N-terminal fusions with GFP were generated by using the Gateway-compatible pAM-PAT-GFP vector (a gift from F. Turck, Max-Planck-Institut, Cologne, Germany).
Transient Transformation of Arabidopsis Protoplasts. Protoplasts were prepared from an Arabidopsis Col-0 cell suspension culture (30). Then 50 ml of a 1-wk-old culture (generally at 80 g/liter) was digested with 20 μl of enzyme solution [2% cellulase Onozuka R-10/0.5% Macerozyme R-10 (Serva)]. All of the subsequent steps were performed as described (31). After a 36-h incubation, transformed protoplasts were observed by using a confocal microscope (Leica Microsystems, Heidelberg). The laser settings were the following: 488 nm at 37% of maximal power and 543 nm at 100% of maximal power. The photomultiplicator (PMT)1 was set by using a 500- to 530-nm window to collect the GFP fluorescence whereas the PMT2 was set by using a 620- to 700-nm window to detect the RFP fluorescence. Because of the nature of the excitation peak, the RFP could be excited by the shorter-wave laser. We verified that the PMT1 signal corresponded only to the GFP emission and the PMT2 signal only to the RFP emission. For each construct, at least 10 different transformed Arabidopsis protoplasts were observed.
Results
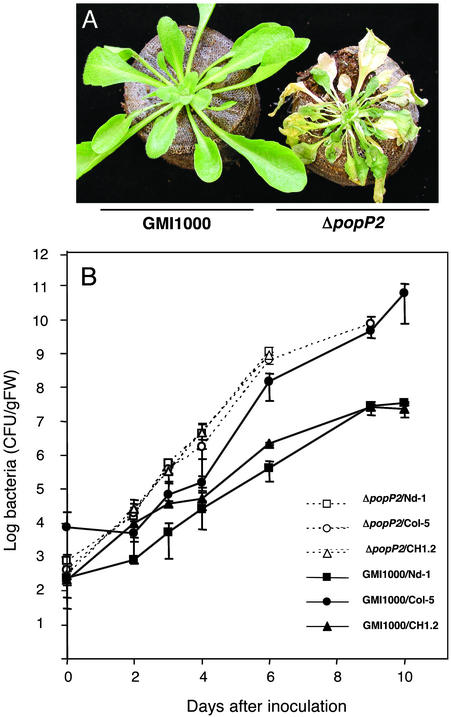
R. solanacearum popP2 Encodes the Avirulence Determinant Matching the Arabidopsis RRS1-R Gene. The recent completion of the R. solanacearum genome sequence allowed the identification of >50 genes encoding candidate TTSS-dependent effectors, including several proteins homologous to Avr proteins described in other bacterial plant pathogens (32). A functional genomic approach aimed at establishing the complete repertoire of R. solanacearum TTSS-effectors was initiated with the systematic disruption analysis of these candidate genes (S. Cunnac and S.G., unpublished data). To identify the Avr determinant recognized by RRS1-R, a set of disruption mutants generated in strain GMI1000 were screened for their pathogenicity on Arabidopsis Nd-1 (resistant) plants. Among 50 candidate genes tested, a single strain disrupted in the popP2 gene caused the wilting of Nd-1 plants up to 5 days after inoculation. As expected, no wilting was observed after inoculation of the GMI1000 parental strain (Fig. 1A). Bacterial concentrations were determined in Nd-1 plants challenged with the ΔpopP2 strain and were found to be similar to those detected in susceptible Col-5 plants inoculated with the GMI1000 strain (Fig. 1B). The introduction of a functional popP2 gene cloned on the pLAFR6 plasmid in the ΔpopP2 mutant strain reversed the virulent phenotype observed on Nd-1 plants to avirulent (Fig. 1B). A similar complementation assay using the pLAFR6::popP2 plasmid was also performed with the R. solanacearum strain Rd15, a natural isolate virulent both on Col-5 and Nd-1 ecotypes (21): an incompatible interaction was observed on resistant plants (data not shown). Finally, the observation that the ΔpopP2 mutant strain triggered wilting in the CH1–2 transgenic Col-5 plants expressing the RRS1-R gene (22) further demonstrated that the popP2/RRS1-R genes behave as in a typical gene-for-gene relationship (Fig. 1B).
Fig. 1.
popP2 is the avr gene corresponding to the RRS1-R R gene. (A) Phenotypical responses of Nd-1 plants to the GMI1000 (avirulent) and ΔpopP2 (virulent) strains 5 days after root inoculation. (B) Internal bacterial growth curves of R. solanacearum strains GMI1000 and ΔpopP2 in Arabidopsis Nd-1 (▪ and □, respectively), Col-5 (• and ○, respectively), and CH1–2 Col-5 transgenic plants expressing the RRS1-R gene (▴ and ▵, respectively). Root inoculations were performed as described (21). CFU/gFW, colony-forming units per gram of fresh weight.
The popP2 gene encodes a 52.8-kDa protein (National Center for Biotechnology Information accession no. CAD14570) that is a member of the YopJ/AvrRxv family of effector proteins translocated through the TTSS of both mammalian and plant pathogens (33, 34). These pathogenicity effectors have been proposed to act as cysteine proteases because they all harbor three conserved amino acids at positions that are characteristic of the active site of these enzymes (34); these essential residues are also conserved in PopP2, except for a conservative change of E to D at position 279. The analysis of the relationship between PopP2 and other members of this family revealed that PopP2 has no close homolog in other bacterial species, the most closely related protein (42% similarity) being the Xanthomonas campestris TTSS-effector XopJ (35). PopP2 is also evolutionarily distinct from the popP1 gene product, another member of the YopJ/AvrRxv family found in strain GMI1000, which acts as an avirulence determinant toward certain Petunia lines (28).
A popP2-lacZ transcriptional fusion was generated to monitor the expression of the gene. The popP2 gene displayed an expression pattern similar to hrp genes in culture (i.e., induction in minimal vs. complete medium). The expression of the popP2-lacZ fusion was dependent upon the presence of a functional hrpB gene, which encodes the key regulator controlling expression of hrp genes and several TTSS substrates (data not shown). The coregulation at the transcriptional level of popP2 with hrp genes thus further suggested that popP2 encodes a likely substrate of the TTSS translocation machinery.
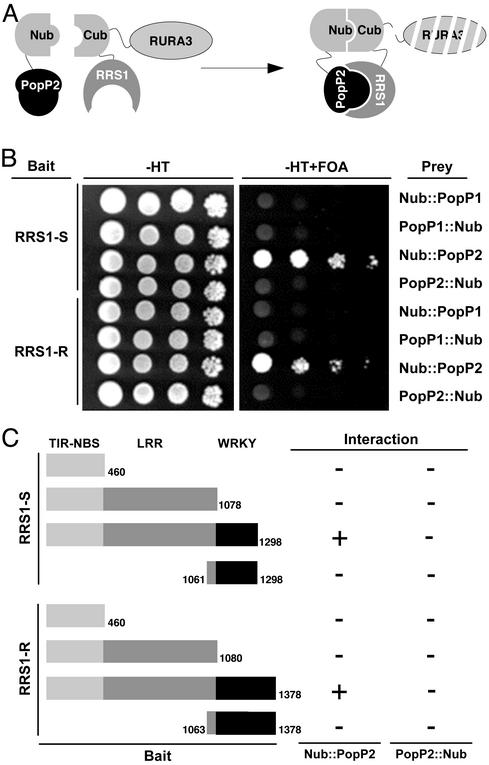
RRS1-R Interacts Specifically with PopP2 in a Split-Ubiquitin Assay. So far, a direct interaction between an NBS-LRR R protein and the corresponding Avr protein was reported in only one case (5). To check whether RRS1-R and RRS1-S proteins were able to bind PopP2, the yeast split-ubiquitin two-hybrid system was used. It is based on the fusion of the prey and the bait to the N- and C-terminal halves of ubiquitin (Nub and Cub, respectively), which are then able to form a native-like ubiquitin upon interaction (36). Ubiquitin-specific proteases recognize the reconstituted ubiquitin and cleave off a reporter protein, URA3, linked to the C terminus of Cub and whose degradation results in uracil auxotrophy and 5-FOA resistance (Fig. 2A).
Fig. 2.
Interaction of the RRS1-R and RRS1-S proteins with PopP2 in the split-ubiquitin system. (A) Schematic representation of the split-ubiquitin two-hybrid system in yeast. (B) Dilution series of yeast JD53 cells expressing both bait fusions (RRS1-R::Cub-URA3 or RRS1-S::-Cub-URA3) and prey fusions (PopP1 or PopP2 fused to the N or C terminus of Nub) were grown on yeast synthetic medium minus histidine and tryptophan (-HT) or on a minimal medium containing uracil and 5-FOA as indicated at the top. (C) Schematic diagram of the different combinations of the RRS1-S and RRS1-R domains used as baits. The size of the corresponding proteins is indicated on the left side of each construct. Results of the various interactions obtained in the yeast split-ubiquitin system between Nub fused to the N or C terminus of PopP2 are summarized on the right (+, interaction; -, no interaction).
The RRS1-S and RRS1-R full-length cDNAs were fused to the Cub domain coding sequence followed by the reporter gene URA3 (RRS1-R/S::Cub-URA3) for use as baits. The popP2 coding sequence was fused to either the C or the N terminus of Nub (Nub::PopP2 and PopP2::Nub) for use as preys (Fig. 2B). Expression of either the RRS1-R/S::Cub-URA3 construct or the Nub::PopP2 and PopP2::Nub constructs alone did not lead to expression of the uracil auxotrophy and 5-FOA resistance reporter.
Coexpression of RRS1-R::Cub-URA3 and Nub::PopP2 conferred a resistance to 5-FOA phenotype, indicating an interaction between the RRS1-R and PopP2 proteins. Interestingly, a similar result was obtained by using RRS1-S::Cub-URA3 as a bait. The RRS1–PopP2 interaction was also tested in a reciprocal combination with PopP2 fused to Cub-URA3 for use as a bait and RRS1-S and RRS1-R fused to the C and the N terminus of Nub. In this situation, no interaction between PopP2::Cub-URA3 and the RRS1 prey fusions was detected (Fig. 2B). The use of PopP2::Nub as a prey did not confer any 5-FOA-resistance phenotype, indicating that the conformation of this fusion protein might not be suitable for its interaction with the RRS1-R/S::Cub-URA3 constructs.
To identify domains of the RRS1 proteins involved in the interaction with PopP2, various constructs using the TIR-NBS, TIR-NBS-LRR, and WRKY domains of the RRS1 proteins were generated and used as baits (Fig. 2C). No interaction was detected when these constructs were coexpressed with either Nub::PopP2 or Pop2::Nub, suggesting that a proper protein folding of the full-length RRS1 proteins is required for the binding to PopP2.
Another member of the YopJ/AvrRxv protein family identified in R. solanacearum, PopP1, conferring avirulence on Petunia, was also used as a prey. Cells coexpressing the RRS1-R::Cub-UR A3 (or RRS1-S::Cub-UR A3) and Nub::PoP1/PopP1::Nub were phenotypically uracil prototrophic and 5-FOA sensitive. This indicated that no interaction between PopP1 and RRS1 proteins could be detected with this assay (Fig. 2B). The stable expression of the various PopP1 and PopP2 prey proteins in yeast cells was confirmed by Western blot analysis (data not shown).
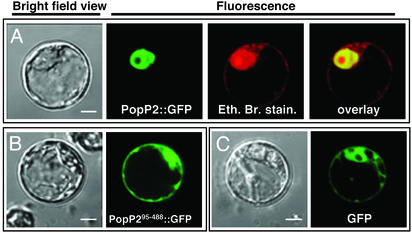
RRS1 and PopP2 Proteins Colocalize to the Plant Nucleus. The presence of a putative bipartite nuclear localization signal (RRRRX11RRQRQ at positions 34–53) in the N-terminal domain of PopP2 prompted us to check whether this effector protein was targeted to the plant nucleus. Results presented in Fig. 3A demonstrate that the PopP2::GFP (or RFP) fusion is localized in the nucleus in Arabidopsis protoplasts after transient expression. To establish that the N-terminal region of PopP2 carries a functional nuclear localization signal (NLS), a shorter protein, PopP295-488, which lacks the first 94 N-terminal amino acids, was generated. The PopP295-488::GFP fusion protein was found to be located both in the nucleus and in the cytoplasm, exhibiting an expression pattern similar to that of the control GFP fusion protein (Fig. 3 B and C). Taken together, these data establish that the PopP2 effector is specifically targeted to the plant nucleus.
Fig. 3.
PopP2 is targeted to the nucleus of Arabidopsis protoplasts. Shown are confocal laser scanning micrographs of Arabidopsis protoplasts transfected with PopP2::GFP (A), PopP295-488::GFP (B), or GFP::GFP (C). For each transfection, bright-field views of the protoplasts are presented. The GFP and the ethidium bromide signals are shown as green and red, respectively. The overlay signal of GFP and ethidium bromide appears as yellow. (The white scale bar in the bright-field view represents 8 μm.)
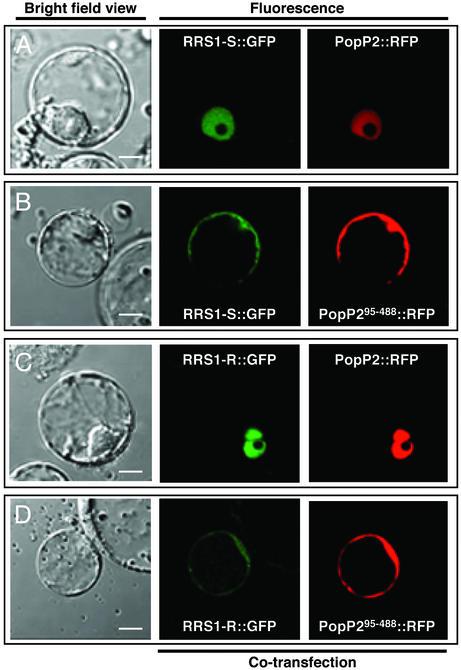
The RRS1 proteins are atypical R proteins possessing a putative transcriptional activity attributable to the WRKY domain. To determine the cellular localization of these proteins within plant cells, fusion proteins, between GFP (or RFP) and the full-length RRS1 proteins, were generated and used in transient assays in Arabidopsis protoplasts. No signal could be detected. Cotransfection experiments using PopP2::GFP and RRS1::RFP fusions (or PopP2::RFP and RRS1::GFP fusions) were then performed. As expected, the PopP2::GFP (or RFP) fusions were located in the nucleus but, in the presence of PopP2, both RRS1-R and RRS1-S fusion proteins were also detectable in this cellular compartment (Fig. 4 A and C). We then checked whether the NLS present in the avirulence protein was required for the nuclear localization of the RRS1 proteins. Similar cotransfection experiments using PopP295-488::GFP and RRS1::RFP fusions (or PopP295-488::RFP and RRS1::GFP fusions) showed that the locations of both Avr and RRS1 fusion proteins were similar to that of the GFP alone (Fig. 4 B and D). These results indicated that a full-length popP2 gene is required for the visualization of both RRS1-R and RRS1-S proteins in the nucleus.
Fig. 4.
The nuclear localization of the RRS1 proteins is dependent on the presence of PopP2. Shown are confocal laser scanning micrographs of Arabidopsis protoplasts cotransfected with RRS1-S::GFP and PopP2::RFP (A), RRS1-S::GFP and PopP295-488::RFP (B), RRS1-R::GFP and PopP2::RFP (C), or RRS1-R::GFP and PopP295-488::RFP (D). Bright-field views of the protoplasts are presented in Left. The GFP and RFP signals are shown as green and red, respectively. (The white scale bar in the bright-field view represents 8 μm.)
Discussion
We could demonstrate that PopP2, a R. solanacearum type III effector, which belongs to the YopJ/AvrRxv family (33, 34), is an avirulence protein that interacts in the yeast two-hybrid system with RRS1-R, an atypical TIR-NBS-LRR R protein conferring resistance to several strains of the soil-borne pathogen. That PopP2 is the avirulence protein corresponding to RRS1-R was demonstrated by several means: Disruption of the popP2 gene rendered strain GMI1000, normally avirulent on Nd-1, virulent on this ecotype. The strain containing an inactivated popP2 version regained its avirulence function on Nd-1 when complemented with popP2. Strain Rd-15, naturally virulent on Nd-1, also became avirulent after introduction of the popP2 gene. The avirulence protein does not promote pathogen virulence, because the severity of wilt disease symptoms is comparable after inoculation of susceptible Col-5 plants with bacterial strains carrying or lacking the popP2 gene.
TTSS-effectors from the YopJ/AvrRxv family are widespread among several animal and plant pathogenic bacteria, as well as in some plant symbionts (33). These proteins have been proposed to act as cysteine proteases on the basis of structural features (34). In mammals, the Yersinia YopJ effector inhibits the host immune response and induces apoptosis by blocking different signaling pathways, including the NF-κB pathway in the infected cell. The substrates for YopJ are conserved ubiquitin-like molecules (34) but the substrates for plant pathogens YopJ/AvrRxv members are not yet known. Search and characterization of PopP2-interacting plant partners might further illustrate mechanistic conservations between animal and plant responses to bacterial pathogens.
The popP2 gene has several interesting features: First, its G+C content of 59.8% is significantly lower than the mean of the R. solanacearum genome (67%), and it is surrounded by bacteriophage sequences, which is suggestive of a horizontal acquisition. Second, it encodes one of the largest members of the YopJ/AvrRxv family (488 aa). Experimental evidence showing that PopP2 possesses a functional NLS and is targeted to the host cell nucleus has never been reported to date for any member of the YopJ/AvrRxv family, although some bacterial type III effectors are known to be targeted to this organelle (37, 38). Because NLS motifs are undetectable in various YopJ/AvrRxv family members, it is tempting to speculate that the members of this family may be targeted to different cellular compartments, where they function as cysteine proteases for potentially diverse host substrates. This speculation is also supported by the low level of sequence identity between several members of this family outside of the core central domain containing the conserved amino acid triad predicted to be essential for enzymatic activity (34).
The interaction detected by the yeast two-hybrid system between RRS1-R and PopP2 required the full-length R protein. Indeed, all of the domains of this modular protein failed by themselves to bind the avirulence protein. In contrast, the leucine-rich domain of Pi-ta, the only other NBS-LRR protein shown so far to interact with its corresponding avirulence protein, Avr-Pita, was sufficient for binding Avr-Pita, whereas the whole Pi-Ta protein interacts only poorly with the avirulence protein in the yeast two-hybrid system (5). RRS1-R belongs to another family of R proteins, the TIR-NBS-LRR subclass, although the WRKY domain is a unique feature of this protein. The TIR domain absent in Pi-ta and directly implicated as a specificity determinant of the flax rust resistance genes L6 and L7 (39) may play a similar role in the recognition of the pathogen ligand PopP2. Based on the yeast experiments, this domain was not by itself capable of binding RRS1-R. Thus, a particular folding of the full-length R protein, allowing exposure of specific LRR and/or TIR residues, may be required for the R/Avr complex formation.
No interaction was detected between the resistance protein and PopP1, another member of the YopJ/AvrRxv family carried by strain GMI1000. The specificity of interaction between the R/Avr proteins in yeast is also supported by the absence of PopP2 binding to the truncated forms of the RRS1 proteins. Taken together, these results suggest a direct physical interaction of the R/Avr proteins, but the possibility that a highly conserved eukaryotic protein in yeast could mediate the binding of the RRS1/PopP2 proteins cannot be completely excluded.
Of particular interest was the interaction in the yeast two-hybrid system between RRS1-S, a protein highly similar to RRS1-R and present in the susceptible ecotype, and PopP2. Nonproductive R/Avr complex formation has already been observed by coimmunoprecipitation experiments between AvrB, the Pseudomonas syringae avirulence protein “recognized” by RPM1, and RPS2, an R gene whose corresponding avirulence gene is avrRpt2 (6). These observations suggest that a direct or indirect R/Avr interaction is not sufficient per se to confer gene-for-gene specificity: the formation of active and specific complexes probably requires other component(s).
The most striking feature of the RRS1 proteins is the presence of a putative NLS and of the strictly conserved WRKY domain, both undetected in any other R protein identified so far (22). These original findings have hinted toward the nucleus as a possible new “battlefield of plant defense” as recently commentated by Lahaye (40). Our present results strengthen this suggestion. As shown above, the PopP2::GFP (or RFP) fusion proteins are located in the nucleus, and this localization requires the presence of a functional NLS in the avirulence protein. In contrast, the RRS1::GFP (or RFP) proteins expressed alone were not detectable by using a similar approach. The observation that full-length RRS1 proteins cannot be visualized may indicate that the conformation of the RRS1::GFP (or RFP) fusions limit the fluorescent emission, preventing their detection. Alternatively, the fusion protein may be unstable and degraded rapidly. Our results show that the fluorescence of the RRS1::GFP proteins could be detected in the nucleus or in the cytoplasm only upon expression of PopP2 or PopP295-488, respectively. PopP2 and PopP295-488 may either stabilize these proteins or provoke conformational changes leading to the detection of the GFP. These possibilities support a direct interaction in vivo between PopP2 and the RRS1 proteins and also imply a direct interaction between PopP295-488 and the full-length RRS1 proteins (Fig. 4 B and D), which was originally detected by using the split-ubiquitin system in yeast (data not shown). Although the truncated avirulence protein is still capable of binding to the RRS1 proteins (but not to the various domains of these proteins), it does not promote their nuclear localization. In a simple model, RRS1 and PopP2 proteins colocalize, as do most matching R/Avr partners, and their interaction that occurs in the cytoplasm is required for the translocation of the RRS1 proteins into the nucleus. Binding of the Avr protein by means of the LRR and/or the TIR domain(s) may then activate the WRKY domain, leading to its interaction with its cognate cis-elements, termed W-boxes, which are present in many pathogen-responsive promoters (41, 23).
Future experiments aimed at the understanding of the biological function of the catalytic triad of PopP2 and of the role of the putative NLS motifs present in the RRS1 proteins should also provide further information on this initial R/Avr recognition step.
To explain the apparent genetic recessiveness of RRS1-R (22), we proposed that RRS1-R and RRS1-S might compete for bacterial or plant components essential for pathogen perception and/or signaling (22). The observation that both RRS1 proteins are capable of binding PopP2 and have a nuclear localization suggests that they may compete for the binding to their DNA target(s). Because the RRS1 proteins differ mainly in their C-terminal transcription factor domains (22), they may lead to distinct transcriptional readouts resulting, in the case of RRS1-R, in plant resistance. It is tempting to speculate that such mechanisms may constitute the molecular basis of some recessive resistances.
The guard model postulates that NBS-LRR proteins interact with virulence targets of Avr proteins (9). The original structure of the RRS1 proteins suggests a dual function in pathogen perception and downstream signaling (22). How do these unique features of this R protein, as well as the demonstration of a RRS1/PopP2 binding, fit with the guard hypothesis? In a purely speculative conception, RRS1-R may fulfill, by means of its NBS-LRR domains, the function of a cytoplasmic guard protein able to “intercept” the PopP2 effector, preventing the latter from reaching its nuclear target. The RRS1 TIR-NBS-LRR domain may be required both for the recognition of the Avr factor and for the “sequestration” of the WRKY domain in the cytoplasm. The disease-resistance protein may then reach the nucleus by a “piggyback mechanism” upon its interaction with an NLS-bearing effector protein. The transcriptional activity of the WRKY domain on defense-related gene expression would then occur only upon detection of the PopP2 protein. Search and characterization of PopP2- and RRS1-R-interacting proteins as well as the identification of the target genes of RRS1-R should shed some light on the molecular mechanisms leading to this rather atypical resistance.
Acknowledgments
We thank Dr. F. Turck (Max-Planck-Institut) and I. Small (Unité de Recherche en Génomique Végétale, Evry, France) for providing the pAM-PAT-GFP and pGR0029RFP2 vectors, respectively. We thank also A. Jauneau (Institut Fédératif de Recherche 40) for technical assistance in the microscopy studies.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TIR, Toll-IL-1 receptor; NBS, nucleotide-binding site; LRR, leucine-rich repeat region; TTSS, type III secretion system, 5-FOA, 5-fluoroorotic acid; RFP, red fluorescent protein; Nub and Cub, N- and C-terminal halves, respectively, of ubiquitin; NLS, nuclear localization signal.
References
- 1.Flor, H. H. (1997) Annu. Rev. Phytopathol. 9, 275-296. [Google Scholar]
- 2.Baker, B., Zambryski, P., Staskawicz, B. & Dinesh-Kumar, S. P. (1997) Science 276, 726-733. [DOI] [PubMed] [Google Scholar]
- 3.Scofield, S. R., Tobias, C. M., Rathjen, J. P., Chang, J. F., Lavelle, D. T., Michelmore, R. W. & Staskawicz, B. J. (1996) Science 274, 2063-2065. [DOI] [PubMed] [Google Scholar]
- 4.Tang, X., Frederick, R. D., Zhou, J., Halterman, D. A., Jia, Y. & Martin, G. B. (1996) Science 274, 2060-2063. [DOI] [PubMed] [Google Scholar]
- 5.Jia, Y., McAdams, S. A., Bryan, G. T., Hershey, H. P. & Valent, B. (2000) EMBO J. 19, 4004-4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leister, R. T. & Katagiri, F. (2000) Plant J. 22, 345-354. [DOI] [PubMed] [Google Scholar]
- 7.Rivas, S., Romeis, T. & Jones, J. D. G. (2002) Plant Cell 14, 689-702. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Dixon, M. S., Golstein, C., Thomas, C. M., van der Biezen, E. A. & Jones, J. D. G. (2000) Proc. Natl. Acad. Sci. USA 97, 8807-8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dangl, J. L. & Jones, J. D. G. (2001) Nature 411, 826-833. [DOI] [PubMed] [Google Scholar]
- 10.Nimchuk, Z., Rohmer, L., Chang, J. H. & Dangl, J. L. (2001) Curr. Opin. Plant Biol. 4, 288-294. [DOI] [PubMed] [Google Scholar]
- 11.Hueck, C. J. (1998) Microbiol. Mol. Biol. 62, 379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Biezen, E. A. & Jones, J. D. G. (1998) Trends Biochem. Sci. 23, 454-456. [DOI] [PubMed] [Google Scholar]
- 13.Mackey, D., Holt, B. F., II, Wiig, A. & Dangl, J. L. (2002) Cell 108, 743-754. [DOI] [PubMed] [Google Scholar]
- 14.Kobe, B. & Deisenhofer, J. (1995) Curr. Opin. Struct. Biol. 5, 409-416. [DOI] [PubMed] [Google Scholar]
- 15.Kajava, A. V. (1998) J. Mol. Biol. 277, 519-527. [DOI] [PubMed] [Google Scholar]
- 16.Jones, D. A. & Jones, J. D. G. (1997) Adv. Bot. Res. Incorp. Adv. Plant Pathol. 24, 90-167. [Google Scholar]
- 17.Banerjee, D., Zhang, X. C. & Bent, J. A. (2001) Genetics 158, 439-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saraste, M., Sibbald, P. R. & Wittinghofer, A. (1990) Trends Biochem. Sci. 15, 430-434. [DOI] [PubMed] [Google Scholar]
- 19.Aravind, L., Dixit, V. M. & Koonin, E. V. (1999) Trends Biochem. Sci 24, 47-53. [DOI] [PubMed] [Google Scholar]
- 20.van der Biezen, E. A. & Jones, J. D. G. (1998) Curr. Biol. R226-R227. [DOI] [PubMed]
- 21.Deslandes, L., Pileur, F., Liaubet, L., Camut, S., Can, C., Williams, K., Holub, E., Beynon, J., Arlat, M. & Marco, Y. (1998) Mol. Plant–Microbe Interact. 11, 659-667. [DOI] [PubMed] [Google Scholar]
- 22.Deslandes, L., Olivier, O., Theulières, F., Hirsch, J., Feng, D. X., Bittner-Eddy, P., Beynon, J. & Marco, Y. (2002) Proc. Natl. Acad. Sci. USA 99, 2404-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eulgem, T., Rushton, P. J., Robatzek, S. & Somssich, I. E. (2000) Trends Plant Sci. 5, 199-206. [DOI] [PubMed] [Google Scholar]
- 24.Wittke, S., Lewke, N., Müller, S. & Johnsson, N. (1999) Mol. Biol. Cell 10, 2519-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guthrie, C. & Fink, G. R. (1991) Guide to Yeast Genetics and Molecular Biology (Academic, San Diego), p. 194.
- 26.Ausubel, F., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. G. & Struhm, K. (1987) Current Protocols in Molecular Biology (Greene & Wiley, New York).
- 27.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 28.Lavie, M., Shillington, E., Eguiluz, C., Grimsley, N. & Boucher, C. (2002) Mol. Plant Pathol. Interact. 10, 1058-1068. [DOI] [PubMed] [Google Scholar]
- 29.Hellens, R. P., Edwards, E. A., Leyland, N. R., Bean, S. & Mullineaux, P. M. (2000) Plant Mol. Biol. 42, 819-832. [DOI] [PubMed] [Google Scholar]
- 30.Axelos, M., Curie, C., Mazzolini, L., Bardet, C.& Lescure, B. (1992) Plant Physiol. Biochem. 30, 123-128. [Google Scholar]
- 31.Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W. L., Gomez-Gomez, L., Boller, T., Ausubel, F. M. & Sheen, J. (2002) Nature 415, 977-983. [DOI] [PubMed] [Google Scholar]
- 32.Salanoubat, M., Genin, S., Artiguenave, F., Gouzy, J., Mangenot, S., Arlat, M., Billault, A., Brottier, P., Camus, J. C., Cattolico, L., et al. (2002) Nature 415, 497-502. [DOI] [PubMed] [Google Scholar]
- 33.Staskawicz, B. J., Mudgett, M. B., Dangl, J. L. & Galan, J. E. (2001) Science 292, 2285-2289. [DOI] [PubMed] [Google Scholar]
- 34.Orth, K. (2002) Curr. Opin. Microbiol. 5, 38-43. [DOI] [PubMed] [Google Scholar]
- 35.Noel, L., Thieme, F., Nennstiel, D. & Bonas, U. (2001) Mol. Microbiol. 41, 1271-1281. [DOI] [PubMed] [Google Scholar]
- 36.Stagljar, I., Korostensky, C., Johnsson, N. & te Heesen, S. (1998) Proc. Natl. Acad. Sci. USA 95, 5187-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van den Ackerveken, G., Marois, E. & Bonas, U. (1996) Cell 87, 1307-1316. [DOI] [PubMed] [Google Scholar]
- 38.Szurek, B., Rossier, O., Hause, G. & Bonas, U. (2002) Mol. Microbiol. 46, 13-23. [DOI] [PubMed] [Google Scholar]
- 39.Luck, J. E., Lawrence, G. J., Dodds, P. N., Shepherd, K. W. & Ellis, J. G. (2000) Plant Cell 12, 1367-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lahaye, T. (2002) Trends Plant Sci. 7, 425-427. [DOI] [PubMed] [Google Scholar]
- 41.Maleck, K., Levine, A., Eulgem, T., Morgan, A., Schmid, R., Lawton, K. A., Dangl, J. L. & Dietrich, R. A. (2000) Nat. Genet. 26, 403-410. [DOI] [PubMed] [Google Scholar]