Abstract
Mus81-Mms4 and Rad1-Rad10 are homologous structure-specific endonucleases that cleave 3′ branches from distinct substrates and are required for replication fork stability and nucleotide excision repair, respectively, in the yeast Saccharomyces cerevisiae. We explored the basis of this biochemical and genetic specificity. The Mus81-Mms4 cleavage site, a nick 5 nucleotides (nt) 5′ of the flap, is determined not by the branch point, like Rad1-Rad10, but by the 5′ end of the DNA strand at the flap junction. As a result, the endonucleases show inverse substrate specificity; substrates lacking a 5′ end within 4 nt of the flap are cleaved poorly by Mus81-Mms4 but are cleaved well by Rad1-10. Genetically, we show that both mus81 and sgs1 mutants are sensitive to camptothecin-induced DNA damage. Further, mus81 sgs1 synthetic lethality requires homologous recombination, as does suppression of mutant phenotypes by RusA expression. These data are most easily explained by a model in which the in vivo substrate of Mus81-Mms4 and Sgs1-Top3 is a 3′ flap recombination intermediate downstream of replication fork collapse.
Homologous recombination is required during DNA replication to repair double-strand breaks (DSBs) and restart forks that have stalled because of DNA damage (10, 39). This is best understood in Escherichia coli, where the stability of stalled replication forks (RFs) requires the recF recombination pathway (9). In eukaryotes, several proteins have been implicated in this process, including homologs of the bacterial RecQ DNA helicase, which functions in the recF recombination pathway. Loss of the RecQ family proteins human BLM, Schizosaccharomyces pombe Rqh1, and S. cerevisiae Sgs1 results in DNA damage sensitivity and hyperrecombination phenotypes consistent with a role in restarting stalled RFs (6). These 3′-to-5′ DNA helicases function as part of a complex with DNA topoisomerase III (Top3) (2, 17, 19, 24, 47), an enzyme specific for relaxing of negatively supercoiled DNA (28). Still unclear, however, is the nature of the in vivo substrate for the eukaryotic RecQ-Top3 complexes (39, 45).
MUS81 and MMS4 were identified in S. cerevisiae in a screen for genes that are essential for viability in the absence of SGS1 or TOP3 (34). They have also been isolated in yeast two-hybrid screens with scRad54, scMek1, and Cds1 from S. pombe (4, 12, 23). On their own, mutations in MUS81 and MMS4 confer identical phenotypes, including strong MMS sensitivity, weak UV sensitivity, and defects in sporulation (4, 12, 23, 34, 48). The similarity of these phenotypes to those of sgs1 top3 mutants suggested that they function in overlapping pathways for the repair of spontaneous or induced DNA damage (34), and epistasis tests placed MUS81 and MMS4 in the same genetic pathway (12, 34). An additional similarity between the two pathways is that elimination of meiotic recombination suppresses the sporulation defects of both mus81 mms4 (3, 12, 27) and top3 mutant strains (18). Thus, both pathways appear to play a role in resolving recombination intermediates in meiosis.
The C terminus of Mus81 shares a 200-amino-acid domain with the yeast Rad1 and human XPF endonucleases (23, 34) that are required for nucleotide excision repair (NER) and single-strand annealing. The yeast Rad1-Rad10 and human XPF-ERCC1 complexes are heterodimeric endonucleases that cleave the 3′ single-stranded DNA (ssDNA) tail from partially duplex DNA substrates containing splayed nonhomologous ssDNA arms (simple Y substrate) (1, 11). Like the NER enzymes, Mus81 and Mms4 form a heterodimeric structure-specific endonuclease (27). Although Mus81-Mms4 cleaves the 3′ ssDNA tail from a simple Y, like the NER enzymes, it is more active on duplex DNA with a 3′ ssDNA branch (3′ flap substrate) or an RF substrate (27). On the other hand, it has been reported that Mus81 from S. pombe or human cell extracts is part of a complex that is specific for resolving Holliday junctions (HJs), a putative intermediate in the process of stabilizing stalled RFs (3, 7). Although the Mus81-associated endonuclease cleaves HJs with apparent symmetry at multiple sites within the homologous core of a branch-migratable HJ, its products cannot be ligated (3, 7) and the human enzyme prefers 3′ flap substrates (8). Thus, the symmetry of HJ cleavage relates only to the population of molecules as a whole, not individual products of HJ cleavage. The mechanism by which these products might be processed into the nicked linear-duplex forms typical of prokaryotic and mitochondrial resolvases remains to be demonstrated.
To better understand the function of Mus81-Mms4 and its relationship to Rad1-Rad10 (Rad1-10), we compared their in vitro activities and in vivo phenotypes. The Mus81-Mms4 and Rad1-Rad10 endonuclease activities are biochemically distinct, with little overlap in substrate specificity. Mus81-Mms4 nicks the flap-containing strand upstream of the branch point to generate duplex products with a 5-nucleotide (nt) gap. In contrast to Rad1-Rad10, where the site of cleavage is determined by the duplex-ssDNA junction, the site of Mus81-Mms4 cleavage is determined by the 5′ end of the strand at the flap junction. The requirement for the 5′ end of this downstream strand limits Mus81-Mms4 activity to flap structures as opposed to simple Ys or other branched substrates. In vivo studies showed that, unlike mutations in RAD1-RAD10, mutations in MUS81-MMS4 confer sensitivity to camptothecin (CPT), a compound known to produce replication-dependent DSBs. This sensitivity to CPT is shared by sgs1-top3 mutants. DSBs are further implicated in the mechanism of these two pathways given that the synthetic lethality of mus81 sgs1 mutants requires the RAD52 DSB repair pathway. Taken together, the data suggest that Mus81-Mms4 and Sgs1-Top3 are required to process recombination intermediates that form downstream of collapsed RFs.
MATERIALS AND METHODS
Strains and plasmids.
The S. cerevisiae strains used in this study are isogenic derivatives of W303-1a (MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100). Strain construction, growth, and transformation were performed by using standard procedures (38). Mutants were constructed by the one-step gene disruption method by using PCR fragments marked with kanamycin (pUG6) (21) or hygromycin B (pAG32) (20). Strains CHY125 (wild type), JMY375 (mms4::KAN), and JMY380 (mus81::KAN) have been described previously (34) and were used to create strains WFY1559 (rad1::KAN), WFY1560 (mms4::KAN rad1::HGR), and WFY1561 (mus81::KAN rad1::HGR), respectively. AMR60 (top3 sgs1) and JMY531 (sgs1) have been described previously (34). JMY512 is MATaade2-1 ade3::hisG ura3-1 his3-11,15 trp1-1 leu2-3,112 top3-11::KAN loxP. Proper integrative transplacement was verified by analytical PCR. Suppression of sgs1 synthetic lethality was determined by crossing sgs1 slx double mutants carrying plasmid pJM500 (URA3::SGS1) (34) with HKY615-1A (rad1), HKY1039-4D (rad51), HKY614-10B (rad52), HKY624 (rad54), HKY597-2C (rad55), or HKY598-8B (rad57) (29). Meiotic segregants were identified and tested for suppression by growth on 5-fluoroorotic acid.
To express Rad1-Rad10 endonuclease, the S. cerevisiae RAD1 and RAD10 genes were first cloned into the expression vectors pET28a and pET11a, respectively. The T7 expression cassettes were then combined essentially as previously described (27) to create a bicistronic plasmid expressing both His6-Rad1 and Rad10 (pNJ6954). Plasmid pKR6980 was constructed for constitutive RusA expression in S. cerevisiae by placing the RusA open reading frame (an NdeI/BamHI PCR fragment) downstream of the SGS1 promoter in pSM100 (33). The RusA open reading frame in pKR6980 was verified by DNA sequencing.
Expression and purification of recombinant proteins.
Mus81-(His6)Mms4 was expressed and purified as previously described (27). The His6-MMS4 allele functionally complements sgs1 mms4 synthetic lethality (data not shown).
Expression and purification of (His6)Rad1-Rad10 were done by using procedures similar to those used for Mus81-(His6)Mms4. Briefly, BL21-RIL cells containing pNJ6954 were grown at 37°C in Luria-Bertani broth plus ampicillin to an optical density at 595 nm of 0.5 and cooled to 25°C. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to 0.4 mM, and growth was continued for 6 h at 25°C. Cells were pelleted, frozen once, thawed, and resuspended at 20 ml/liter of culture in A buffer (25 mM Tris [pH 7.5], 1 mM EDTA, 0.01% NP-40, 10% glycerol, 0.1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM dithiothreitol [DTT]) plus 1% NP-40, 150 mM NaCl, and protease inhibitors. Extract preparation and chromatographic purification were identical to those of Mus81-(His6)Mms4 (27), except that (His6)Rad1-Rad10 eluted from the phosphocellulose resin between 525 and 975 mM NaCl. This pool was bound directly to Ni-Probond resin (Invitrogen) in the presence of 10 mM imidazole. The 200 mM imidazole wash contained highly purified Rad1-Rad10 and two triplets of Rad1 breakdown products at 45 and 80 kDa. This identification was based on an antihemagglutinin immunoblot and a (His6)Rad1(hemagglutinin)-Rad10 expression system.
Nuclease assays.
Nuclease assays were performed essentially as previously described (27), except that Mus81-Mms4 reactions were performed with 20 mM Tris (pH 8.0)-100 mM NaCl-0.2 mM DTT-5% glycerol-10 mM MgCl2 and incubation for 30 min at 30°C. Rad1-Rad10 reactions were performed with 50 mM Tris (pH 8.5)-5 mM MgCl2-5 mM DTT and incubation for 60 min at 37°C. The data were analyzed by phosphorimager with IPLab gel densitometry software to quantitate band intensities. Reaction mixtures analyzed by using sequencing gels contained 2 nM Mus81-Mms4 and 2 fmol of substrate in a 20-μl reaction volume, and the reactions were terminated by addition of EDTA to a final concentration of 5 mM.
DNA substrates.
DNA structures were constructed from 5′ 32P-end-labeled oligonucleotides that were annealed and purified as previously described (46). The oligonucleotides used for this study were as follows: 888 (49 nt), GACGCTGCCGAATTCTGGCGTTAGGAGATACCGATAAGCTTCGGCTTAA;1128 (50 nt), ATCGATGTCTCTAGACAGCACGAGCCCTAACGCCAGAATTCGGCAGCGTC; 994 (25 nt), GCTCGTGCTGTCTAGAGACATCGAT; 1038 (24 nt), CTCGTGCTGTCTAGAGACATCGAT; 1039 (22 nt), CGTGCTGTCTAGAGACATCGAT; 1040 (20 bp), TGCTGTCTAGAGACATCGAT; 1078 (45 nt), GACGCTGCCGAATTCTGGCGTTAGGAGATACCGATAAGCTTCGGC; 1079 (40 nt), GACGCTGCCGAATTCTGGCGTTAGGAGATACCGATAAGCT; 1080 (35 nt), GACGCTGCCGAATTCTGGCGTTAGGAGATACCGAT; 1081 (30 nt), GACGCTGCCGAATTCTGGCGTTAGGAGATA; 1082 (28 nt), GACGCTGCCGAATTCTGGCGTTAGGAGA; 1083 (26 nt), GACGCTGCCGAATTCTGGCGTTAGGA; 1084 (25 nt), GACGCTGCCGAATTCTGGCGTTAGG; 1115 (24 nt), GACGCTGCCGAATTCTGGCGTTAG; 1116 (23 nt), GACGCTGCCGAATTCTGGCGTTA; 1117 (22 nt), GACGCTGCCGAATTCTGGCGTT; 1118 (21 nt), GACGCTGCCGAATTCTGGCGT; 1119 (20 nt), GACGCTGCCGAATTCTGGCG; 1122 (26 nt), GGCCGTGCTGTCTAGAGACATCGAT; 1123 (28 nt), AGGGCTCGTGCTGTCTAGAGACATCGAT; 1124 (30 nt), TTAGGGCTCGTGCTGTCTAGAGACATCGAT; 1125 (23 nt), TCGTGCTGTCTAGAGACATCGAT; 1126 (21 nt), GTGCTGTCTAGAGACATCGAT; 1127 (49 nt), TTAAGCCGAAGCTTATCGGTATCTGCTCGTGCTGTCTAGAGACATCGAT.
RESULTS
Substrate specificity of Mus81-Mms4 and Rad1-Rad10.
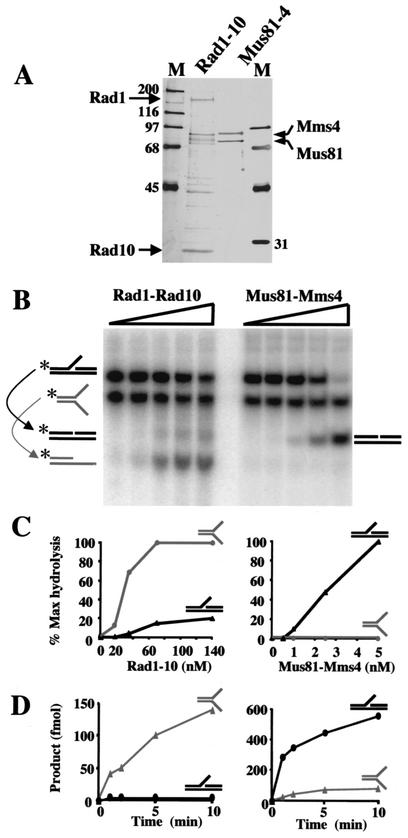
To enzymatically distinguish Mus81-Mms4 from Rad1-Rad10, we compared their endonuclease activities in vitro. The Rad1-Rad10 complex was expressed and purified with a recombinant expression system similar to that used to obtain Mus81-Mms4 (27). Purified Rad1-Rad10 consisted primarily of 160- and 27-kDa polypeptides, along with some Rad1 breakdown products, while Mus81-Mms4 consisted of 72- and 80-kDa proteins (Fig. 1A). To determine the extent of substrate overlap, we assayed both enzymes on simple Y or 3′ flap substrates. These substrates have highly related DNA sequences: they contain the same 5′-end-labeled oligonucleotide (oligonucleotide 888, 49 nt) and an unlabeled complement (oligonucleotide 1128, 50 nt), which pair to form the simple Y substrate (Fig. 1B, gray substrate). A third, unlabeled, oligonucleotide (oligonucleotide 994, 25 nt) anneals to create the 3′ flap substrate (Fig. 1B, black substrate). An equimolar mixture of the two substrates with the same specific activity was incubated with increasing concentrations of each enzyme. With this protocol, Rad1-Rad10 exhibited a fivefold preference for the simple Y substrate over the 3′ flap substrate. This result is consistent with similar studies with the XPF-ERCC1 endonuclease (11). Mus81-Mms4 preferred the 3′ flap over the simple Y by 100-fold (Fig. 1B and C). Substrate specificity was more pronounced with a kinetic analysis and individual substrates. Under these conditions, each enzyme preferred its specific substrate over the other by at least 2 orders of magnitude (Fig. 1D). We note that, on the preferred substrates, Mus81-Mms4 is approximately 20- to 30-fold more active than Rad1-Rad10. The specific activity of the enzymes used here is similar to that of previously published preparations from human and yeast sources (1, 11, 27, 42). We conclude that the in vitro activities of Rad1-Rad10 and Mus81-Mms4 are distinct with respect to substrate preference.
FIG. 1.
Purification and substrate preference of Rad1-Rad10 and Mms4-Mus81. (A) Five hundred nanograms of Rad1-10 and 250 ng of Mms4-Mus81 were subjected to sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis and visualized by silver staining. The positions of the individual subunits His6-Rad1 (160 kDa), Rad10 (27 kDa), His6-Mms4 (80 kDa), and Mus81 (72 kDa) are shown. Molecular size markers (lanes M) are indicated in kilodaltons. (B) Increasing concentrations of Rad1-10 (0, 18, 35, 70, and 140 nM) or Mms4-Mus81 (0, 0.5, 1.2, 2.5, and 5.0 nM) were incubated with 50 nM (each) 32P-labeled simple Y and 3′ flap substrates, and the products were resolved on a 10% native polyacrylamide gel prior to autoradiography. (C) The data in panel B were quantitated and are presented as percent maximum (Max) hydrolysis for Rad1-Rad10 (left) and Mus81-Mms4 (right). (D) Sixty-seven nanomolar Rad1-10 (left) or 5 nM Mus81-Mms4 (right) was incubated in the presence of a single 32P-labeled substrate (50 nM). Eighteen-microliter aliquots were removed at the indicated times and analyzed as described above, and the product contained inthe aliquot was plotted as a function of time. Simple Y and 3′ flap substrates were assembled from oligonucleotides *888/1128 and *888/1128/994, respectively. The asterisks indicate 5′ 32P labeling.
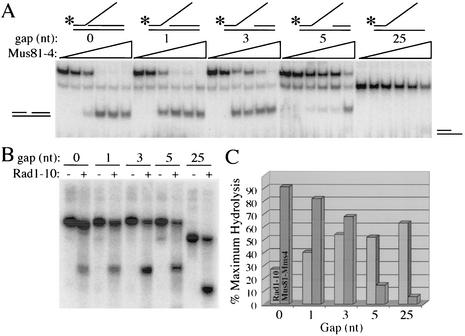
To investigate the nature of this substrate specificity, we constructed a series of branched substrates that were intermediate between the simple Y and the 3′ flap. As shown in Fig. 2A (top), these substrates contained increasing amounts of ssDNA immediately 3′ of the branch junction. Each substrate was incubated with increasing concentrations of Mus81-Mms4 and analyzed by native gel electrophoresis. Intermediate amounts of Mus81-Mms4 were sufficient to completely digest the 3′ flap substrate (gap = 0) or the substrate containing a 1-nt gap (Fig. 2A). However, cleavage efficiency declined as the gap size increased to 3 nt and was drastically reduced with a gap of 5 nt. As before, the Y substrate (i.e., gap = 25 nt) was virtually resistant to cleavage under these conditions (Fig. 2A). The efficiency of cleavage obtained with 1.6 nM Mus81-Mms4 was determined for each substrate (Fig. 2A, lanes 5) and plotted as a function of gap size (Fig. 2C).
FIG. 2.
Inverse activities of Mms4-Mus81 and Rad1-Rad10. (A) Mms4-Mus81 (0, 0.2, 0.4, 0.8, 1.6, or 3.2 nM) was incubated in the presence of a 0.1 nM concentration of the indicated substrate, and the products were analyzed on a 10% native polyacrylamide gel. Gap refers to the number of unpaired nucleotides 3′ of the flap. (B) Duplicate reactions were incubated with (+) or without (−) 70 nM Rad1-10 endonuclease in the presence of a 50 nM concentration of the indicated gapped substrate, as illustrated in panel A. (C) Following quantitation, the product obtained with 1.6 nM Mms4-Mus81 in panel A and 70 nM Rad1-Rad10 in panel B was plotted as a function of substrate gap size. Substrates were assembled as follows: 3′ flap, 0-bp gap (*888/1128/994), 1-bp gap (*888/1128/1038), 3-bp gap (*888/1128/1039), or 5-bp gap (*888/1128/1040); simple Y, 25-bp gap (*888/1128).
These five substrates were then incubated with 70 nM Rad1-Rad10 and analyzed as described above (Fig. 2B). Although the overall efficiency of cleavage was less than that obtained with Mus81-Mms4, a greater fraction of the substrate was hydrolyzed by Rad1-Rad10 if it contained a larger gap downstream of the branch point. This information was quantified as described above and plotted (Fig. 2C). This presentation confirms that there is an inverse relationship between Rad1-Rad10 and Mus81-Mms4 with respect to substrate specificity. We also conclude that flap substrates containing a gap of 5 nt or more are cleaved poorly by Mus81-Mms4.
Mapping of the Mus81-Mms4 cleavage site.
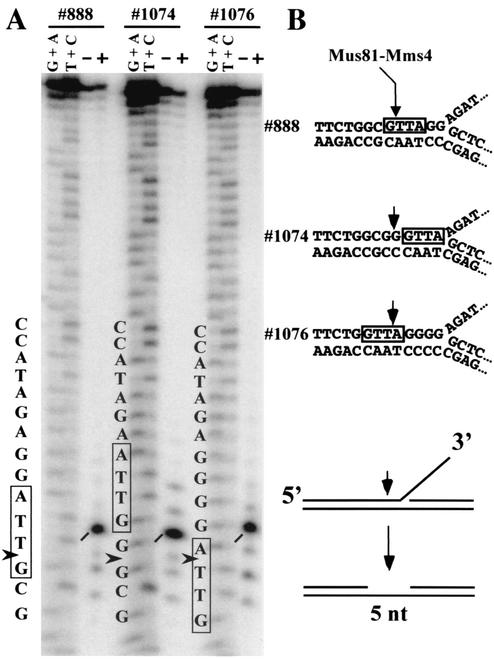
To determine the site of cleavage by Mus81-Mms4, we used a flap substrate and subjected its reaction products to sequencing gel analysis alongside a chemical sequencing ladder of the labeled oligonucleotide. The flap-containing strand (oligonucleotide 888) was cleaved by Mus81-Mms4 to yield a single intense band with significantly weaker bands distributed ± 3 nt (Fig. 3A). This cleavage product terminates in a 3′ OH, as it can be extended by a proofreading-deficient DNA polymerase (data not shown). In this gel system, DNA strands with 3′ hydroxyls interdigitate with the corresponding Maxam-Gilbert ladder. Because chemical sequencing products retain a charged 3′ phosphate, the Mus81-Mms4 cleavage product from the oligonucleotide 888 substrate is judged to comigrate with the faster-migrating band corresponding to the first T residue of the sequence 5′-GTTA-3′ (boxed in Fig. 3). Given that chemical sequencing eliminates the 3′-terminal base, this alignment is consistent with hydrolysis of the substrate by Mus81-Mms4 5′ to this residue (i.e., 5′-GˇTTA-3′). To test whether there is any sequence specificity at this cleavage site, we designed and assayed two new substrates in which the sequence 5′-GTTA-3′ was moved two bases in either the 3′ (oligonucleotide 1074) or the 5′ (oligonucleotide 1076) direction. Analysis of the reaction products revealed that cleavage occurred at the same position relative to the branch point (Fig. 3A), 5 nt 5′ of the flap, independent of the DNA sequence (Fig. 3B). On the basis of these and additional homopolymeric substrates (data not shown), we conclude that hydrolysis by Mus81-Mms4 is structure specific and sequence independent. Similar results have been reported for Rad1-Rad10 and XPF-ERCC1 cleavage of simple Y's, except that there are two major sites of cleavage by the NER enzymes, 2 and 3 nt 5′ of the duplex-ssDNA junction (1, 11, 41). In the case of Mus81-Mms4, the cleavage product is expected to be duplex DNA with a 5-nt gap (Fig. 3B, bottom).
FIG. 3.
The cleavage site of Mus81-Mms4 is sequence independent. (A) 3′ flap junctions were incubated in the presence (+) or absence (−) of Mus81-Mms4, and the products were resolved by sequencing gel electrophoresis alongside a chemical sequencing ladder of the labeled oligonucleotide. As indicated, the Maxam-Gilbert ladder of products runs faster than corresponding fragments terminating in a 3′ OH. (B) Sequences, structures, and cleavage sites of the substrates used in the experiment whose results are shown in panel A.
Mechanism of Mus81-Mms4 cleavage site selection.
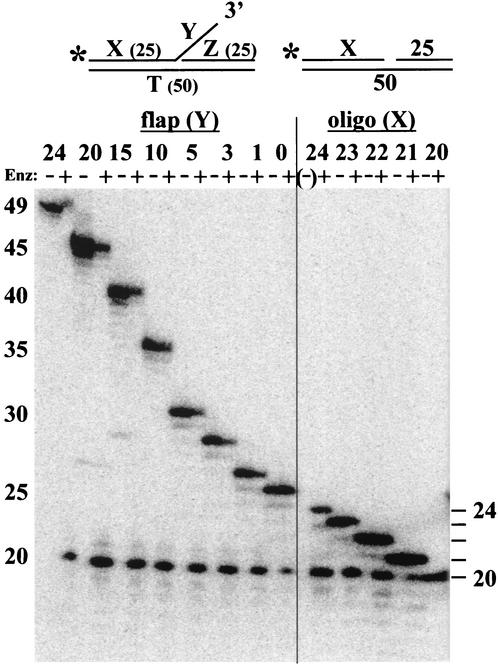
To define further the DNA structural requirements for Mus81-Mms4 activity, we constructed a series of flap substrates with decreasing flap sizes. After incubation with Mus81-Mms4, the reaction products were analyzed with a sequencing gel. The original substrate containing a 24-nt flap (Y) and a 25-bp duplex region (X) was hydrolyzed to produce a 20-nt product consistent with cleavage 5 nt 5′ to the flap (Fig. 4, left). As the flap size decreased, the site of cleavage remained unchanged. Surprisingly, the substrate with a flap size of 0 nt (i.e., nicked DNA) was still hydrolyzed 5 nt upstream of the nick. We note, however, that the product yield with nicked substrates was reduced somewhat compared to that with flap-containing substrates. The cleavage of a nicked substrate suggested that a crucial determinant for Mus81-Mms4 endonuclease activity is recognition of the nick at the flap junction. To determine whether the enzyme recognized the 3′ or 5′ side of the nick, substrates were designed in which the upstream oligonucleotide (X) was truncated at its 3′ end to produce gapped substrates for treatment with Mus81-Mms4 (Fig. 4, right). As oligonucleotide X was truncated from 24 to 21 nt, the site of hydrolysis remained unchanged to consistently yield a 20-nt product. As expected, a substrate with a gap of 5 nt was not digested (Fig. 4, right; X = 20 nt).
FIG. 4.
Upstream determinants for Mus81-Mms4 cleavage. Various 3′ flap (left) or gapped duplex (right) substrates were designed with the indicated flap (Y) or upstream (X) sizes given in nucleotides. Substrates were incubated with (+) or without (−) Mus81-Mms4, and the products were resolved on a 10% sequencing gel. Substrates were assembled as follows: 3′ flap with a 24-nt branch (*888/1128/994) or 20- to 0-nt branches (*1078-1084/1128/994); gapped-duplex substrates (*1115-1119/1128/994). A minus sign indicates a lane in which the untreated substrate remained in the well. The values on the left and right are numbers of nucleotides. Enz, enzyme; oligo, oligonucleotide; T, template.
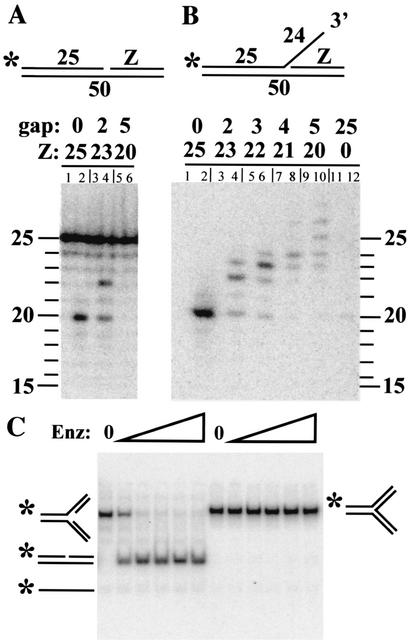
Our results are consistent with the hypothesis that Mus81-Mms4 recognizes the positions of the two oligonucleotides to generate a product with a 5-nt gap. We tested this idea by truncating the downstream oligonucleotide (Z) of the nicked substrate by 2 nt to produce a 2-nt gapped substrate (Fig. 5A). As expected, the digestion product correspondingly increased in size by 2 nt (Fig. 5A, lane 4), although a weak 20-nt band remained. This result is consistent with the idea that Mus81-Mms4 detects the 5′ end of the downstream oligonucleotide (Z) and cleaves 5 nt upstream. The unexpected heterogeneity may have resulted from the use of suboptimal substrates lacking a flap.
FIG. 5.
Downstream determinants of Mus81-Mms4 cleavage. Various duplex (A) or 3′ flap (B) substrates were designed with the indicated downstream oligonucleotides (Z). Substrates were incubated with (even-numbered lanes) or without (odd-numbered lanes) Mus81-Mms4, and the products were resolved on a 10% sequencing gel. (C) RF (*888/1128/992/994) or Y-junction (*888/1128/1127) substrates with identical sequences were assembled and incubated at 0.2 nM with 75, 150, 300, 600, 1,200 pM Mus81-Mms4. Reaction mixtures were analyzed by native gel electrophoresis. Reaction substrates and products are illustrated. Enz, enzyme.
To confirm the role of the 5′ end of the downstream oligonucleotide (Z), we tested the preferred flap substrate and truncated its downstream oligonucleotide. Sequencing gel analysis indicated that the original flap substrate, containing no gap, was cleaved precisely to yield a single 20-nt product (Fig. 5B, lane 2). When truncated versions of oligonucleotide Z were present downstream of the flap, the substrate was cleaved with less specificity, generating small distributions of products. However, the center of each distribution moved in tandem with the downstream oligonucleotide and was localized precisely 5 nt upstream of the 5′ end of oligonucleotide Z. That is, when the gap between oligonucleotide Z and the flap was 2 nt, the most abundant product was increased from 20 to 22 nt (Fig. 5B, lane 4); with a gap of 3 nt, the product was 23 nt (lane 6); with a gap of 4 nt, the product was 24 nt (lane 8); and with a gap of 5 nt, the product was 25 nt (lane 10). In the absence of any downstream oligonucleotide (i.e., a gap of 25 nt), a weak band of 20 nt was detected (lane 12; product = 7% of that obtained in lane 2). Taken together, these results indicate that the primary determinant of Mus81-Mms4 cleavage site selection involves recognition of the 5′ end of the strand present at the flap junction.
To confirm the requirement for a 5′ end, as opposed to adjacent duplex DNA, we designed a fully duplex Y junction and assayed it relative to an RF substrate with the same sequence. As shown in Fig. 5C, when these substrates were incubated with increasing amounts of Mus81-Mms4, the RF substrate was cleaved efficiently while the duplex Y junction lacking a 5′ end was resistant to a 16-fold excess of Mus81-Mms4. Analysis of these reaction products by denaturing gel electrophoresis confirmed that the labeled strand was undigested (data not shown). The requirement for a 5′ end at the flap junction has also been reported for partially purified preparations of human Mus81-associated endonuclease (8).
MUS81-MMS4 functional analysis.
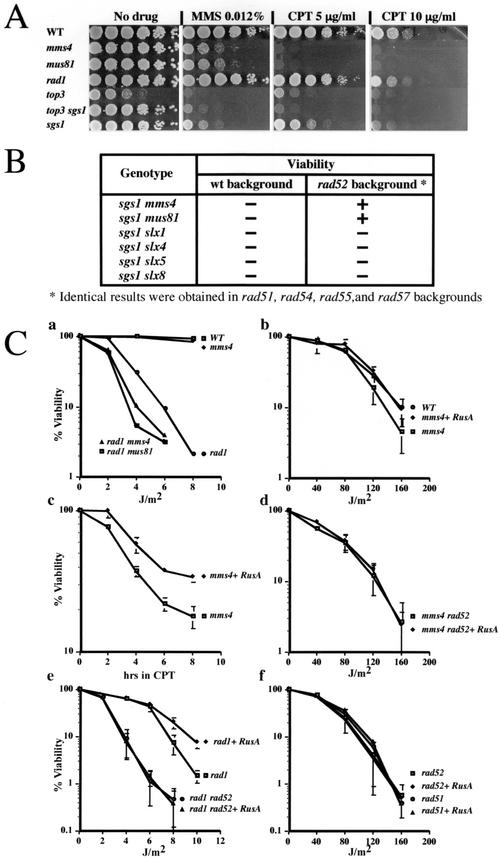
Inactivation of any one of the genes encoding Mus81-Mms4 or Rad1-Rad10 results in sensitivity to radiomimetic DNA-damaging agents. However, rad1 rad10 mutants are sensitive to low levels of UV light and high concentrations of the DNA-methylating agent methyl methanesulfonate while mus81 mms4 mutants are sensitive to high levels of UV and low levels of methyl methanesulfonate (23, 34, 48). We found that the rad1 mms4 mutant strain was more sensitive than either single mutant to low doses of UV, as was the rad1 mus81 double mutant (Fig. 6C, part a; reference 23; data not shown). These epistasis tests indicate that MUS81-MMS4 and RAD1-RAD10 function in separate pathways with respect to DNA damage tolerance.
FIG.6.
MUS81-MMS4 functions downstream of homologous recombination. (A) S. cerevisiae strains with the indicated genotypes were concentrated to an optical density at 600 nm of 3.0 and serially diluted 10-fold, and approximately 5-μl volumes were spotted on YPD (38) plates containing the indicated drug. Plates were photographed following 3 days of growth at 30°C. WT, wild type. (B) The indicated double and triple mutants were constructed by genetic crosses between appropriately marked strains, some of which carried a complementing SGS1/URA3 plasmid (34). Following sporulation and tetrad dissection, the viability of the meiotic segregants was determined by attempting to eliminate the SGS1/URA3 plasmid by growth on medium containing 5-fluoroorotic acid. wt, wild type. (C, part a) Yeast cells with the indicated genotypes were spread on YPD plates and irradiated with UV light, and viability was determined following 3 days of growth at 30°C. (C, parts b to f) Yeast cells carrying an empty vector (pRS415) or the RusA expression plasmid (pKR6980) were spread on the appropriate selective plates and treated as in the experiment whose results are shown in part a. For CPT sensitivity testing, yeast cells were grown in selective medium containing CPT at 5 μg/ml. After the indicated times, aliquots of cells were removed, washed once, and plated on selective plates lacking CPT. Viability was then determined following 3 days of growth at 30°C. WT, wild type.
To identify a unique function for Mus81-Mms4, we searched for DNA-damaging agents that could distinguish between the MUS81-MMS4 and NER pathways in a qualitative way. The mus81 and mms4 mutants were found to be sensitive to CPT at 5 μg/ml, as were sgs1 and top3 mutant cells (Fig. 6A). CPT, an inhibitor of DNA Top1, is known to induce replication-dependent DSBs as forks replicate across Top1/CPT-induced DNA lesions (31, 36). The CPT-sensitive phenotype of these mutants has recently been reported by others (30, 44) and is conserved in S. pombe rqh1, eme1, and mus81 mutants (13). However, CPT sensitivity is not observed in S. cerevisiae rad1 mutant cells, even at higher concentrations (10 μg/ml), nor is it observed in four other slx mutants isolated in the sgs1 synthetic-lethal screen (Fig. 6A and data not shown). We conclude that Sgs1-Top3 and Mus81-Mms4 play specific roles in the repair of replication-dependent DSBs induced by CPT.
Homologous recombination is known to play a critical role in the repair of CPT-induced DSBs (15, 37). The idea that Sgs1-Top3 and Mus81-Mms4 act in this recombination-mediated repair pathway is consistent with evidence that they function in meiotic recombination: Specifically, the sporulation defects of mus81 mms4 and sgs1 top3 mutant cells can be suppressed by eliminating the initiation of meiotic recombination (3, 12, 18, 27). To explore this idea, we tested whether mus81 sgs1 synthetic lethality could be suppressed by eliminating recombination in vegetative cells. As documented in Fig. 6B, the viability of mus81 sgs1 or mms4 sgs1 mutants was restored by eliminating any one of five RAD52 epistasis group genes (RAD51, RAD52, RAD54, RAD55, or RAD57). The ability of these mutations to suppress sgs1 synthetic lethality was not a general one, however, as none of the other four slx sgs1 combinations was suppressed (Fig. 6B). These results are in complete agreement with a similar analysis recently reported by Fabre and colleagues (16). The simplest interpretation of this result is that Mus81-Mms4 and Sgs1-Top3 have the same in vivo substrate that is the product of homologous recombination. We conclude that Mus81-Mms4 and Sgs1-Top3 act following the initiation of homologous recombination in vegetative cells.
To distinguish further the functions of MUS81-MMS4 and RAD1-RAD10, we expressed RusA, the bacterial HJ resolvase, in mutant cells. Expression of RusA in S. pombe partially suppresses the phenotypes of mus81 mutant cells and the UV and hydroxyurea sensitivity of rqh1 mutant strains (3, 13, 14). Consistent with the S. pombe results, an mms4 mutant strain carrying a constitutive RusA expression plasmid showed complete suppression of its UV sensitivity (Fig. 6C, part b) and partial suppression of its CPT sensitivity (Fig. 6C, part c). RAD52 was required for suppression of UV sensitivity by RusA (Fig. 6C, part d), in agreement with the idea that Mus81-Mms4 function is dependent on homologous recombination. We next tested whether RusA is capable of complementing defects in RAD1/RAD10 function. RusA partially suppressed the UV sensitivity of rad1 mutant cells (Fig. 6C, part e), as well as S. pombe swi10 mutant cells (Swi10 is the ortholog of Rad10/ERCC1; data not shown). As in the mms4 background, suppression of rad1 by RusA expression also required RAD52 function.
It has been proposed that the suppression of UV sensitivity by RusA expression is due to its ability to cleave HJs that arise when RFs, stalled at sites of UV-induced DNA damage, undergo fork regression (3, 5, 13, 14). Cleavage of regressed forks by resolvase should create DSBs that are dependent on subsequent recombination for efficient repair. We tested the prediction that cleavage of damage-induced HJs by RusA would be detrimental to a strain lacking homologous recombination. However, as shown in Fig. 6C, part f, expression of RusA did not alter the viability of rad51 or rad52 mutant cells after exposure to UV. We conclude that, under these conditions, RusA expression does not contribute significantly to the formation of DSBs.
DISCUSSION
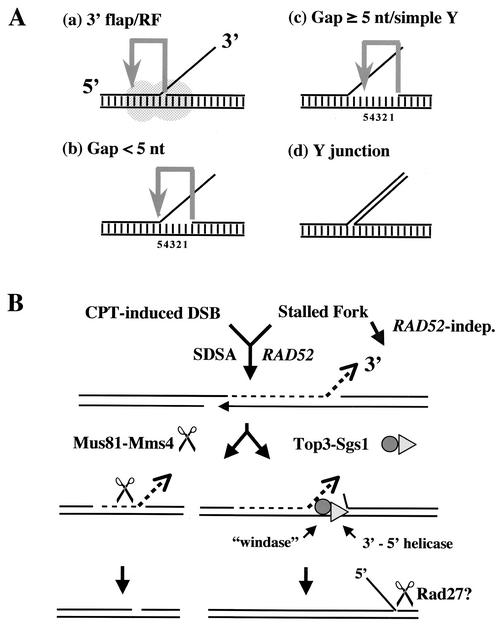
This study has demonstrated that the homologous structure-specific endonucleases Rad1-Rad10 and Mus81-Mms4 have distinct substrate preferences. Kinetic analyses indicate that each of these enzymes has an at least 100-fold preference for its specific substrate over that of the other. The presence of a 5′ end of DNA at the flap junction appears to be sufficient to explain this difference, as well as the mechanism of cleavage site selection by Mus81-Mms4. As shown in Fig. 7A, Mus81-Mms4 recognizes the 5′ end of the nicked DNA to direct cleavage 5 nt upstream (gray arrow). Such a model explains several features of Mus81-Mms4 activity. A nuclease that specifically recognizes the 5′ end at a flap junction would be expected to bind both 3′ flap and RF substrates (Fig. 7A, part a). Since cleavage occurs in the duplex region 5 nt upstream of this end, these substrates should be cleaved with the same efficiency, which was demonstrated previously (27). As shown here, the minimal substrate requirement for Mus81-Mms4 cleavage is a 3′ flap with a gap of 4 nt or less (Fig. 7A, part b). Although flap substrates with small gaps are cleaved by Mus81-Mms4, these events are imprecise and generate a distribution of products. This heterogeneity may be due to the difficulty in nicking substrates close to the flap junction if the duplex is susceptible to transient denaturation. Substrates with gaps of 5 nt or more, like simple Y's, are relatively resistant to cleavage (Fig. 7A, part c), as are duplex substrates lacking a 5′ end, like a Y junction (Fig. 7A, part d). On the latter two substrates, Mus81-Mms4 displays a minor cleavage activity upstream of the branch. This nonspecific in vitro activity may be a general property of XPF family endonucleases that recognize substrates containing duplex-ssDNA junctions (1, 11). On the basis of their nonoverlapping phenotypes, however, it appears that this minor cleavage activity is not biologically significant. Finally, we noted that while the length of the 3′ flap did not appear to affect Mus81-Mms4 nuclease activity in vitro, cleavage efficiency was compromised on substrates containing only a nick (Fig. 4). Thus, we suggest that 3′ flap structures are the relevant biological substrate of Mus81-Mms4.
FIG. 7.
Mechanism of Mus81-Mms4 cleavage and its functional overlap with Sgs1-Top3. (A) Mus81-Mms4 (transparent gray dimer) recognizes the 5′ end of the DNA at a flap junction and nicks the flap strand 5 nt upstream within the DNA duplex (gray arrow). Conditions a through d illustrate Mus81-Mms4 activity on various substrates as described in the text. (B) Model of DNA repair following fork stalling or a CPT-induced DSB illustrating the formation of a 3′ flap substrate following SDSA. This substrate is appropriate for cleavage by Mus81-Mms4 to create a 5-nt gap (left). Alternatively, Sgs1-Top3 is proposed to process this substrate by displacing the downstream strand and annealing the newly synthesized flap DNA (right). indep., independent.
Earlier studies could not address the question of whether the 3′ flap or the RF is the in vivo substrate for Mus81-Mms4 since the enzyme is equally active on these two substrates. We had therefore suggested that Mus81-Mms4 might cleave stalled RFs in order to reinitiate replication via homologous recombination (27). In light of the present results, there are several reasons to believe that the in vivo substrate of this enzyme is a 3′ flap structure. First, the requirement for Mus81-Mms4 and Sgs1-Top3 to tolerate CPT-induced DNA damage suggests that these activities are needed in response to RF collapse, not to catalyze their collapse. Second, the requirement for a 5′ end to be within 4 nt of the flap junction is inconsistent with the extensive ssDNA that is likely to be present at a fork. With an RF as the substrate, the 5′ end of the DNA at the flap junction would belong to an RNA-primed Okasaki fragment on the lagging strand template. It is unlikely that Okasaki fragments are synthesized at this proximity to the branch given the requirement for proteins such as RPA on the lagging-strand template and the idea that the lagging strand is an extended loop. Finally, suppressor analysis indicates that Mus81-Mms4 and Sgs1-Top3 act downstream, not upstream, of the initiation of recombination.
Unlike Rad1-Rad10, Mus81-Mms4 is important for tolerating CPT-induced DNA damage. It should be noted that Rad1-Rad10 may play a role in repairing CPT-induced DNA damage in certain genetic backgrounds, however (30, 44). The CPT sensitivity of mus81 mms4 mutants is likely to be significant since sgs1 top3 mutant cells share this phenotype, as do the corresponding S. pombe mutants (13), and it may provide a clue regarding the overlapping functions of these enzymes. It is known that the major pathway for the repair of CPT-induced DSBs requires homologous recombination (15, 35, 37). It is also known that, in the case of UV-induced DNA damage, recombinational repair utilizes sister chromatids as a template, occurs largely during DNA replication, and may be distinct from the repair of DSBs induced by ionizing radiation in G2 (25, 26). Thus, the CPT sensitivity of cells lacking Mus81-Mms4 or Sgs1-Top3 argues for a role for these enzymes in recombinational DNA repair of collapsed RFs, most likely involving sister chromatids. The synthetic lethality resulting from the loss of both Mus81-Mms4 and Sgs1-Top3 then suggests that these enzymes are required to repair forks that collapse spontaneously, as well as those that collapse in response to CPT.
An alternative role for Mus81-Mms4 and Sgs1-Top3 is the stabilization of stalled RFs. In a variation of the template switch model (22), it has been argued that stalled RFs regress into a more stable HJ. This HJ could then be reversed by a branch-migrating DNA helicase, like Sgs1 or BLM, or it could be cleaved by HJ resolvase to create a DSB. Evidence of fork regression has been obtained under lethal conditions in bacteria lacking replicative DNA helicase activity (40) or yeast cells lacking checkpoint control (32). Evidence that regression plays a role in DNA damage tolerance under less severe conditions in eukaryotes is lacking. The ability of RusA expression to suppress the phenotypes of S. pombe rqh1 and mus81 mutants has been taken as evidence of such a mechanism (3, 13, 14), and we have confirmed that RusA expression can suppress mms4-induced phenotypes in S. cerevisiae. But the interpretation that RusA suppression involves the cleavage of regressed forks is highly dependent on the specificity of RusA for HJs. Interestingly, two recent studies found that the specificity of RusA for cleavage of HJs was high but less than absolute; RusA was observed to cleave RFs and other branched substrates in vitro (5, 13). Although it has been argued that RusA is only active on HJs in E. coli (5), its lack of specificity in vitro suggests that it may be similarly promiscuous in a eukaryotic nucleus. A simpler and more direct explanation for RusA suppression of the phenotypes caused by mus81, mms4, and rqh1 is that it can cleave 3′ flaps in vivo. Even though RusA is a potent HJ resolvase, a 3′ flap endonuclease activity, albeit minor in vitro, may be significant when overexpressed in yeast. This explanation eliminates the problem of why fission yeast's endogenous resolvase was insufficient to suppress the rqh1 mutant phenotypes (14) and is consistent with the failure of RusA expression to show any detrimental effect in budding yeast lacking RAD52. In that regard, it is possible that RAD52 is needed for fork regression since it is required for HJs to form in the rRNA gene of replication mutants (49). However, HJ formation in this system did not require RAD51 (49). Since RusA expression had no effect in rad51 cells either, it seems unlikely that failure to regress can explain the lack of DSBs caused by RusA. Indeed, given the potency of RusA resolvase activity in vitro, we argue that fork regression occurs rarely, if at all, in viable cells. RusA may partially complement the rad1-induced phenotype by the same mechanism. The increase in fork stalling, or collapse, that occurs because of unrepaired thymine dimers in NER mutants may overwhelm the RAD52-dependent Mus81-Mms4 pathway, which could benefit from excess RusA flap endonuclease activity.
Finally, we consider a more parsimonious model for repair of DNA damage by Mus81-Mms4 and Sgs1-Top3 that does not invoke HJ intermediates. On the basis of a previous model of Mus81-Mms4 function in meiosis (12), we suggest that stalled or collapsed RFs attempt repair with synthesis-dependent strand annealing (SDSA; Fig. 7B). SDSA requires the RAD52 epistasis group genes, and in this case, we suggest that it is initiated by a DSB, although other initiating lesions are conceivable (16). Following invasion of the sister chromatid by a broken 3′ end, a fraction of molecules extended by DNA polymerase during SDSA will be overreplicated such that, following displacement and reannealing, the product will contain a 3′ flap that can be cleaved by Mus81-Mms4. In the absence of Mus81-Mms4, Sgs1-Top3 drives the annealing step of SDSA to completion by virtue of the ability of the helicase to displace the downstream strand. This model proposes that Top3 acts as a windase to assist what is essentially a strand exchange reaction. Although Top3 may not be required for this reaction in vitro, it may be needed to overcome topological constraints resulting from extreme overreplication in vivo. The product of this reaction would contain a 5′ ssDNA branch that is an appropriate substrate for Rad27/FEN1. In addition to addressing the redundancy of MMS4-MUS81 and SGS1-TOP3 downstream of a RAD52-dependent reaction, this model accounts for the requirement for Top3 in Sgs1 function and the fact that Top3 relaxes negatively, not positively, supercoiled DNA. In addition, the slow-growth and hyperrecombinational phenotypes of top3 mutants would be explained by the generation of ssDNA by the Sgs1 helicase. Finally, this model predicts that mutations in MUS81 and RAD27 should be synthetically lethal, which has been previously reported (43). As shown in Fig. 7B, the ability of RAD52 mutations to suppress sgs1 mus81 synthetic lethality implies the existence of a RAD52-independent repair mechanism for stalled or collapsed forks. The identification of players in this pathway could provide a test of this model.
Acknowledgments
S.A.B.-S. and W.M.F. contributed equally to this work.
We gratefully acknowledge Tony Carr, Greg Freyer, Hannah Klein, Nancy Walworth, and Matthew Whitby for contributing strains and plasmids. We also thank Val Carabetta for expert technical assistance. Camptothecin was a gift from the Drug Synthesis & Chemistry Branch, DTP, DCTD, NCI.
This work was supported by grant AG16637 from the National Institutes of Health.
REFERENCES
- 1.Bardwell, A. J., L. Bardwell, A. E. Tomkinson, and E. C. Friedberg. 1994. Specific cleavage of model recombination and repair intermediates by the yeast Rad1-Rad10 DNA endonuclease. Science 265:2082-2085. [DOI] [PubMed]
- 2.Bennett, R. J., M. F. Noirot-Gros, and J. C. Wang. 2000. Interaction between yeast Sgs1 helicase and DNA topoisomerase III. J. Biol. Chem. 275:26898-26905. [DOI] [PubMed] [Google Scholar]
- 3.Boddy, M. N., P. H. Gaillard, W. H. McDonald, P. Shanahan, J. R. Yates III, and P. Russell. 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107:537-548. [DOI] [PubMed] [Google Scholar]
- 4.Boddy, M. N., A. Lopez-Girona, P. Shanahan, H. Interthal, W. D. Heyer, and P. Russell. 2000. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 20:8758-8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolt, E. L., and R. G. Lloyd. 2002. Substrate specificity of RusA resolvase reveals the DNA structures targeted by RuvAB and RecG in vivo. Mol. Cell 10:187-198. [DOI] [PubMed] [Google Scholar]
- 6.Chakraverty, R. K., and I. D. Hickson. 1999. Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. Bioessays 21:286-294. [DOI] [PubMed] [Google Scholar]
- 7.Chen, X. B., R. Melchionna, C. M. Denis, P. H. Gaillard, A. Blasina, I. Van de Weyer, M. N. Boddy, P. Russell, J. Vialard, and C. H. McGowan. 2001. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell 8:1117-1127. [DOI] [PubMed] [Google Scholar]
- 8.Constantinou, A., X. B. Chen, C. H. McGowan, and S. C. West. 2002. Holliday junction resolution in human cells: two junction endonucleases with distinct substrate specificities. EMBO J. 21:5577-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Courcelle, J., C. Carswell-Crumpton, and P. C. Hanawalt. 1997. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc. Natl. Acad. Sci. USA 94:3714-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox, M. M. 2001. Historical overview: searching for replication help in all of the rec places. Proc. Natl. Acad. Sci. USA 98:8173-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Laat, W. L., E. Appeldoorn, N. G. Jaspers, and J. H. Hoeijmakers. 1998. DNA structural elements required for ERCC1-XPF endonuclease activity. J. Biol. Chem. 273:7835-7842. [DOI] [PubMed] [Google Scholar]
- 12.de los Santos, T., J. Loidl, B. Larkin, and N. M. Hollingsworth. 2001. A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics 159:1511-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doe, C. L., J. S. Ahn, J. Dixon, and M. C. Whitby. 2002. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 25:25.. [DOI] [PubMed] [Google Scholar]
- 14.Doe, C. L., J. Dixon, F. Osman, and M. C. Whitby. 2000. Partial suppression of the fission yeast rqh1− phenotype by expression of a bacterial Holliday junction resolvase. EMBO J. 19:2751-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eng, W. K., L. Faucette, R. K. Johnson, and R. Sternglanz. 1988. Evidence that DNA topoisomerase I is necessary for the cytotoxic effects of camptothecin. Mol. Pharmacol. 34:755-760. [PubMed] [Google Scholar]
- 16.Fabre, F., A. Chan, W. D. Heyer, and S. Gangloff. 2002. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA 99:16887-16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fricke, W. M., V. Kaliraman, and S. J. Brill. 2001. Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J. Biol. Chem. 276:8848-8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gangloff, S., B. de Massy, L. Arthur, R. Rothstein, and F. Fabre. 1999. The essential role of yeast topoisomerase III in meiosis depends on recombination. EMBO J. 18:1701-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gangloff, S., J. P. McDonald, C. Bendixen, L. Arthur, and R. Rothstein. 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14:8391-8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 21.Guldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgins, N. P., K. Kato, and B. Strauss. 1976. A model for replication repair in mammalian cells. J. Mol. Biol. 101:417-425. [DOI] [PubMed] [Google Scholar]
- 23.Interthal, H., and W. D. Heyer. 2000. MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet. 263:812-827. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, F. B., D. B. Lombard, N. F. Neff, M. A. Mastrangelo, W. Dewolf, N. A. Ellis, R. A. Marciniak, Y. Yin, R. Jaenisch, and L. Guarente. 2000. Association of the Bloom syndrome protein with topoisomerase IIIα in somatic and meiotic cells. Cancer Res. 60:1162-1167. [PubMed] [Google Scholar]
- 25.Kadyk, L. C., and L. H. Hartwell. 1993. Replication-dependent sister chromatid recombination in rad1 mutants of Saccharomyces cerevisiae. Genetics 133:469-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadyk, L. C., and L. H. Hartwell. 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132:387-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaliraman, V., J. R. Mullen, W. M. Fricke, S. A. Bastin-Shanower, and S. J. Brill. 2001. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15:2730-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, R. A., and J. C. Wang. 1992. Identification of the yeast TOP3 gene product as a single strand-specific DNA topoisomerase. J. Biol. Chem. 267:17178-17185. [PubMed] [Google Scholar]
- 29.Klein, H. 2001. Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2Δ with other DNA repair genes in Saccharomyces cerevisiae. Genetics 157:557-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, C., J. J. Pouliot, and H. A. Nash. 2002. Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc. Natl. Acad. Sci. USA 99:14970-14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, L. F., P. Duann, C. T. Lin, P. D'Arpa, and J. Wu. 1996. Mechanism of action of camptothecin. Ann. N. Y. Acad. Sci. 803:44-49. [DOI] [PubMed] [Google Scholar]
- 32.Lopes, M., C. Cotta-Ramusino, A. Pellicioli, G. Liberi, P. Plevani, M. Muzi-Falconi, C. S. Newlon, and M. Foiani. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412:557-561. [DOI] [PubMed] [Google Scholar]
- 33.Mullen, J. R., V. Kaliraman, and S. J. Brill. 2000. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics 154:1101-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mullen, J. R., V. Kaliraman, S. S. Ibrahim, and S. J. Brill. 2001. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157:103-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nitiss, J., and J. C. Wang. 1988. DNA topoisomerase-targeting antitumor drugs can be studied in yeast. Proc. Natl. Acad. Sci. USA 85:7501-7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nitiss, J. L., and J. C. Wang. 1996. Mechanisms of cell killing by drugs that trap covalent complexes between DNA topoisomerases and DNA. Mol. Pharmacol. 50:1095-1102. [PubMed] [Google Scholar]
- 37.Pouliot, J. J., C. A. Robertson, and H. A. Nash. 2001. Pathways for repair of topoisomerase I covalent complexes in Saccharomyces cerevisiae. Genes Cells 6:677-687. [DOI] [PubMed] [Google Scholar]
- 38.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Rothstein, R., B. Michel, and S. Gangloff. 2000. Replication fork pausing and recombination or “gimme a break.” Genes Dev. 14:1-10. [PubMed] [Google Scholar]
- 40.Seigneur, M., V. Bidnenko, S. D. Ehrlich, and B. Michel. 1998. RuvAB acts at arrested replication forks. Cell 95:419-430. [DOI] [PubMed] [Google Scholar]
- 41.Sijbers, A. M., W. L. de Laat, R. R. Ariza, M. Biggerstaff, Y. F. Wei, J. G. Moggs, K. C. Carter, B. K. Shell, E. Evans, M. C. de Jong, S. Rademakers, J. de Rooij, N. G. Jaspers, J. H. Hoeijmakers, and R. D. Wood. 1996. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell 86:811-822. [DOI] [PubMed] [Google Scholar]
- 42.Tomkinson, A. E., A. J. Bardwell, L. Bardwell, N. J. Tappe, and E. C. Friedberg. 1993. Yeast DNA repair and recombination proteins Rad1 and Rad10 constitute a single-stranded-DNA endonuclease. Nature 362:860-862. [DOI] [PubMed] [Google Scholar]
- 43.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 44.Vance, J. R., and T. E. Wilson. 2002. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc. Natl. Acad. Sci. USA 99:13669-13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, J. C. 1991. DNA topoisomerases: why so many? J. Biol. Chem. 266:6659-6662. [PubMed] [Google Scholar]
- 46.White, M. F., and D. M. Lilley. 1996. The structure-selectivity and sequence-preference of the junction-resolving enzyme CCE1 of Saccharomyces cerevisiae. J. Mol. Biol. 257:330-341. [DOI] [PubMed] [Google Scholar]
- 47.Wu, L., S. L. Davies, P. S. North, H. Goulaouic, J. F. Riou, H. Turley, K. C. Gatter, and I. D. Hickson. 2000. The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 275:9636-9644. [DOI] [PubMed] [Google Scholar]
- 48.Xiao, W., B. L. Chow, and C. N. Milo. 1998. Mms4, a putative transcriptional (co)activator, protects Saccharomyces cerevisiae cells from endogenous and environmental DNA damage. Mol. Gen. Genet. 257:614-623. [DOI] [PubMed] [Google Scholar]
- 49.Zou, H., and R. Rothstein. 1997. Holliday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell 90:87-96. [DOI] [PubMed] [Google Scholar]