Abstract
Biological electron transfer is designed to connect catalytic clusters by chains of redox cofactors. A review of the characterized natural redox proteins with a critical eye for molecular scale measurement of variation and selection related to physiological function shows no statistically significant differences in the protein medium lying between cofactors engaged in physiologically beneficial or detrimental electron transfer. Instead, control of electron tunnelling over long distances relies overwhelmingly on less than 14 Å spacing between the cofactors in a chain. Near catalytic clusters, shorter distances (commonly less than 7 Å) appear to be selected to generate tunnelling frequencies sufficiently high to scale the barriers of multi-electron, bond-forming/-breaking catalysis at physiological rates. We illustrate this behaviour in a tunnelling network analysis of cytochrome c oxidase. In order to surmount the large, thermally activated, adiabatic barriers in the 5–10 kcal mol−1 range expected for H+ motion and O2 reduction at the binuclear centre of oxidase on the 103–105 s−1 time-scale of respiration, electron access with a tunnelling frequency of 109 or 1010 s−1 is required. This is provided by selecting closely placed redox centres, such as haem a (6.9 Å) or tyrosine (4.9 Å). A corollary is that more distantly placed redox centres, such as CuA, cannot rapidly scale the catalytic site barrier, but must send their electrons through more closely placed centres, avoiding direct short circuits that might circumvent proton pumping coupled to haems a to a3 electron transfer. The selection of distances and energetic barriers directs electron transfer from CuA to haem a rather than a3, without any need for delicate engineering of the protein medium to ‘hard wire’ electron transfer. Indeed, an examination of a large number of oxidoreductases provides no evidence of such naturally selected wiring of electron tunnelling pathways.
Keywords: electron and nuclear tunnelling, oxidoreduction catalysis, cytochrome oxidase
1. Introduction
Darwin discovered the basic principles of natural selection and evolution at the scale of the organism and its readily visible parts using scientific tools that were no more sophisticated than a net, callipers and a magnifying glass. Technological advances such as electron microscopes, synchrotrons and X-ray diffractometers have allowed access to biological design at finer scales, culminating in the molecular scale at the angstrom level. The family of oxidoreductases that have been structurally resolved at this level is large enough and sufficiently characterized to allow us to get some statistics and begin to look at the natural selection at this scale.
2. Natural selection of distance
We can see from a survey of crystal structures that distances between redox elements in electron transfer chains are, with a few exceptions, less than 14 Å (Page et al. 1999). It appears that most of the natural electron transfer proteins are selected for catalytic turnover of times at least in the milliseconds domain and the maximum distance seen is that which allows a minimal driving force (ΔG°) to promote sub-millisecond electron tunnelling rates (figure 1). To understand where this 14 Å may come from, we can consider the following simplest possible one-parameter expression to estimate the electron tunnelling rate in a protein where the distance between the redox centres, but little else, is known; all other parameters must take generic values.
| (2.1) |
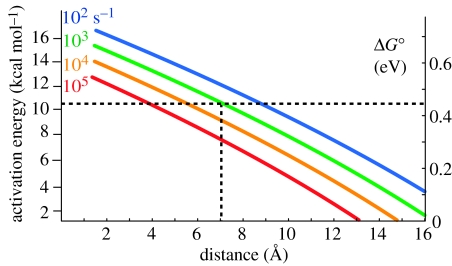
Figure 1.
Maximum distances required for tunnelling at certain rates for given activation energies (left scale). Corresponding driving force for endergonic electron transfer is shown on the right scale. With an activation energy barrier of 10 kcal mol−1, a distance of less than 7 Å will be required to achieve electron transfer in a millisecond or less.
R is the edge-to-edge distance in Å and the rate constant is given in s−1. This simple expression for a generic near zero driving force serves to illustrate that the distance must be less than 16 Å for the electron tunnelling rates to be faster than 103.
Often, we know of one more item, namely the driving force ΔG° (in eV); hence, we can consider driving forces different from the generic near zero value rather common in biology, in a two-parameter expression:
| (2.2a) |
| (2.2b) |
The first expression applies when ΔG° is exergonic and the second when ΔG° is endergonic, reflecting a ΔG° penalty for uphill electron transfer. Uphill electron transfers are typical at the multi-electron catalytic clusters of natural enzymes. Single-electron oxidation and reduction are endergonic, while two or more electron transfers (sometimes with associated proton transfer) are exergonic for typical substrates. Catalytic sites can also add non-tunnelling adiabatic barriers associated with bond making and breaking to simple electron-tunnelling barriers. In order to surmount these barriers in a sufficiently short period of time, tunnelling distances need to shrink as the barrier gets larger. Figure 1 illustrates the typical relationship between activation energy barrier and maximum distance of several common catalytic rates using equation (2.2b) that includes an explicit Hopfield-like temperature dependence (Moser et al. 1992). An example of natural reaction on the edge of this range is well characterized in photosynthetic reaction centres, where electron tunnelling through 14 Å from QA to the QB site promotes quinone to hydroquinone reduction. Driven by a modest free energy, electron tunnelling occurs at 104–105 s−1; when the apparently small chemical barrier is added to this electron-tunnelling barrier, catalysis is in milliseconds. For a more common 10 kcal mol−1 endergonic barrier, cofactors need to be separated by not more than 7 Å in order to achieve catalysis faster than a millisecond. These considerations are critical in considering the way the electron transfer cofactor chain is engineered to drive proton pumping and oxygen reduction catalysis in cytochrome oxidase.
3. Selection of driving force
Redox midpoint potential (Em) and hence the driving force (ΔG°) between redox cofactors of oxidoreductases sometimes appear to be selected. Redox cofactor Em values found in oxidoreductases are usually in the general range set by the Em values of the enzymes–substrates. Many oxidoreductases are readily reversible and hence substrate-to-substrate ΔG values are often very small. This means that the driving forces are also constrained to be small (as assumed in the very simple one-parameter tunnelling equation (2.1)) and much less than the values needed to maximize the electron-tunnelling rate (i.e. driving force matches the reorganization energy as described later). Thus, most oxidoreductases are forced to use sufficiently short distances between multiple redox cofactors in electron tunnelling chains to promote the high-tunnelling rates supporting millisecond catalytic turnover. Indeed, it is evident that distances between the successive redox cofactors in electron transfer chains leading to and from catalytic sites can themselves become short enough to accommodate significant endergonic tunnelling, while remaining fast enough for catalysis. This greater freedom in the choice of Em values is seen in dramatic swings in the Em values of redox cofactors in the extended linear chains very familiar in water-soluble and membrane-bound oxidoreductases (Page et al. 2003). The largest amplitude Em value swing discovered so far is 0.6 V in the five FeS clusters of nitrate reductase (Bertero & Rothery 2003). There is much to learn about the roots of these roller coaster profiles of cofactor redox potentials, but it seems clear that Em values in cofactor chains are not selected in nature for optimized rates of tunnelling over long distances. While the wide range of individual Em values may express drift due to a lack of evolutionary pressure, in some cases, the wide range of Em values in a chain may reflect a useful design that pools multiple electrons in an enzyme as a prelude to multiple electron delivery to the catalytic site.
4. Selection of reorganization energy
Another parameter that can influence electron tunnelling rates in protein is the reorganization energy (λ) required to distort the equilibrium nuclear geometry of the donor and acceptor before electron transfer, to resemble the equilibrium nuclear geometry after electron transfer. Equations (4.1a) and (4.1b) provide three-parameter empirical expressions that show how λ combines with ΔG° in setting the electron-tunnelling rates.
| (4.1a) |
| (4.1b) |
Unfortunately, there are relatively few protein systems in which systematic attempts to measure reorganization energy have been pursued. So far, these have been limited mostly to the laser-activated tunnelling reactions of photosynthetic reaction centres (Moser et al. 1992; Itoh et al. 2001; Haffa et al. 2002) or ruthenium-derived proteins (Gray & Winkler 2003), in which tunnelling rate constants are followed as the ΔG° value is systematically changed over hundreds of millielectron volt by cofactor modification or replacement or mutagenesis. The better-studied cases have been studied over a wide temperature range and led to a quantized version of the Marcus expression used in equations (2.2a)–(5.1b) (Moser et al. 1992). The measured values of reorganization energy of clear tunnelling reactions range from the remarkably low 0.25–0.5 eV of the pico- to nanosecond initial charge separation deep in the reaction centres (Iwaki & Itoh 1989; Haffa et al. 2002) to the high 1.1–1.5 eV for the more polar QB site reoxidation associated with proton exchange (Moser et al. 2000) as well as for coordinated ruthenium derivatives of haem proteins exposed to water. The majority of reorganization energies for tunnelling within proteins with cofactors excluded from the aqueous phase are in the range 0.6–0.9 eV. In the absence of the determined value, we use a generic value of 0.7 eV consistent with this majority. Alternative methods to determine reorganization energy values by measuring the temperature dependencies of electron-transfer rate constants and fitting to the Marcus classical expression are subject to several sources of considerable uncertainty (Moser et al. 1997). If the nuclei are quantized, the classical expression yields significant underestimates of the reorganization energy.
Despite the lack of data, it appears that the selection of reorganization energy is not very strong, with the possible exception of the initial charge separation reactions in photosynthetic systems in which reorganization energy appears unusually small, and may provide a barrier against short-circuit charge recombination (Moser et al. 2000; Haffa et al. 2002).
5. Selection of the tunnelling medium
The effect of selection on distance is clear and dominant. The distance ranging from van der Waals contact to 14 Å, which accommodates 95% of distances between cofactors performing physiologically productive electron tunnelling through protein, offers seven orders of magnitude from which a rate is selected. However, our work demonstrates that intervening tunnelling medium is heterogeneous and can influence the rate. An empirical four-parameter tunnelling expression that includes the influence of the packing (ρ) of the intervening protein medium is given by equations (5.1a) and (5.1b):
| (5.1a) |
| (5.1b) |
As we have described, ρ can vary ideally between 0 for vacuum (presenting a tunnelling barrier of 8 eV) and 1.0 for a completely packed medium (tunnelling barrier 2 eV). Protein heterogeneity ranges around an average ρ value of 0.76 (coinciding with 0.6 coefficient for R in equations (2.1)–(4.1b)) with a standard deviation of about 0.10. Thus, at 7, 10 and 14 Å, a standard deviation change for ρ represents 3.7-, 6.3- and 13-fold rate modulation from the average. Greater than order of magnitude changes in rate due to medium packing should be observable. However, we have found that the structure of the protein medium between the natural redox cofactors is statistically the same for physiologically productive and unproductive natural electron transfer (Moser et al. 1992; Page et al. 1999). There is no evidence that intervening polypeptide structural motifs have been adjusted by natural selection to speed up or slow down electron tunnelling for specific functions. Any variance in the packing density of the tunnelling medium appears unrelated to function, whether it is electron transfer through extended cofactor chains or at the interface between electron-transfer chains and cofactor clusters of catalytic sites.
Another approach for considering the influence of the intervening protein medium involves the connection of redox centres with mostly bonded, favoured electron tunnelling pathways (Beratan et al. 1992). This approach in itself is neutral with respect to whether the structure of the protein medium has been naturally selected (Tan et al. 2004; Skourtis et al. 2005) and in general the two approaches yield the same view of the protein structure, with some exceptions (Jones et al. 2002). However, some who have used this method profess to see the evidence of pathway selection and consider it vital (Regan et al. 1998; Medvedev et al. 2000), especially in cytochrome oxidase, where pathways are said to be needed not only to guide electrons from substrate cytochrome c to CuA and haem a, but also to make necessary ‘hard-wired’ inputs and outputs into the catalytic binuclear centre for efficient coupling of electrons and protons in the proton pump.
6. Example of a tunnelling network analysis: cytochrome c oxidase
The present uncertainties in distance, driving force and reorganization energy generate tunnelling rate uncertainties typically less than an order of magnitude. This is enough to address some controversies over what sort of electron tunnelling is possible or not possible on a given time-scale, whether certain electron transfers are likely to be tunnelling limited or limited by other barriers, and to gain insight into the natural design of electron transfer proteins with an idea of how finely tuned or robustly they have been constructed under the influence of genetic variance and natural selection.
Tunnelling network analysis has been successful with several systems, such as the photosystems I and II (Moser & Dutton in press; Moser et al. 2005) and purple bacterial reaction centres (Moser et al. 2003), but the simulations are equally appropriate for non-photosynthetic systems, such as the respiratory membrane protein cytochrome bc1 (Osyczka et al. 2004). Cytochrome c oxidase has been chosen because: (i) it is of central importance in respiration; (ii) this system has been intensely studied, with many experimentally determined rates taking advantage of relatively easy light activation with the ensuing reactions examined by a variety of spectroscopies in the nanosecond to seconds time-scale; and (iii) it offers a highly complex enzyme with many outstanding experimental disagreements and interpretive controversies and no settled-upon mechanism for how the single-electron tunnelling chain of cytochrome c, CuA and haem a is coupled to proton motion and transmembrane vectorial pumping and to the four-electron reduction of dioxygen at the haem a3/CuB (tyrosine) catalytic cluster.
7. Structure, cofactor interactions and free energies
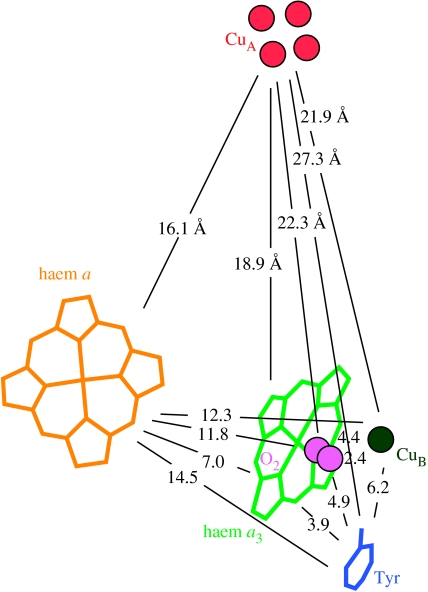
Figure 2 illustrates crystal structure-derived distances between redox centres in cytochrome c oxidase that provide the most important parameter in estimating electron-tunnelling rates. Table 1 provides an estimate of the redox potentials of the cofactors of oxidase that may be relevant for estimating the driving force of electron transfer between the centres. Haems a and a3 and CuB are sufficiently close to each other that electrostatic interactions affecting the driving force for the electron transfer are measurable. It also seems likely that ligand and internal protonation state changes near the redox centres during the catalytic cycle can affect the midpoint potentials (Wikstrom & Verkhovsky in press). It has been estimated that there are about 35–40 mV interaction energies between the redox centres, such that the reduction of any one makes it harder to reduce the others (Blair et al. 1986). The same report suggests a similar interaction energy between the more distant CuA and haem a. A possible set of midpoint potentials between many haem a/haem a3/CuB redox microstates that is consistent with spectral redox titrations has been provided by Nicholls & Wrigglesworth (1988). Except for endergonic electron transfer reactions, the influence of small changes in the driving force on electron tunnelling rates is minor. Instead of initially complicating the tunnelling analysis with a host of variable electronic microstate redox midpoint potentials of uncertain significance, we begin our examination with a simple average of the values of Nicholls & Wrigglesworth and the value of CuA reported by Moody & Rich (1990). We also need to make a rough estimate of the unknown ferryl redox potential of haem a3. It is clear that under certain experimental conditions, the tyrosine, which is identified in the structure as post-translationally modified, becomes oxidized during oxygen reduction (Proshlyakov et al. 2000). Although it is not clear whether this is formed directly or is just the final resting place of the radical, we model it as a redox centre that is not likely to be far away from the relevant radical centre, with a potential similar to other redox active tyrosines (Tommos 1999).
Figure 2.
Distances between redox centres in cytochrome c oxidase, as described in the Protein Data Bank (PDB) crystal structure 1QLE (Harrenga & Michel 1999). Diatomic oxygen is modelled in the position of carbon monoxide as in the structure 1V54 (Tsukihara 2003).
Table 1.
Redox potential estimates of cytochrome oxidase. (For haems a, a3 and CuB, the numbers are averages of those reported by Nicholls & Wrigglesworth (1988). The numbers in parenthesis are estimates of elevated redox potentials when the system is mostly oxidized.)
Water redox couples are known in solution (table 2), but unknown in the protein. It is believed that an important role that protein plays in catalysis is to moderate the barriers to electron transfer relative to solution (Babcock & Wikstrom 1992), to some extent ‘levelling out’ the redox couples in a multi-electron transfer reaction. An extreme example of ‘levelling’ has each oxygen couple the same as the average of all the four couples, 0.81 V. The last line in table 2 provides an example of possible redox couple midpoints if the solution values have been only gently ‘levelled out’ by 5%.
Table 2.
Redox potential estimates of O2 in aqueous solution, with complete ‘levelling’ by the catalytic site of cytochrome oxidase and with a more moderate 5% levelling.
| O2 redox couple, electrons added | 0/I | I/II | II/III | III/IV |
|---|---|---|---|---|
| Em H2O, pH 7 (V)a | −0.33 | 0.94 | 0.305 | 2.33 |
| Em estimate oxidase, 100% levelling | 0.81 | 0.81 | 0.81 | 0.81 |
| Em estimate oxidase, 5% levelling | −0.27 | 0.93 | 0.33 | 2.25 |
8. Expected individual tunnelling rates versus experimental electron-transfer rates
The rate constants of individual physiologically productive electron transfer from substrate ferrocytochrome c to CuA and the following electron transfer to haem a and then to the catalytic binuclear cluster haem a3 and CuB, as well as the potentially unproductive electron transfer between CuA and haem a3, have received considerable attention, both experimentally and theoretically. Table 3 compares the measured electron-transfer rate constants with the calculated rate constants using ΔG° from table 1 and a generic (default) reorganization energy of 0.7 eV using either equations (4.1a) and (4.1b) with the default ρ of 0.76 for average protein, or equations (5.1a) and (5.1b) including explicit ρ values.
Table 3.
Calculated tunnelling rates and some experimental rates for forward electron transfer between some redox centres in cytochrome oxidase. (Driving forces use the average midpoints of table 1. The default generic reorganization energy is 0.7 eV. The relevant packing densities in oxidase are higher than the generic protein default value of 0.76.)
9. Expected rates of CuA to haem a
For the 16 Å electron transfer between the nearest copper of CuA and edge of the conjugated ring of haem a, the calculated rate constant using equations (4.1a) and (4.1b) (default ρ) is sevenfold below the measured rate of 3.0×104 s−1 (Adelroth et al. 1995). Inclusion of the measured packing density of 0.87 in equations (5.1a) and (5.1b) raises the calculated value to fourfold faster than the measured rate. This strongly suggests that this reaction occurs by simple electron tunnelling without evident coupling to any rate-limiting adiabatic chemistry steps.
The 0.7 eV generic reorganization energy differs markedly from the unusually low 0.3 eV value derived from the temperature dependency of the rate constant of this reaction (Brzezinski 1996). This 0.3 eV value, used in several calculations in table 3, appears to be another example of the inappropriate application of temperature dependencies of electron-transfer rates to classical Marcus theory (Marcus & Sutin 1985) to determine reorganization energies, rather than non-classical nuclear vibrational modes, as is well documented in photosynthetic reaction centres (Gunner & Dutton 1989). Figure 1 shows that the activation energy derived from equations (5.1a) and (5.1b) for a low driving force electron tunnelling reaction with a generic reorganization energy of 0.7 eV is about 1.4 kcal mol−1 or 60 meV, matching the 53 meV generic reorganization energy reported for the CuA to haem a electron transfer (Adelroth et al. 1995); hence, there is no need for a low reorganization energy. If Mendeved et al. (2000), George et al. (2001) and Tan et al. (2004) used the 0.7 eV λ, which is consistent with the measured activation energy and non-classical Marcus theory, the measured versus calculated rate gap would widen to as much as 100-fold. Table 3 records an even smaller reorganization energy of 0.09 eV (Winkler et al. 1995). This extremely low value is unprecedented to date, more than twofold smaller than the lowest measured so far, and warrants further examination since the difference in the calculated rate between 0.09 and 0.7 eV is 70-fold. It seems that these pathway-based calculations underestimate the electronic coupling between CuA and haem a and, without low reorganization energies, lead to the significant shortfall in the calculated rate constants.
10. Expected rates of haems a to a3
Table 3 presents our calculated electron tunnelling rate constant for the 6.9 Å spacing between the porphyrin edges from haems a to a3, again assuming a default generic reorganization energy of 0.7 eV. The calculation of a rate constant of 6×108 s−1 (using equations (4.1a) and (4.1b) and a default ρ) became possible when the cytochrome oxidase crystal structure provided a good distance estimate. This rate excelled for a decade by being 200-fold faster than the 3 μs measured electron transfer equilibrium between haems a3 and a after activation by CO photolysis from the mixed valence state of cytochrome oxidase (Oliveberg & Malmstrom 1991; Verkhovsky et al. 1992), and 2000-fold faster than the 30 μs characteristic time of O2 reduction at the haem a3/CuB binuclear centre (Adelroth et al. 1998). The difference has been considered (Regan et al. 1998) as an error rooted in our empirically derived definition of the tunnelling distance between the haems as the shortest distance between the edges of the conjugated porphyrin macrocycle.
This contrast in rates led us to consider that the expected intrinsically nanosecond tunnelling time between closely placed cofactor edges, such as those of haems a and a3, can be obscured by the coupling to catalytic chemistry (Page et al. 1999). Thus, the finding that the tunnelling between haems a and a3 is coupled to uphill movement of protons as a first step in the pumping mechanism (Verkhovsky et al. 1995) or to thermal barriers associated with haem a3 reduction and ensuing O2 reduction with the effect of decreasing the rate of electron transfer compared to the intrinsic tunnelling rate is not unexpected (Page et al. 1999), as others have noted (Wikstrom et al. 2003; McMahon 2004; Faxen et al. 2005). In the simplest case of using the rule-of-thumb in figure 1, an observed rate of 3×105 s−1 suggests a catalytic tunnelling barrier of about 0.2 eV or ca 6 kcal mol−1 for the reduction of haem a3 to get a 2000-fold deceleration to the observed 30 μs time-scale. An example of an activation barrier measured between the O2 bound state and heterolytically cleaved ‘Pr’ state, has been measured in this range (Karpefors et al. 2000). In sharp contrast, table 3 presents two early through-bond polypeptide pathway models that link the Fe atoms of the two haems and provide a match with the measured 3×105 s−1 rate constants if the reorganization energy is as small as 0.44 eV. This calculation would be 20-fold too slow (Medvedev et al. 2000) if the published λ estimate of 0.76 eV was used (Adelroth et al. 1995). The long pathways selected (Regan et al. 1998) and the slow tunnelling rates derived, in our view, would not surmount the adiabatic barriers mentioned earlier in the familiar time-scales of respiratory turnover (Page et al. 1999).
The first hints of nanosecond components to the kinetics of electron transfer between haem a and a3 appeared in 2001 (Verkhovsky et al. 2001). Recently, ultrafast CO photolysis examination of the mixed valence state has uncovered a 1.2 ns electron equilibration between haems a and a3 (Pilet et al. 2004) in keeping with the expected tunnelling rates. Remarkably, it appears that this CO photolysis reaction leaves tunnelling between haems a and a3 without the barrier apparently associated with the O2 reduction reactions in the catalytic cluster. The temperature dependences of these rapid kinetics are very weak. This has also been used to derive reorganization energy for electron transfer between haems a and a3 that is unusually small (less than 0.2 eV; Jasaitis et al. 2005). However, while Jasaitis et al. appropriately consider the coupling of electron tunnelling to quantized nuclear vibrations, their reorganization energy estimate relies critically on a small, 3 mV change in driving force as the temperature changed from 2 to 32 °C. In fact, if there is no change in the driving force, then a 0.15 eV vibrational mode coupled to electron tunnelling gives an essentially temperature-independent rate that varies by 1% or less, even if the reorganization energy is as high as the generic 0.7 eV. Thus, the reorganization energy of this reaction remains unknown. Our calculations, including the generic reorganization energy of 0.7 eV with an average generic ρ (equations (4.1a) and (4.1b)) or the measured ρ (equations (5.1a) and (5.1b)) still yield rates of 7×108 and 1.4×109 s−1, respectively, remarkably close to the recently measured 3×108 s−1 tunnelling rate between the haems a and a3 (Pilet et al. 2004).
Recent pathway calculations of Tan et al. (2004) based on a dynamic model have abandoned the long pathways that gave slow rates for this reaction and recently found a direct tunnelling through space jump between the haems as the best pathway, which with a small λ of 0.3 eV is consistent with the new nanosecond rate (table 4).
Table 4.
Calculated tunnelling rates for cytochrome oxidase by various groups over time. (Numbers in parenthesis are the λ estimates in eV.)
| reaction | measured electron tunnelling rate (s−1) | calculated rate | |||||
|---|---|---|---|---|---|---|---|
| average protein | pathways based analysis | packing density | |||||
| Moser et al. (1992) with 1995 structurea | Regan et al. (1998) | Medvedev et al. (2000) | George (2001) | Tan et al. (2004) | this work | ||
| CuA→a | 2×104b | 3×103 (0.7) | 2×104 (0.09) | 3.3×102 (0.3) | 1.3×104 (0.3) | 6×104 (0.3) | 8.7×104 (0.7) |
| a→a3 pre-2001 | 3×105 | 6.5×108 (0.7) | 3×105 (0.44) | 5.9×104 (0.76)b | |||
| a→a3 post-2001 | 7×108b | 3×108 (0.76) | 1.4×109 (0.7) | ||||
| CuA→a3 | 4c | 90 (0.7) | 40 (0.5) | 0.17 (0.8) | 250 (0.7) | ||
11. Expected rate of CuA to haem a3
Electron transfer directly from CuA to haem a3 is often considered a physiological short circuit (Babcock & Callahan 1983; Papa et al. 1998; Wikstrom et al. 2003). An estimate of direct electron-transfer rate from CuA to a3 has been provided by mutant work (Kannt et al. 1999), although this has been challenged (Jasaitis et al. 2001). Applying equations (5.1a) and (5.1b) to the 18.9 Å electron tunnelling between the CuA copper and the haem a3 edge including the measured ρ of 0.79, with a near zero driving force with a default reorganization energy of 0.7 eV, generates an expected rate of 250 s−1. This is about 3% of the productive rate from CuA to haem a. If there is a substantial energy coupling of proton pumping to electron tunnelling from haem a to a3 as discussed earlier, this short-circuit rate is large enough to be a genuine engineering concern in the design of cytochrome oxidase. Some sort of barrier for a3 reduction by CuA needs to be added to slow down this tunnelling rate. The simplest solution is the finding that the reorganization energy for this reaction is increased from the default 0.7 eV to a value around 1.3 eV (table 3), a value often found when one or both tunnelling cofactors are located in polar amino acid environments or with access to water. Alternatively, electron transfer to the catalytic cluster may be associated with barriers accompanying liganding changes, oxygen reactions or even proton transfers, as has been suggested previously (Verkhovsky et al. 1995; Brzezinski 1996).
12. Tunnelling network analysis
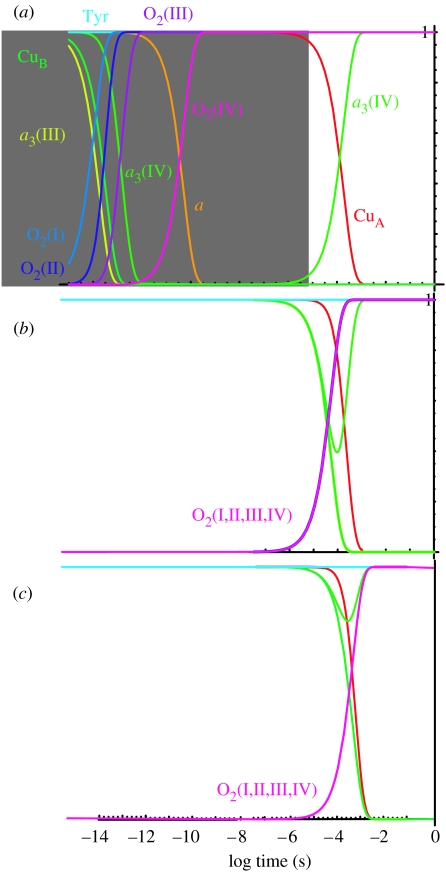
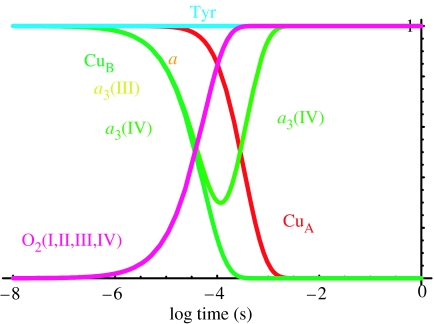
With the estimates of distances, driving forces and the quantized tunnelling expression of equations (4.1a)–(5.1b), it becomes possible to make a tunnelling network analysis of oxidoreductases which combines all the calculated tunnelling rates between all the redox centres, including both physiologically productive and unproductive reactions, into a set of differential equations to simulate the electron transfer kinetics of the system. This analysis aims to understand the engineering of cytochrome oxidase as an electron transfer protein and deliberately keeps the number of free parameters to a minimum; thus, we choose generic reorganization energies, packing densities and average redox midpoint potentials unless non-default values seem to be required. Figure 3a shows a tunnelling network simulation of the experiment of completely reduced oxidase reacting with O2 using completely averaged Em values for water reduction in oxidase (table 2). We have not included the time for diffusion of O2 to the oxidase (about 10 μs), but have looked at the inherent tunnelling time. We can immediately see that electron transfer time from haems a to a3 and O2 are inherently picoseconds to nanoseconds and are too fast compared to the observed O2 reduction rates of about 30 μs. O2 reduction will slow down if there is a barrier. We will consider two ways to introduce such a barrier: (i) O2 redox couples that are not so heavily levelled relative to the solution values; and (ii) a significant, non-tunnelling limited barrier for electron transfer due to bond breaking or protein reorganization, possibly associated with proton pumping.
Figure 3.
Electron tunnelling simulation of oxidase using different estimates of the midpoint potentials of the individual redox couples of O2. (a) Completely averaged O2 redox couple values as if 100% moderated by the enzyme environment; the picosecond electron tunnelling reactions in this simulation would not be observed experimentally as they would be limited by the 10 μs O2 diffusion rate (grey shading). (b) O2 redox couple midpoints as in aqueous solution, but moderate 5% levelling by the protein. (c) O2 redox couples as reported in aqueous solution, pH 7. Default reorganization energies for electron tunnelling are 0.7 eV throughout.
Figure 3b shows a simulation with water redox couples for O2 reduction, moderated to 5% by the protein towards the average O2/H2O redox couple. If there is no moderation, then electron tunnelling would be too slow to fit the experiment (figure 3c). These simulations fit well with the experiment on the fully reduced oxidase on O2 reduction and CuA oxidation (approx. 30 and 300 μs). Note that the close placing of haems a to a3 means that the uphill barrier for the first reduction of O2 can be scaled in the 30 μs time-scale.
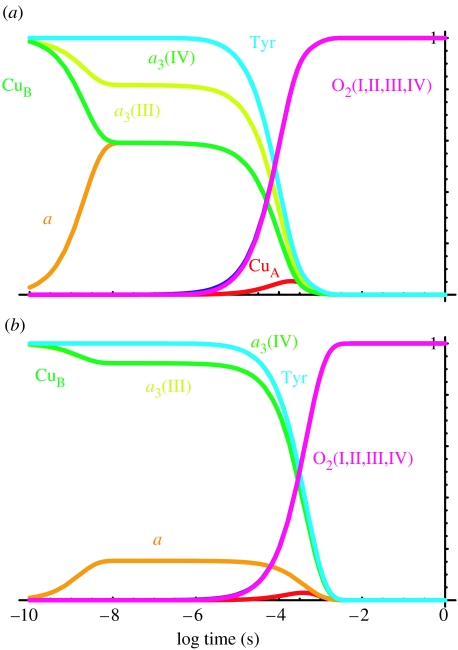
This barrier for O2 reduction will not slow down the electron transfer of direct CuA to haem a3. Although the distance (18.9 Å) between CuA and haem a3 is significantly longer than the distance (16.1 Å) between CuA and haem a, this direct rate may compete with the indirect electron transfer to a certain extent, potentially decreasing the efficiency of energy coupling of cytochrome oxidase. An argument has been made that only this sort of inefficiency may be advantageous for regulation (Papa et al. 1998). If oxidase was designed to keep the efficiency as high as possible, some other sort of barrier for electron transfer to haem a3, either a large reorganization energy for a3 reduction by CuA or a non-tunnelling chemical barrier for a3 reduction, would be needed to make electron transfer from CuA through haem a to haem a3 much faster than the direct electron transfer from CuA. An easy way to examine the effect of reorganization energy on CuA to haem a3 direct electron transfer is to perform a tunnelling simulation in a hypothetical haem a knockout version of oxidase. Figure 4 simply removes haem a from the simulation, analogous to the knockout experiments in cytochrome bc1 complex (Osyczka et al. 2004). Direct electron transfer from CuA to haem a3 can be as slow as desired (e.g. minutes) by adjusting the reorganization energy, while easily keeping electron transfer through haem a to haem a3 at the physiologically observed rate. Figure 5 replaces haem a in these knockout simulations using the middle reorganization energy value for CuA to catalytic cluster electron transfer. The rates are still comparable to the experiment.
Figure 4.
Electron tunnelling network simulation in a haem a knockout allows an examination of direct CuA to catalytic cluster electron transfer, which may short circuit important proton pumping action. Using the default reorganization energy and barrier for cluster reduction of 0.7 eV, CuA to cluster electron tunnelling is 0.4 ms (fine dashed lines). Increasing the cluster reduction reorganization energy of 1.7 eV slows this electron tunnelling to 0.2 s (medium dashed lines). A 2.5 eV reorganization energy slows this potential short circuit to about a minute.
Figure 5.
Electron tunnelling network of completely reduced oxidase, with haem a present, using the 1.7 eV reorganization barrier for cluster reduction by haem a3 as in figure 4. Oxygen reduction still takes place at the observed 30 μs rate.
13. Mixed valence state electron transfer
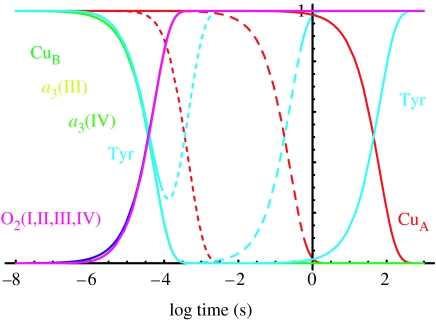
The simulations presented so far have begun with completely reduced oxidase with O2 bound and ready for electron transfer, and have been compared with experiments in which completely reduced oxidase has been prepared in the CO-ligated state, mixed with O2 and photolysed. Cytochrome oxidase is often studied in a mixed valence state, in which only haem a3 and CuB are reduced and CO bound before mixing with O2 and photolysed to start electron transfer. Under these conditions, O2 reduction is slowed to about 300 μs, compared to 30 μs, and the Tyr radical appears and is relatively stable in the so-called Pm state. Figure 6a shows that with the average redox midpoint values for the haem a3 and CuB, there is the expected nanosecond back flow of electrons into haem a, but the O2 reduction rate in the mixed valence state is too fast, being very similar to that of the completely reduced state. Even though the driving force for electron transfer from tyrosine is less than that from haem a, the tyrosine is sufficiently close that electron transfer is not rate limiting. However, if the redox midpoints of haem a3 and CuB are raised to 320 mV, which is close to the values expected for a partly reduced oxidase (Nicholls & Wrigglesworth 1988), this increases the barrier for O2 reduction and slows the rate to 310 μs, comparable to the observed deceleration of O2 reduction in the mixed valence experiment.
Figure 6.
Tunnelling simulation of the mixed valence state (i.e. CuA and haem a initially oxidized). (a) With average redox midpoint potentials, O2 reduction is comparable to the O2 reduction rate in the reduced system and not 10 times slower as experimentally observed. Electron back flow to haem a and a small amount of CuA are observed. (b) Raising the redox midpoint of haem a3 and CuB to 320 mV, as expected for a partly reduced system, slows the O2 reduction rate to 300 μs, comparable to the observed mixed valence rate. Only a small amount of haem a is reduced on the nanosecond time-scale.
14. Conclusion
Natural selection at the molecular level acts on electron transfer protein design principally through the selection of distances of 14 Å or less between redox centres to assure adequately fast tunnelling down of the electron transfer chains, and even smaller distances, typically 7 Å or less, to assure adequately fast electron transfer with multi-electron catalytic clusters, where there are barriers to single-electron oxidation and reduction. Cytochrome oxidase is a case in point with a clustering of redox centres within 7 Å to one another at the four-electron catalytic site of oxygen reduction, and the positioning of chain element CuA at 16 Å. As a chain element, CuA may well be placed at the long end of the chain distance range by natural selection for the benefit of making direct electron transfer to the cluster less likely, assuring that energy-coupling events associated with electron transfer from haems a to a3 are not short circuited. It appears that such short-circuit prevention would benefit from the introduction of an additional barrier. At this point, we do not know exactly what such a barrier might be—proton movement as a prelude to pumping, charge compensation and large reorganizations in a polar catalytic site filled with water and proton and hydroxyl transfers, or possible bond breaking and making. A resolution to these questions and an examination of reorganization energies and barriers will rely on mutant work and possibly cofactor changes that dramatically change the energetics of the oxidase system, as has been done in the photosynthetic reaction centres.
As a part of the present work, we reviewed other approaches to the calculation of electron tunnelling rate constants in cytochrome oxidase. A future stress on experiments that measure reorganization energies will also help to resolve issues surrounding the orders of magnitude range of rates obtained by pathway model analyses. We noted that these methods tend to underestimate electron-tunnelling rates. This may arise from several sources. The earliest (Regan et al. 1998) may lack the additional parameters derived for dynamic protein structures (rather than static crystal structures), which were considered to assist tunnelling taken for calculation (Tan et al. 2004). A pathway method that overemphasizes the importance of bonded paths through space gaps runs the risk of constructing a tortuous path that underestimates rates. It appears that pathway analyses have often been done directly on published crystal structural models that often contain voids around unresolved occupants; this was commented in Medvedev et al. (2000). This will accentuate the importance of the bonded pathway and diminish the choices for shorter high-coupling pathways. In contrast, the packing density model has no choice but the shortest tunnelling distance between defined edges (Page et al. 1999, 2003) in the static crystal structure, and has always filled large voids with water, using a packaged subroutine (Sybyl). The calculated rates using the packing density model on cytochrome oxidase seem appropriate and indicate that the 0.7 eV value for the default reorganization energy may not be far from the mark when renewed efforts meet the challenge of measuring this parameter. We are also aware that the use of the default value of ρ (0.76 for average packing of a protein) may provide a useful estimate of the tunnelling rate, especially if it is below 10 Å distance.
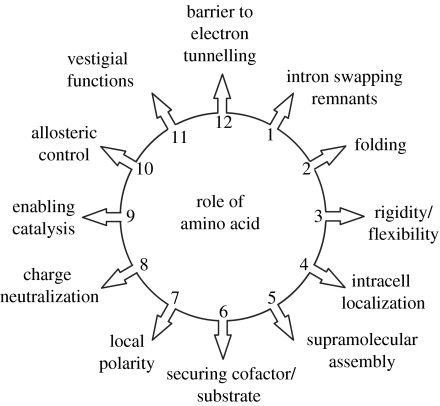
Compared to distance, the influence of the variation of packing density on tunnelling rates is small. This observation together with the lack of correlation between packing density and physiological function (Page et al. 1999), strongly suggests that structure of the polypeptide medium between tunnelling cofactors has not generally been naturally selected in the engineering of oxidoreductases. The good correlation between the packing density and pathway methods of approach suggests that this conclusion also applies to a variety of pathway-based models (Kawatsu et al. 2001; Jones et al. 2002). Despite this, users of protein pathways tunnelling models sometimes conclude that a pathway tracked in a protein between cofactors has been naturally selected and even optimized for the purpose. For instance, Medvedev et al. (2000) quantify the highly conserved pathways and side chains between haems a and a3 as indicators of strong natural selection for electron tunnelling in cytochrome c oxidase. Such a high degree of conservation is probably the consequence of many parallel and concurrent aspects allied to the construction, manifold functions and demolition of cytochrome c oxidase as illustrated in the molecular scale view of Darwin's principle of multiple utility (Darwin 1872) shown in figure 7. All the different utilities are accommodated within the structure and compromise any tendency of the different functions becoming optimized to varying degrees (Page et al. 2003).
Figure 7.
According to a molecular scale of version of Darwin's principle of multiply utility (Darwin 1872), each amino acid in a protein may serve many functions, and is not in general optimized for any single function of a particular scientist's interest.
Instead of focusing on the effects of protein medium on tunnelling (Gray & Winkler 2003, 2005), it seems clear that there is now a need to focus on how nature exploits distance and energetic barriers as a robust sort of engineering underlying oxidoreductase catalysis. The tunnelling network analysis presented here shows how distance is selected to set the tunnelling frequency needed to accommodate two types of barriers: the simple tunnelling barriers evident in cofactor chains that are set by distance, Em and λ, and the more complex barriers found at the interface of chains and catalytic sites associated with multiple electron transfer, and the non-tunnelling adiabatic barriers associated with bond making and breaking. The promise of simple tunnelling theory to help probe the energetics at the catalytic sites, which is the present topic of theoretical developments (Siegbahn et al. 2003; Kim et al. 2005; Olsson et al. 2005) and the main interest of this issue of Royal Society Transactions, is described in this paper.
Acknowledgements
Supported by US Public Health Service.
This paper is dedicated to Jerry Babcock (1956–2000), who wished to remind us that electron tunnelling gets much more interesting when its doing real chemistry.
One contribution of 16 to a Discussion Meeting Issue ‘Quantum catalysis in enzymes—beyond the transition state theory paradigm’.
References
- Adelroth P, Brzezinski P, Malmstrom B.G. Internal electron transfer in cytochrome c oxidase from Rhodobacter sphaeroides. Biochemistry. 1995;34:2844–2849. doi: 10.1021/bi00009a014. doi:10.1021/bi00009a014 [DOI] [PubMed] [Google Scholar]
- Adelroth P, Ek M, Brzezinski P. Factors determining electron-transfer rates in cytochrome c oxidase: investigation of the oxygen reaction in the R. sphaeroides enzyme. Biochim. Biophys. Acta. 1998;1367:107–117. doi: 10.1016/s0005-2728(98)00142-x. doi:10.1016/S0005-2728(98)00142-X [DOI] [PubMed] [Google Scholar]
- Babcock G.T, Callahan P.M. Redox-linked hydrogen-bond strength changes in cytochrome a: implications for a cytochrome oxidase proton pump. Biochemistry. 1983;22:2314–2319. doi: 10.1021/bi00279a002. doi:10.1021/bi00279a002 [DOI] [PubMed] [Google Scholar]
- Babcock G.T, Wikstrom M. Oxygen activation and the conservation of energy in cell respiration. Nature. 1992;356:301–309. doi: 10.1038/356301a0. doi:10.1038/356301a0 [DOI] [PubMed] [Google Scholar]
- Beratan D.N, Betts J.N, Onuchic J.N. Tunneling pathway and redox-state-dependent electronic couplings at nearly fixed distance in electron-transfer proteins. J. Phys. Chem. 1992;96:2852–2855. doi:10.1021/j100186a014 [Google Scholar]
- Bertero M.G, Rothery R.A. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat. Struct. Biol. 2003;10:681–687. doi: 10.1038/nsb969. doi:10.1038/nsb969 [DOI] [PubMed] [Google Scholar]
- Blair D.F, Ellis W.R, Wang H, Gray H.B, Chan S.I. Spectroelectrochemical study of cytochrome c oxidase: pH and temperature dependences of the cytochrome potentials. Characterization of site–site interactions. J. Biol. Chem. 1986;261:1524–1537. [PubMed] [Google Scholar]
- Brzezinski P. Internal electron-transfer reactions in cytochrome oxidase. Biochemistry. 1996;35:5611–5615. doi: 10.1021/bi960260m. doi:10.1021/bi960260m [DOI] [PubMed] [Google Scholar]
- Darwin C. Earlton House; New York, NY: 1872. Origin of species by means of natural selection. [Google Scholar]
- Faxen K, Gilderson G, Adelroth P, Brzezinski P. A mechanistic principle for proton pumping by cytochrome oxidase. Nature. 2005;437:286–289. doi: 10.1038/nature03921. [DOI] [PubMed] [Google Scholar]
- George S.D, et al. A quantitative description of the ground-state wave function of CuA by X-ray absorption spectroscopy: comparison to plastocyanin and relevance to electron transfer. J. Am. Chem. Soc. 2001;123:5757–5767. doi: 10.1021/ja004109i. doi:10.1021/ja004109i [DOI] [PubMed] [Google Scholar]
- Gray H.B, Winkler J.R. Electron tunneling through proteins. Q. Rev. Biophys. 2003;36:341–372. doi: 10.1017/s0033583503003913. doi:10.1017/S0033583503003913 [DOI] [PubMed] [Google Scholar]
- Gray H.B, Winkler J.R. Long-range electron transfer. Proc. Natl Acad. Sci. USA. 2005;102:3534–3539. doi: 10.1073/pnas.0408029102. doi:10.1073/pnas.0408029102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunner M.R, Dutton P.L. Temperature and -ΔG° dependence of the electron transfer from BPh- to QA in reaction center protein from Rhodobacter sphaeroides with different quinones as QA. J. Am. Chem. Soc. 1989;111:3400–3412. doi:10.1021/ja00191a043 [Google Scholar]
- Haffa A.L.M, Lin S, Katilius E, Williams J.C, Taguchi A.K.W, Allen J.P, Woodbury N.W. The dependence of the initial electron-transfer rate on driving force in Rhodobacter sphaeroides reaction centers. J. Phys. Chem. B. 2002;106:7376–7384. doi:10.1021/jp0257552 [Google Scholar]
- Harrenga A, Michel H. The cytochrome c oxidase from Paracoccus denitrificans does not change the metal center ligation upon reduction. J. Biol. Chem. 1999;274:33 296–33 299. doi: 10.1074/jbc.274.47.33296. doi:10.1074/jbc.274.47.33296 [DOI] [PubMed] [Google Scholar]
- Itoh S, Iwaki M, Ikegami I. Modification of photosystem I reaction center by the extraction and exchange of chlorophylls and quinones. Biochim. Biophys. Acta. 2001;1507:115–138. doi: 10.1016/s0005-2728(01)00199-2. doi:10.1016/S0005-2728(01)00199-2 [DOI] [PubMed] [Google Scholar]
- Iwaki M, Itoh S. Electron transfer in spinach photosystem I reaction center containing benzoquinone, naphthoquinone and anthraquinones in place of phylloquinone. FEBS Lett. 1989;256:11–16. doi:10.1016/0014-5793(89)81708-9 [Google Scholar]
- Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. doi:10.1038/376660a0 [DOI] [PubMed] [Google Scholar]
- Jasaitis A, Backgren C, Morgan J.E, Puustinen A, Verkhovsky M.I, Wikstrom M. Electron and proton transfer in the arginine-54–methionine mutant of cytochrome c oxidase from Paracoccus denitrificans. Biochemistry. 2001;40:5269–5274. doi: 10.1021/bi002948b. [DOI] [PubMed] [Google Scholar]
- Jasaitis A, Rappaport F, Pilet E, Liebl U, Vos M.H. Activationless electron transfer through the hydrophobic core of cytochrome c oxidase. Proc. Natl Acad. Sci. USA. 2005;102:10 882–10 886. doi: 10.1073/pnas.0503001102. doi:10.1073/pnas.0503001102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.L, Kurnikov I.V, Beratan D.N. The nature of tunneling pathway and average packing density models for protein-mediated electron transfer. J. Phys. Chem. A. 2002;106:2002–2006. doi:10.1021/jp0133743 [Google Scholar]
- Kannt A, Pfitzner U, Ruitenberg M, Hellwig P, Ludwig B, Mäntele W, Fendler K, Michel H. Mutation of Arg-54 strongly influences heme composition and rate and directionality of electron transfer in Paracoccus denitrificans cytochrome c oxidase. J. Biol. Chem. 1999;274:37 974–37 981. doi: 10.1074/jbc.274.53.37974. doi:10.1074/jbc.274.53.37974 [DOI] [PubMed] [Google Scholar]
- Karpefors M, Adelroth P, Namslauer A, Zhen Y.J, Brzezinski P. Formation of the “peroxy” intermediate in cytochrome c oxidase is associated with internal proton/hydrogen transfer. Biochemistry. 2000;39:14 664–14 669. doi: 10.1021/bi0013748. doi:10.1021/bi0013748 [DOI] [PubMed] [Google Scholar]
- Kawatsu T, Kakitani T, Yamato T. Worm model for electron tunneling in proteins: consolidation of the pathway model and the Dutton plot. J. Phys. Chem. B. 2001;105:4424–4435. doi:10.1021/jp003918l [Google Scholar]
- Kim J, Mao J, Gunner M.R. Are acidic and basic groups in buried proteins predicted to be ionized? J. Mol. Biol. 2005;348:1283–1298. doi: 10.1016/j.jmb.2005.03.051. doi:10.1016/j.jmb.2005.03.051 [DOI] [PubMed] [Google Scholar]
- Marcus R.A, Sutin N. Electron transfers in chemistry and biology. Biochim. Biophys. Acta. 1985;811:265–322. [Google Scholar]
- McMahon B.H, et al. FTIR studies of internal proton transfer reactions linked to inter-heme electron transfer in bovine cytochrome c oxidase. Biochim. Biophys. Acta. 2004;1655:321–331. doi: 10.1016/j.bbabio.2004.01.007. doi:10.1016/j.bbabio.2004.01.007 [DOI] [PubMed] [Google Scholar]
- Medvedev D.M, Daizadeh I, Stuchebrukhov A.A. Electron transfer tunneling pathways in bovine heart cytochrome c oxidase. J. Am. Chem. Soc. 2000;122:6571–6582. doi:10.1021/ja0000706 [Google Scholar]
- Moody A.J, Rich P.R. The effect of pH on redox titrations of haem a in cyanide-liganded cytochrome c oxidase: experimental and modelling studies. Biochim. Biophys. Acta. 1990;1015:205–215. doi: 10.1016/0005-2728(90)90022-v. doi:10.1016/0005-2728(90)90022-V [DOI] [PubMed] [Google Scholar]
- Moser, C. C. & Dutton, P. L. In press. Photosystem I. The plastocyanin: ferredoxin oxidoreductase in photosynthesis (ed. D. Bruce). Academic Press.
- Moser C.C, Keske J.M, Warncke K, Farid R.S, Dutton P.L. Nature of biological electron transfer. Nature (London) 1992;355:796–802. doi: 10.1038/355796a0. doi:10.1038/355796a0 [DOI] [PubMed] [Google Scholar]
- Moser C.C, Page C.C, Chen X, Dutton P.L. Biological electron tunneling through native protein media. J. Biol. Inorg. Chem. 1997;2:393–398. doi:10.1007/s007750050149 [Google Scholar]
- Moser C.C, Page C.C, Chen X, Dutton P.L. Electron transfer in natural proteins: theory and design. Subcell. Biochem. 2000;35:1–30. doi: 10.1007/0-306-46828-x_1. [DOI] [PubMed] [Google Scholar]
- Moser C.C, Page C.C, Cogdell R.J, Barber J, Wraight C.A, Dutton P.L. Length, time, and energy scales of photosystems. Adv. Protein Chem. 2003;63:71–109. doi: 10.1016/s0065-3233(03)63004-4. [DOI] [PubMed] [Google Scholar]
- Moser C.C, Page C.C, Dutton P.L. Tunneling in PSII. Photochem. Photobiol. Sci. 2005;4:933–939. doi: 10.1039/b507352a. doi:10.1039/b507352a [DOI] [PubMed] [Google Scholar]
- Nicholls P, Wrigglesworth J.M. Routes of cytochrome a3 reduction: the neoclassical model revisited. Ann. NY Acad. Sci. 1988;550:59–67. doi: 10.1111/j.1749-6632.1988.tb35323.x. [DOI] [PubMed] [Google Scholar]
- Oliveberg M, Malmstrom B.G. Internal electron transfer in cytochrome c oxidase: evidence for a rapid equilibrium between cytochrome a and the bimetallic site. Biochemistry. 1991;39:7053–7057. doi: 10.1021/bi00243a003. doi:10.1021/bi00243a003 [DOI] [PubMed] [Google Scholar]
- Olsson M.H.M, Sharma P.K, Warshel A. Simulating redox coupled proton transfer in cytochrome c oxidase: looking for the proton bottleneck. FEBS Lett. 2005;579:2026–2034. doi: 10.1016/j.febslet.2005.02.051. doi:10.1016/j.febslet.2005.02.051 [DOI] [PubMed] [Google Scholar]
- Osyczka A, Moser C.C, Daldal F, Dutton P.L. Reversible redox energy coupling in electron transfer chains. Nature. 2004;427:607–612. doi: 10.1038/nature02242. doi:10.1038/nature02242 [DOI] [PubMed] [Google Scholar]
- Page C.C, Moser C.C, Chen X, Dutton P.L. Natural engineering principles of electron tunneling in biological oxidation–reduction. Nature (London) 1999;402:47–52. doi: 10.1038/46972. doi:10.1038/46972 [DOI] [PubMed] [Google Scholar]
- Page C.C, Moser C.C, Dutton P.L. Mechanism for electron transfer within and between proteins. Curr. Opin. Chem. Biol. 2003;7:1–6. doi: 10.1016/j.cbpa.2003.08.005. doi:10.1016/j.cbpa.2003.08.005 [DOI] [PubMed] [Google Scholar]
- Papa S, Capitanio N, Villani G. A cooperative model for protonmotive heme–copper oxidases. The role of heme a in the proton pump of cytochrome c oxidase. FEBS Lett. 1998;439:1–8. doi: 10.1016/s0014-5793(98)01305-2. doi:10.1016/S0014-5793(98)01305-2 [DOI] [PubMed] [Google Scholar]
- Pilet E, Jasaitis A, Liebl U, Vos M.H. Electron transfer between hemes in mammalian cytochrome c oxidase. Proc. Natl Acad. Sci. USA. 2004;101:16 198–16 203. doi: 10.1073/pnas.0405032101. doi:10.1073/pnas.0405032101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proshlyakov D.A, Pressler M.A, DeMaso C, Leykam J.F, DeWitt D.L, Babcock G.T. Oxygen activation and reduction in respiration: involvement of redox-active tyrosine 244. Science. 2000;290:1588–1591. doi: 10.1126/science.290.5496.1588. doi:10.1126/science.290.5496.1588 [DOI] [PubMed] [Google Scholar]
- Regan J.J, Ramirez B.E, Winkler J.R, Gray H.B, Malmstrom B.G. Pathways for electron tunneling in cytochrome c oxidase. J. Bioenerg. Biomembr. 1998;30:35–39. doi: 10.1023/a:1020551326307. doi:10.1023/A:1020551326307 [DOI] [PubMed] [Google Scholar]
- Siegbahn P.E.M, Blomberg M.R.A, Blomberg M.L. Theoretical study of the energetics of proton pumping and oxygen reduction in cytochrome oxidase. J. Phys. Chem. B. 2003;107:10 946–10 955. doi:10.1021/jp035486v [Google Scholar]
- Skourtis S.S, Balabin I.A, Kawatsu T, Beratan D.N. Protein dynamics and electron transfer: electronic decoherence and non-Condon effects. Proc. Natl Acad. Sci. USA. 2005;102:3552–3557. doi: 10.1073/pnas.0409047102. doi:10.1073/pnas.0409047102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M.L, Balabin I, Onuchic J.N. Dynamics of electron transfer pathways in cytochrome c oxidase. Biophys. J. 2004;86:1813–1819. doi: 10.1016/S0006-3495(04)74248-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommos C, et al. De novo proteins designed to study properties of redox-active amino acids. J. Inorg. Biochem. 1999;74:316. [Google Scholar]
- Tsukihara T, et al. The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process. Proc. Natl Acad. Sci. USA. 2003;100:15 304–15 309. doi: 10.1073/pnas.2635097100. doi:10.1073/pnas.2635097100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhovsky M.I, Morgan J.E, Wikstrom M. Intramolecular electron transfer in cytochrome c oxidase: a cascade of equilibria. Biochemistry. 1992;31:11 860–11 863. doi: 10.1021/bi00162a026. doi:10.1021/bi00162a026 [DOI] [PubMed] [Google Scholar]
- Verkhovsky M.I, Morgan J.E, Wikstrom M. Control of electron delivery to the oxygen reduction site of cytochrome c oxidase: a role for protons. Biochemistry. 1995;34:7483–7491. doi: 10.1021/bi00022a023. doi:10.1021/bi00022a023 [DOI] [PubMed] [Google Scholar]
- Verkhovsky M.I, Jasaitis A, Wikstrom M. Ultrafast haem–haem electron transfer in cytochrome c oxidase. Biochim. Biophys. Acta. 2001;1506:143–146. doi: 10.1016/s0005-2728(01)00220-1. doi:10.1016/S0005-2728(01)00220-1 [DOI] [PubMed] [Google Scholar]
- Wikstrom, M. & Verkhovsky, M. I. In press. Towards the mechanism of proton pumping by the haem–copper oxidases. Biochim. Biophys. Acta [DOI] [PubMed]
- Wikstrom M, Verkhovsky M.I, Hummer G. Water-gated mechanism of proton translocation by cytochrome c oxidase. Biochim. Biophys. Acta. 2003;1604:61–65. doi: 10.1016/s0005-2728(03)00041-0. doi:10.1016/S0005-2728(03)00041-0 [DOI] [PubMed] [Google Scholar]
- Winkler J.R, Malmstrom B.G, Gray H.B. Rapid electron injection into multisite metalloproteins: intramolecular electron transfer in cytochrome oxidase. Biophys. Chem. 1995;54:199–209. doi: 10.1016/0301-4622(94)00156-e. doi:10.1016/0301-4622(94)00156-E [DOI] [PubMed] [Google Scholar]