Abstract
Human RECQL1 and RECQL5 belong to the RecQ family that includes Bloom syndrome, Werner syndrome, and Rothmund-Thomson syndrome causative genes. Cells derived from individuals suffering from these syndromes show significant levels of genomic instability. However, neither RECQL1 nor RECQL5 has been related to a disease, and nothing is known about the functions of RecQL1 and RecQL5. We generated here RECQL1−/−, RECQL5−/−, RECQL1−/−/RECQL5−/−, RECQL1−/−/BLM−/−, and RECQL5−/−/BLM−/− cells from chicken B-lymphocyte line DT40 cells. Although BLM−/− DT40 cells showed a slow-growth phenotype, a higher sensitivity to methyl methanesulfonate than the wild type, and an ∼10-fold increase in the frequency of sister chromatid exchange (SCE) compared to wild-type cells, RECQL1−/−, RECQL5−/−, and RECQL1−/−/RECQL5−/− cells showed no significant difference from the wild-type cells in growth, sensitivity to DNA-damaging agents, and the frequency of SCE. However, both RECQL1−/−/BLM−/− and RECQL5−/−/BLM−/− cells grew more slowly than BLM−/− cells because of the increase in the population of dead cells, indicating that RecQL1 and RecQL5 are somehow involved in cell viability under the BLM function-impaired condition. Surprisingly, RECQL5−/−/BLM−/− cells showed a higher frequency of SCE than BLM−/− cells, indicating that RecQL5 suppresses SCE under the BLM function-impaired condition.
The Escherichia coli RecQ helicase is one of the proteins involved in the recF recombination pathway that is effective in the recBCD sbcB background (23). In human cells, five genes encoding RecQ helicase homologues have been identified, and three are the causative genes for Bloom syndrome (BS), Werner syndrome (WS), and Rothmund-Thomson syndrome (RTS) (4, 17, 40). The remaining two are RECQL1 and RECQL5 (16, 26, 27).
BS is a rare genetic disorder characterized by retarded growth, sunlight sensitivity, immunodeficiency, male infertility, and predisposition to a wide variety of malignant tumors. The most characteristic feature of BS cells is genomic instability, which is manifested as an elevated frequency of chromosome breaks, interchange between homologous chromosomes, and sister chromatid exchange (SCE) (10). The other two genetic disorders, WS and RTS, also show genomic instability (21).
Before we cloned a cDNA encoding RecQL1, we purified this protein and designated it DNA-dependent ATPase Q1 and later DNA helicase Q1 because of its DNA helicase activity (28, 39). Puranam and Blackshear named the gene encoding this protein RECQL (26). Due to these circumstances and the existence of multiple RecQ homologues in higher eukaryotic cells, we propose to designate this gene RECQL1 (5). Although RecQL1 is a major DNA helicase in human cells, the function of this helicase is not known at all (28).
The fourth and fifth RECQ homologues, originally called RecQ4 and RecQ5 and recently called RECQL4 and RECQL5 (5, 17), were cloned in 1998 after a search for sequences similar to the RecQ helicase motifs in the expressed sequence tag database (16). Mutations in the RECQL4 gene have been found in patients with RTS, a rare genetic disorder characterized by premature aging and a predisposition to cancers such as those typical of WS (17). At present, no genetic disorder has been identified that is caused by mutations in the RECQL1 or RECQL5 gene. This circumstance suggests that RecQL1 and RecQL5 are indispensable for cell viability.
In contrast to human cells, a unicellular eukaryote, Saccharomyces cerevisiae, has a sole gene that encodes a RecQ helicase homologue, SGS1 (slow growth suppressor 1). The sgs1 mutants show multiple phenotypes, such as sensitivity to methyl methanesulfonate (MMS) and hydroxyurea, chromosome missegregation, poor sporulation, premature aging, and checkpoint defects (6, 8, 22, 30, 36, 37). In addition, sgs1 mutants show phenotypes of mitotic hyper-recombination, including interchromosomal homologous recombination, intrachromosomal excision recombination, illegitimate recombination, and unequal sister chromatid recombination (24, 36, 38). Interestingly, the BS gene (BLM) and the WS gene (WRN) partially suppressed the increased homologous and illegitimate recombination in sgs1 mutants (38), suggesting the conservation of function among the proteins encoded by these genes. The SGS1 was originally identified as a suppressor of the slow- growth phenotype of DNA topoisomerase III (TOP3) mutants when it was mutated (8). Indeed, physical interaction of Sgs1 and Top3 has been confirmed (1, 7). In addition, we demonstrated that interaction with Top3 via the N-terminal region of Sgs1 is involved in the complementation of MMS sensitivity and the suppression of hyper-recombination (33). Recent studies revealed that RecQL1, BLM, and RecQL5 interact with DNA Top3α (14, 29), suggesting redundancy of the function of these proteins.
E. coli RecQ can both initiate homologous recombination and disrupt nascent joint molecules in vitro (12). Recently, it was confirmed that SGS1 belongs to the RAD52 recombination repair pathway by an epistasis analysis of MMS sensitivity of sgs1 disruptants (25). In fact, we found that the frequency of interchromosomal recombination that was increased in wild-type cells treated with MMS or UV light was not increased in sgs1 disruptants (9, 25). These results revealed dual functions of Sgs1 to suppress various types of recombination in normal cell growth and to promote homologous recombination under damage-induced conditions. Considering that S. cerevisiae has a sole RecQ homologue, Sgs1, and higher eukaryotic cells have five RecQ homologues, it appears likely that multiple functions of Sgs1 are shared by the multiple RecQL helicases. However, the involvement of RecQ homologues of higher eukaryotic cells in the induction of homologous recombination remains unclear.
To clarify whether RecQL1 and RecQL5 are essential for the viability of cells, we constructed RECQL1−/− cells and RECQL5−/− cells from chicken B lymphocytes, DT40 cells (2), and confirmed that RecQL1 and RecQL5 are not essential for viability. In addition, to clarify the functional redundancy among RecQL1, BLM, and RecQL5 and the involvement of RecQ homologues of higher eukaryotic cells in homologous recombination, we constructed RECQL1−/−/RECQL5−/−, RECQL1−/−/BLM−/−, and RECQL5−/−/BLM−/− cells and examined cell growth, sensitivity to DNA-damaging agents, chromosomal aberrations, targeted integration frequency, and the frequency of SCE.
MATERIALS AND METHODS
Construction of targeting vectors.
A chicken RECQL1 cDNA fragment that corresponded to the helicase domain of the human RECQL1 cDNA sequence was obtained by reverse transcription-PCR (RT-PCR) on chicken testis RNA with primers synthesized based on the human RECQL1 sequence. The C-terminal and N-terminal regions of the chicken RECQL1 gene were obtained by 3′-RACE (rapid amplification of cDNA ends) and 5′-RACE on the chicken testis RNA, respectively. A genomic DNA segment of RECQL1 was amplified by long-range PCR with DT40 genomic DNA as a template. The chicken targeting constructs for RECQL1, RECQL1-histidinolr and RECQL1-blasticidinr, were made by replacing the helicase motif IV with histidinol (His) and blasticidin (Bsr) selection marker cassettes.
A chicken RECQL5 cDNA fragment was isolated during the course of the isolation of chicken BLM cDNA by RT-PCR (34). Full-length RECQL5 cDNA was obtained by 3′-RACE and 5′-RACE. A genomic DNA segment of RECQL5 was amplified by long-range PCR with DT40 genomic DNA as a template. For the chicken targeting constructs for RECQL5, replacing helicase motif Ia with His and Bsr selection marker cassettes yielded RECQL5-histidinolr and RECQL5-blasticidinr.
Cell culture and DNA transfection.
Cells were cultured in RPMI 1640 supplemented with 100 μg of kanamycin/ml, 10% fetal bovine serum, and 1% chicken serum (Sigma, St. Louis, Mo.) at 39.5°C. For gene targeting, 107 DT40 cells were electroporated with 30 μg of linearized targeting constructs by using a Gene Pulser apparatus (Bio-Rad, Hercules, Calif.) at 550 V and 25 μF. Drug-resistant colonies were selected in 96-well plates with medium containing 1 mg of His or 20 μg of Bsr/ml. Genomic DNA was isolated from drug-resistant clones. Gene disruption was confirmed by Southern and Northern blot analyses and RT-PCR. We always isolated several independent clones and used at least two independent clones in each genotype for various experiments to avoid the effects of “clone to clone variation.”
Growth curve.
Cells (2 × 104) were inoculated and cultured at 39.5°C for the specified periods. They were enumerated, and the growth rates were estimated.
Flow cytometry.
The cell cycle distribution and cell death were examined with a FACScan (Becton Dickinson, Mountain View, Calif.). To detect cell death, exponentially growing cells (106) were harvested, washed with phosphate-buffered saline, treated sequentially with trypsin and then RNase A, and then stained with propidium iodide according to the manual of the Cycle Test Plus DNA reagent kit (Becton Dickinson). To analyze the cell cycle distribution, exponentially growing cells (1.5 × 106) were cultured for 15 min in the presence of bromodeoxyuridine (BrdU). The BrdU incorporated into chromosomal DNA and DNA content were detected by fluorescein isothiocyanate-conjugated anti-BrdU monoclonal antibody and 7-amino actinomycin D, respectively, according to the manual of Cycle Test Plus DNA reagent kit. Cells prepared for detection of both cell death and cell cycle distribution were filtered through nylon mesh and analyzed with a FACScan. The obtained data were processed by Cell Quest (Becton Dickinson).
Detection of spontaneously occurring chromosomal aberrations.
Cells were treated with 0.1 μg of Colcemid/ml for 2 h and harvested. The cells were treated with 75 mM KCl for 11 min at room temperature and then fixed with methanol-acetic acid (3:1) for 30 min. The cell suspension was dropped onto ice-cold wet glass slides and air dried. The cells on the slides were stained with 3% Giemsa solution at pH 6.8 for 25 min and examined with a light microscope.
Measurements of sensitivity to MMS, mitomycin C (MMC), and UV irradiation.
To determine sensitivity to MMS, 3 × 102 cells were inoculated into 60-mm dishes containing various concentrations of MMS in a medium supplemented with 1.5% (wt/vol) methylcellulose, 1.5% chicken serum, and 15% fetal bovine serum. To measure sensitivity to MMC, cells were incubated at 39.5°C in a medium containing various concentrations of MMC for 1 h and then washed three times with warm medium. To determine sensitivity to UV, cells were suspended in 1 ml of phosphate-buffered saline, inoculated in six-well plates, and irradiated with various doses of UV. The cells treated with MMC or UV were inoculated into 60-mm dishes containing a medium supplemented with 1.5% (wt/vol) methylcellulose, 1.5% chicken serum, and 15% fetal bovine serum. Colonies were enumerated after 7 to 10 days, and the percent survival was determined relative to the number of colonies of untreated cells.
Measurements of spontaneous and MMC-induced SCE.
Cells (5 × 105) were cultured for two cycle periods with medium containing 10 μM BrdU and pulsed with 0.1 μg of Colcemid/ml for 2 h. The cells were harvested and treated with 75 mM KCl for 11 min at room temperature and then fixed with methanol-acetic acid (3:1) for 30 min. The cell suspension was dropped onto ice-cold wet glass slides and air dried. The cells on the slides were incubated with 10 μg of Hoechst 33258/ml in phosphate buffer (pH 6.8) for 20 min and rinsed with MacIlvaine solution (164 mM Na2HPO4, 16 mM citric acid; pH 7.0). The cells were exposed to a black light (λ = 352 nm) at a distance of 1 cm for 30 min and then incubated in 2× SSC (0.3 M NaCl plus 0.03 M sodium citrate) at 58°C for 20 min and then stained with 3% Giemsa solution for 25 min. To measure MMC-induced SCE, MMC was added at a final concentration of 50 ng/ml to the culture medium 8 h before harvesting.
Measurement of targeted integration frequency.
To analyze the targeted integration at the chicken RECQL1 and RECQL5 loci, a targeting construct, either chicken RECQL1-puromycinr or RECQL5-puromycinr, was transfected into the cells, and then the cells were selected in the medium containing puromycin. Genomic DNA of drug-resistant clones was isolated, and targeted integration was confirmed by PCR or Southern blotting.
Nucleotide sequence accession numbers.
The chicken RECQL1 and RECQL5 cDNA sequences were deposited in GenBank with accession numbers AB074261 and AB074262, respectively.
RESULTS
Isolation of chicken RECQL1 and RECQL5 cDNA.
Chicken RECQL1 and RECQL5 cDNA fragments that encoded peptides consisting of 661 and 446 amino acids, respectively, were isolated from a chicken testis cDNA library. It was reported that there are two splicing variants named α and β in mouse RecQL1 and human RecQL5 (29, 35). Both chicken RECQL1 and RECQL5 cDNA encoded the α type of RecQL1 and RecQL5. The chicken RecQL1 and RecQL5 showed 72.9 and 79.1% identity to the human RecQL1α and RecQL5α at the amino acid level.
Generation of RECQL1−/− and RECQL5−/− cells from DT40 cells.
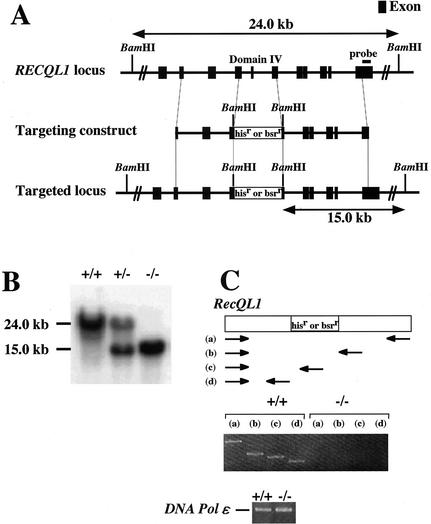
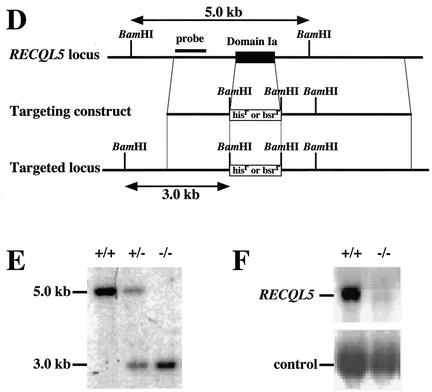
We first generated RECQL1−/− DT40 cells. A RECQL1 genomic DNA fragment of about 8 kb was isolated by long-range PCR with primers that were designed based on the sequence of the chicken RECQL1 cDNA. Chicken RECQL1 targeting constructs, RECQL1-histidinolr and RECQL1-blasticidinr, were designed as shown in Fig. 1A. Targeted integration of these constructs deleted the region from amino acids 281 to 328 which contained domain IV, which is highly conserved among RecQ family helicases, and generated a new 15-kb band when genomic DNA was digested with BamHI and hybridized with the external probe (Fig. 1A). This was confirmed by Southern blotting, as shown in Fig. 1B.
FIG. 1.
Disruption of RECQL1 and RECQL5 genes. (A) Schematic representation of part of the RECQL1 genomic locus, targeting constructs, and configuration of the targeted locus. (B) Southern blot analysis of wild-type cells (+/+), heterozygous RECQL1 cells (+/−), and homozygous RECQL1 cells (−/−). BamHI-digested genomic DNA was hybridized with the probe shown in panel A. (C) RT-PCR analysis of total RNA from wild-type cells (+/+) and homozygous RECQL1 cells (−/−). Primer sets a, b, c, and d were used to detect full-length and truncated forms of RECQL1 mRNA. (D) Schematic representation of part of the RECQL5 genomic locus, targeting constructs, and configuration of the targeted locus. (E) Southern blot analysis of wild-type cells (+/+), heterozygous RECQL5 cells (+/−), and homozygous RECQL5 cells (−/−). BamHI-digested genomic DNA was hybridized with the probe shown in panel D. (F) Northern blot analysis of total RNA with a C-terminal segment of RECQL5 cDNA as a probe.
We could not confirm the expression of RECQL1 mRNA by Northern blotting even in wild-type cells. However, the expression of RECQL1 mRNA in wild-type cells was confirmed by RT-PCR. Neither the full-length form nor the truncated form of RECQL1 mRNA was detected by RT-PCR in RECQL1−/− cells (Fig. 1C).
We next generated RECQL5−/− cells as follows. Briefly, a RECQL5 genomic DNA fragment of about 8 kb was isolated, and the targeting constructs, RECQL5-histidinolr and RECQL5-blasticidinr, were constructed, which deleted the region from amino acids 79 to 99 corresponding to domain Ia (Fig. 1D). Disruption of the RECQL5 gene was confirmed by Southern blot analysis by detecting the 3-kb band, as shown in Fig. 1E. The disruption of the RECQL5 gene was further verified by Northern blotting. Only the 3-kb mRNA, which may correspond to RECQL5α, was detected in the wild-type cells, and this band had completely disappeared in RECQL5−/− cells (Fig. 1F). In addition, the disruption of RECQL1 and RECQL5 genes was expected to disrupt the functions of both RecQL1α and -β and those of both RecQL5α and -β, even if the β forms of the RecQLs were expressed in DT40 cells.
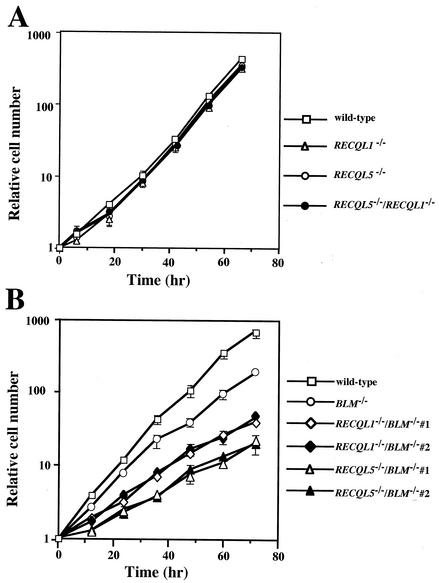
We monitored the growth of RECQL1−/− and RECQL5−/− cells. RECQL1−/− and RECQL5−/− cells proliferated at a rate similar to that of wild-type cells, suggesting that RecQL1 and RecQL5 are not essential for cell growth (Fig. 2A).
FIG. 2.
Growth curves of various RECQL mutants. Cells not stained with trypan blue were counted. The bars indicate standard deviations.
Generation of RECQL1−/−/RECQL5−/−, RECQL1−/−/BLM−/−, and RECQL5−/−/BLM−/− cells.
Since neither RECQL1−/− nor RECQL5−/− cells showed growth defects, it appears likely that functional redundancy exists among the multiple RecQL helicases in higher eukaryotic cells. To investigate the existence of functional redundancy among multiple RecQL helicases, we generated double mutants: RECQL1−/−/RECQL5−/−, RECQL1−/−/BLM−/−, and RECQL5−/−/BLM−/− cells. The RECQL1−/−/RECQL5−/− and RECQL1−/−/BLM−/− cells were constructed by transfecting two RECQL1 targeting vectors sequentially into RECQL5−/− and BLM−/− (34) cells, respectively. To generate RECQL5−/−/BLM−/− cells, we transfected two BLM targeting vectors (34) sequentially into RECQL5−/− cells. Gene disruption was confirmed by Southern blot analysis (data not shown [34]). The cells used in the present study are listed in Table 1.
TABLE 1.
DT40 strains used in this studya
| Strain | Genotype | Resistanceb | Source |
|---|---|---|---|
| 216 (wild type) | HPRT+/− | Neor | S. Takeda |
| RECQL1−/− | HPRT+/−RECQL1−/− | Neor Hisr Bsrr | This study |
| RECQL5−/− | HPRT+/−RECQL5−/− | Neor Hisr Bsrr | This study |
| BLM−/− | HPRT+/−BLM−/− | Neor Hisr Bsrr | W. Wang et al. (34) |
| RECQL1−/−/RECQL5−/− | HPRT+/−RECQL1−/−RECQL5−/− | Neor Hisr Bsrr Hygr Bler | This study |
| RECQL1−/−/BLM−/− | HPRT+/−RECQL1−/−BLM−/− | Neor Hisr Bsrr Hygr Purr | This study |
| RECQL5−/−/BLM−/− | HPRT+/−RECQL5−/−BLM−/− | Neor Hisr Bsrr Hygr Purr | This study |
All strains used in this study are deposited in the Riken cell bank in Japan (http://www.rtc.riken.go.jp/CELL/HTML/RIKEN_Cell_Bank.html).
Neo, neomycin; His, histidinol; Bsr, blasticidin; Hyg, hygromycin; Ble, bleomycin; Pur, puromycin.
RECQL1−/−/BLM−/− and RECQL5−/−/BLM−/− cells but not RECQL1−/−/RECQL5−/− cells show a slow-growth phenotype.
We examined the proliferative properties of RECQL1−/−/RECQL5−/−, RECQL1−/−/BLM−/−, and RECQL5−/−/BLM−/− cells. RECQL1−/−/RECQL5−/− cells proliferated at a rate similar to that of wild-type cells (Fig. 2A), suggesting that both RecQL1 and RecQL5 play no essential role in cell growth. BLM−/− DT40 cells proliferated at a slightly lower rate than wild-type cells, as reported previously (34). As shown in Fig. 2B, RECQL1−/−/BLM−/− and RECQL5−/−/BLM−/− cells proliferated much more slowly than did BLM−/− cells, and the slow-growth phenotype of RECQL5−/−/BLM−/− cells was more severe than that of RECQL1−/−/BLM−/− cells, suggesting that RecQL1 and RecQL5 partially replace the function of BLM under the BLM function-impaired condition.
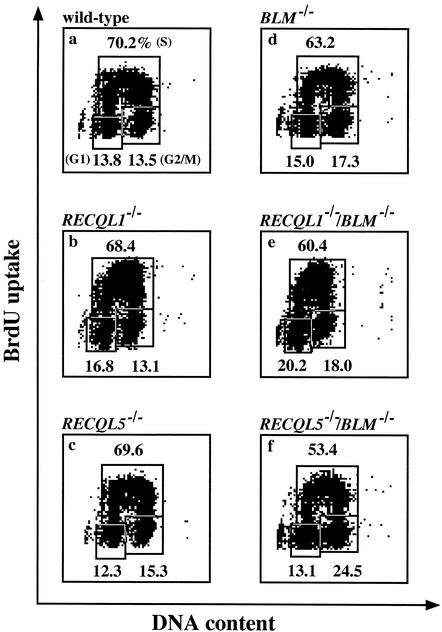
To obtain an insight into the slow-growth phenotype of RECQL1−/−/BLM−/− and RECQL5−/−/BLM−/− cells, we monitored the distribution of cells in the cell cycle by flow cytometry. As shown in Fig. 3, the cell cycle distribution pattern of RECQL1−/−/BLM−/− cells did not differ markedly from that of either wild-type, RECQL1−/−, or BLM−/− cells. In contrast, the G2/M-phase population was slightly increased in the case of RECQL5−/−/BLM−/− cells compared to that of wild-type, RECQL5−/−, or BLM−/− cells, suggesting the occurrence of DNA damage, which prevented the progression of the cell cycle from the G2 to M phase.
FIG. 3.
Cell cycle distribution pattern. DNA content was determined by staining with 7-amino actinomycin, and DNA synthesis was measured by determining the incorporation of BrdU. Cells which contain 2 N and 4 N DNA contents but do not incorporate BrdU correspond to G1- and G2/M-phase populations, respectively. Cells which contain a ca. 2 to 4 N DNA content and incorporate BrdU correspond to an S-phase population. G1, S, and G2/M populations are indicated by a percentage. (a) Wild-type cells; (b) RECQL1−/− cells; (c) RECQL5−/− cells; (d) BLM−/− cells; (e) RECQL1−/−/BLM−/− cells; (f) RECQL5−/−/BLM−/− cells.
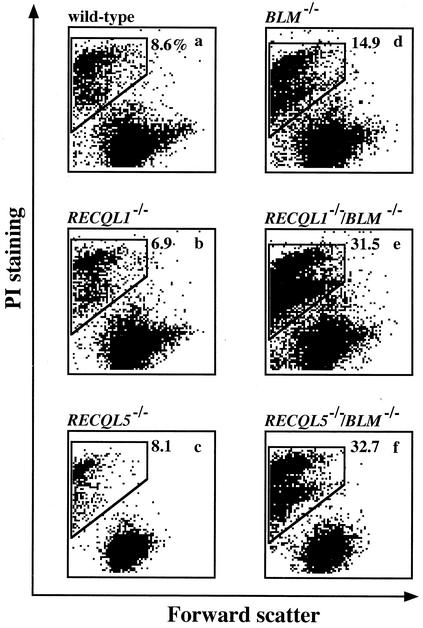
We next measured the frequency of spontaneous cell death. Although the populations of dead cells of RECQL1−/− and RECQL5−/− were similar in wild-type cells, the population of dead cells was slightly increased in BLM−/− cells and was further increased in RECQL1−/−/BLM−/− and RECQL5−/−/BLM−/− cells (Fig. 4). Thus, the slow-growth phenotype of RECQL1−/−/BLM−/− and RECQL5−/−/BLM−/− cells is at least partly explained by the elevated spontaneous cell death.
FIG. 4.
Detection of cell death by flow cytometry. Forward scatter represents the size of cells. Enclosed area, which contains small-sized cells heavily stained with propidium iodide, represents the population of dead cells. The dead-cell populations are indicated as percentages in each panel. (a) Wild-type cells; (b) RECQL1−/− cells; (c) RECQL5−/− cells; (d) BLM−/− cells; (e) RECQL1−/−/BLM−/− cells; (f) RECQL5−/−/BLM−/− cells.
Taking account of the G2/M accumulation and the increased cell death of RECQL1−/−/BLM−/− and RECQL5−/−/BLM−/− cells, we examined the frequency of chromosomal aberrations in the cells of various genotypes. No apparent increase in chromosomal aberrations was observed in RECQL1−/−, RECQL5−/−, BLM−/−, RECQL1−/−/BLM−/−, and RECQL5−/−/BLM−/− cells compared to wild-type cells (Table 2).
TABLE 2.
Chromosome aberrationsa
| Cell clone | Chromatid type (n)
|
Chromosome type (gaps [n]) | ||
|---|---|---|---|---|
| Break | Gap | Exchange | ||
| 216 (wild type) | 1 | 1 | 1 | 4 |
| BLM−/− | 0 | 6 | 1 | 5 |
| RecQL1−/− | 1 | 4 | 0 | 5 |
| RecQL5−/− | 3 | 6 | 0 | 3 |
| RecQL1−/−/BLM−/− | 1 | 7 | 1 | 3 |
| RecQL5−/−/BLM−/− | 2 | 5 | 4 | 3 |
Data are presented as the number (n) of aberrations per 200 cells.
Sensitivity of various RECQL mutants to DNA-damaging agents.
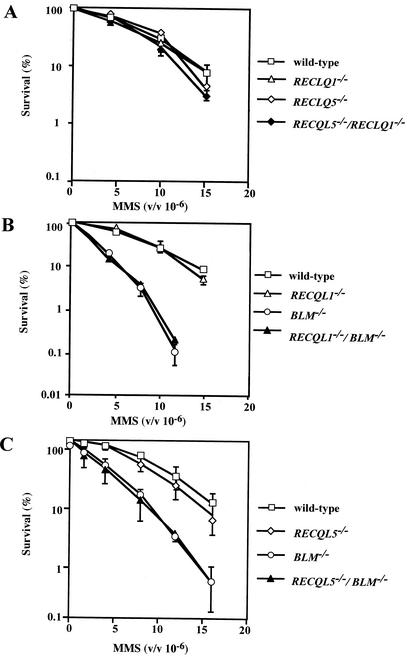
The sensitivity of various RECQL mutants listed in Table 1 to MMS, MMC, and UV irradiation was examined. BLM−/− cells showed a moderate sensitivity to MMS as we reported previously (34), whereas RECQL1−/−, RECQL5−/− single-gene-disrupted cells and RECQL1−/−/RECQL5−/− double-gene-disrupted cells showed no apparent sensitivity to MMS compared to wild-type cells (Fig. 5A). Although RECQL1−/−/BLM−/− and RECQL5−/−/BLM−/− cells showed a slightly more severe growth defect than that of BLM−/− cells, the sensitivity to MMS of either RECQL1−/−/BLM−/− or RECQL5−/−/BLM−/− cells was the same as that of BLM−/− cells (Fig. 5B and C). In addition, all of the mutants, including RECQL1−/−/BLM−/− and RECQL5−/−/BLM−/− cells, showed sensitivities to MMC or UV that were similar to those of wild-type cells (data not shown), suggesting that RecQL1, RecQL5, and BLM play no significant roles in the repair of DNA lesions generated by MMC and UV.
FIG. 5.
Sensitivity of various RECQL mutants to MMS. Cells were treated with the indicated concentration of MMS as described in Materials and Methods. The bars indicate standard deviations.
Targeted integration frequencies in RECQL mutants.
We previously demonstrated that targeted integration frequency was remarkably increased in BLM−/− cells (34). Thus, we examined whether RecQL1 and RecQL5 affect targeted integration events. As shown in Table 3, the targeted integration frequencies of RECQL1−/− and RECQL5−/− cells were essentially the same as those of wild-type cells, suggesting that unlike BLM, RecQL1 and RecQL5 play no essential role in targeted integration.
TABLE 3.
Targeted integration frequency
| Expt | No. of targeted clones/total clones (%)a in strain:
|
|||
|---|---|---|---|---|
| RECQL5
|
RECQL1
|
|||
| Wild type | RECQL1−/− | Wild type | RECQL5−/− | |
| 1 | 9/24 (37.5) | 10/24 (41.7) | 7/24 (29.2) | 11/24 (45.8) |
| 2 | 8/24 (33.3) | 8/24 (33.3) | 8/24 (33.3) | 9/24 (37.5) |
The figures indicate the number of targeted clones at each locus divided by the total number of drug-resistant clones analyzed.
Frequencies of spontaneous and MMC-induced SCE in various RECQL mutants.
E. coli RecQ and S. cerevisiae Sgs1 have dual functions to promote and suppress recombination (12, 25). Sonoda et al. reported that SCE is formed by homologous recombination (31), and we also reported that the majority of increased SCE in BLM−/− cells is formed by homologous recombination (34). To examine whether RecQL1 and RecQL5 affect another type of recombination, we measured the frequencies of SCE in various RECQL mutants. As shown in Table 4, the frequencies of spontaneous SCEs in RECQL1−/−, RECQL5−/−, and RECQL1−/−/RECQL5−/− cells were similar to those of wild-type cells. In addition, disruption of RECQL1 gene in BLM−/− cells did not affect the spontaneously elevated frequency of SCE in BLM−/− cells. In contrast, the frequency of spontaneous SCE in RECQL5−/−/BLM−/− cells was increased by 1.7- and 22-fold compared to those of BLM−/− and wild-type cells, respectively.
TABLE 4.
Spontaneous and MMC-induced SCEs in RECQL mutants
| Cell clone or wild type | Mean no. of SCEs/100 metaphase cells ± SEa
|
|
|---|---|---|
| Spontaneous | MMC induced | |
| Wild type | 2.1 ± 0.14 | 6.6 ± 0.26 |
| RecQL1−/− | 2.0 ± 0.14 | 5.6 ± 0.25 |
| RecQL5−/− | 2.4 ± 0.19 | 7.8 ± 0.29 |
| RecQL1−/−/RecQL5−/− | 2.1 ± 0.15 | 6.9 ± 0.29 |
| BLM−/− | 26.4 ± 0.86 | 46.6 ± 0.89 |
| RecQL1−/−/BLM−/− | 27.5 ± 0.80 | 64.0 ± 3.70b |
| RecQL5−/−/BLM−/− | 45.2 ± 1.22 | NDc |
Spontaneous and MMC-induced SCEs in macrochromosomes (1 to 5 and Z) of 100 metaphase cells of each strain except the RECQL1−/−/BLM−/− strain were counted. The mean number of SCEs per cell and the standard error are shown.
The mean number of SCEs per cell was determined by examining five countable spreads out of 100 metaphase cells.
ND, not determined due to too many SCEs.
MMC increases the frequency of SCE in wild-type cells. Thus, we next examined the frequencies of SCE in various RECQL mutants that were exposed to MMC. As shown in Table 4, the frequencies of SCE in RECQL1−/−, RECQL5−/−, and RECQL1−/−/RECQL5−/− cells were similar to those of wild-type cells. The frequency of MMC-induced SCEs in BLM−/− cells was increased by 1.8-fold compared to that of spontaneous SCEs in BLM−/− cells. As a result of the exposure to MMC, the cells with chromosomes showing too many SCEs to count were increased for BLM−/−, RECQL1−/−/BLM−/−, and RECQL5−/−/BLM−/− cells by 13, 95, and 100% versus metaphase cells, respectively. Since the data of RECQL1−/−/BLM−/− cells (64 SCEs/cell) was obtained with five countable metaphases of RECQL1−/−/BLM−/− cells, the actual SCE frequency of RECQL1−/−/BLM−/− cells should be much higher than 64 SCEs/cell.
DISCUSSION
In the present study, to gain insight into the function of RecQL1 and RecQL5, we generated and characterized RECQL1−/−, RECQL5−/−, RECQL1−/−/RECQL5−/−, RECQL1−/−/BLM−/−, and RECQL5−/−/BLM−/− DT40 cells. To our knowledge, this is the first study reporting intracellular functions of RecQL1 and RecQL5.
RECQL1−/−, RECQL5−/− or RECQL1−/−/RECQL5−/− cells showed no defect in growth, and growth defects due to the dysfunction of RecQL1 or RecQL5 were observed only under BLM function-impaired conditions. The existence of a splicing variant was reported for RecQL1 and RecQL5. The cDNA clones of RECQL1 and RECQL5 encoded the homologue of human RecQL1α (26, 27, 35) and RecQL5α (16, 29). Although the targeting vectors were constructed based on the isolated cDNA, the gene targeting appears to destroy the functions of both isoforms α and β because the vectors for RECQL1 and RECQL5 gene targeting were deleted with helicase motif IV and helicase motif Ia, respectively (11). Thus, it appears unlikely that no phenotype of RECQL1−/−, RECQL5−/− or RECQL1−/−/RECQL5−/− cells in cell growth is due to the compensation of the β form of each RecQL. In addition, no mRNA for RECQL1 and RECQL5 was detected in RECQL1−/− and RECQL5−/− cells by RT-PCR and Northern blotting, respectively, suggesting that these cells are null mutants.
The phenotypes of the recQ homologue mutants of Saccharomyces pombe rqh1− (32) and S. cerevisiae sgs1 are similar to those of BS cells (5), and BLM genes are conserved in nematodes, fruit flies, and vertebrates, including humans, suggesting that BLM plays a very important role in the maintenance of genome stability in multicellular organisms (19). In contrast to BLM, RECQL1 exists in Caenorhabditis elegans but not in Drosophila melanogaster, and RECQL5 exists in D. melanogaster but not in C. elegans (18). Thus, it appears likely that RECQL1 and RECQL5 exist as a backup system for the functions of other RecQ family helicases. In this context, RECQL1−/−/BLM−/− and RECQL5−/−/BLM−/− cells displayed more-severe growth defects than BLM−/− cells, suggesting that RecQL1 and RecQL5 partially replace the function of BLM under the BLM function-impaired condition. However, the growth defect of the double mutants can also be explained as follows. Either RecQL1 or RecQL5 performs a function different from that of BLM, and the effect due to the defect in either the RecQL1 or RecQL5 function is exaggerated under the BLM function-impaired condition.
It was reported that SCE is formed mainly via homologous recombination (31). Previously, we generated BLM−/− and BLM−/−/RAD54−/− DT40 cells, characterized their properties, and suggested that BLM functions to reduce the formation of DNA double-strand breaks (DSB) by disrupting Holliday junctions that are formed during DNA replication and are a potential cause of DSBs (34). In addition, we also suggested that DSB formation increases SCE and that a large portion of increased SCE in BLM−/− cells are formed by homologous recombination.
We examined the frequency of SCE in various RECQL mutants and demonstrated that the frequencies of SCE in RECQL1−/−, RECQL5−/−, and RECQL1−/−/RECQL5−/− cells were similar to that of wild-type cells. Because it is well known that a high incidence of SCE is a characteristic feature of BS cells and there is no evidence suggesting an increase in the frequency of SCE in either WS cells or RTS cells (3, 21), it can be concluded that, among the five RecQLs, only BLM suppresses the formation of SCE. However, RECQL5−/−/BLM−/− cells showed a higher frequency of SCE than that of BLM−/− cells, suggesting that RecQL5 suppresses the formation of SCE under the BLM function-impaired condition. In addition, the frequencies of SCE in RECQL1−/−/BLM−/− and RECQL5−/−/BLM−/− cells were considerably increased by exposure to MMC, and the values were much higher than that of BLM−/− cells exposed to MMC. Thus, both RecQL1 and RecQL5 partially replace BLM in the function to suppress SCE under a certain condition.
Holliday junctions are resolved to result in the formation of either crossover or noncrossover type recombinants, and only the former case can be detected as SCEs by the method used here. Jasin et al. determined the proportion of crossover type unequal sister chromatid recombination to be <3% by physical assay (15). If RecQL5 functions to channel Holliday junctions to yield noncrossover recombinants, it can be suggested that the hyperinduction of spontaneous SCEs in RECQL5−/−/BLM−/− cells is due to the defect in the channeling function of RecQL5. However, the finding that impairing the RecQL5 function did not further increase MMC-induced SCE frequency appears to exclude this possibility (Table 4). Thus, it can be concluded that RecQL5 performs a function similar to that of BLM under the BLM function-impaired condition to result in the further increase in spontaneous and MMC-induced SCE frequencies of BLM−/−cells when the RecQL5 function is impaired. In this context, it is interesting to examine whether WS or RTS affects the formation of SCE in the absence of BLM. Recently, Imamura et al. reported that the elevated SCE frequency of BLM−/− cells was reduced to 2/3 by disrupting the WRN gene, suggesting that WRN promotes sister chromatid-based recombination in the absence of BLM (13). Thus, in the cells lacking BLM, RecQL5 plays a role opposite to that of WRN in the formation of SCE.
Upon exposure to various DNA-damaging agents, WRN−/−/BLM−/− cells showed more severe sensitivity to the DNA-damaging agents, including MMS, than did other single-gene mutants (13). In contrast, all of the RECQL1−/−, RECQL5−/−, and RECQL1−/−/RECQL5−/− cells showed the same sensitivities to MMS, MMC, and UV irradiation compared to wild-type cells. In addition, the sensitivity of BLM−/− cells to MMS was not exaggerated by the disruption of either RECQL1 or RECQL5. These results further supported the notion that both RecQL1 and RecQL5 behave differently from WRN in the absence of BLM.
Finally, if functional redundancy really exists between BLM and RecQL1 and between BLM and RecQL5 in supporting cell growth and suppressing SCE, the next key question is what molecular bases underlie the functional redundancy. Recent biochemical analyses revealed that all of RecQL1α, BLM, and RecQL5β interact with TOP3α in human cells (14, 29). The observation that embryos of Top3α gene knockout mice died at early stages of development suggests that Top3α is essential for viability (20). Thus, it appears likely that RecQLs play an important role by interacting with TOP3α, and the more severe growth defect of RECQL1−/−/BLM−/− and REQCL5−/−/BLM−/− cells than that of BLM−/− cells is due to the reduction of total RecQL-TOP3α function in the cells. However, the function of RecQL-TOP3α in the cell appears to be mainly carried out by BLM-TOP3α because the lack of both RecQL1 and RecQL5 in RECQL1−/−/RECQL5−/− cells did not affect the growth of the cells, the spontaneously occurring and MMC-induced SCEs, and DNA repair.
In conclusion, among five RecQ family helicases in higher eukaryotic cells, only BLM suppresses SCE, and RecQL1 and RecQL5 partially substitute the function of BLM under BLM function-impaired condition.
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research and for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, by Health Sciences Research grants from the Ministry of Health Labor and Welfare of Japan, and by a grant from Takeda Science Foundation.
REFERENCES
- 1.Bennett, R. J., M. F. Noirot-Gros, and J. C. Wang. 2000. Interaction between yeast Sgs1 helicase and DNA topoisomerase III. J. Biol. Chem. 275:26898-26905. [DOI] [PubMed] [Google Scholar]
- 2.Buerstedde, J. M., and S. Takeda. 1991. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell 67:179-188. [DOI] [PubMed] [Google Scholar]
- 3.Darlington, G. J., R. Dutkowski, and W. T. Brown. 1981. Sister chromatid exchange frequencies in Progeria and Werner syndrome patients. Am. J. Hum. Genet. 33:762-766. [PMC free article] [PubMed] [Google Scholar]
- 4.Ellis, N. A., J. Groden, T. Z. Ye, J. Straughen, D. J. Lennon, S. Ciocci, M. Proytcheva, and J. German. 1995. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell 83:655-666. [DOI] [PubMed] [Google Scholar]
- 5.Enomoto, T. 2001. Functions of RecQ family helicases: possible involvement of Bloom's and Werner's syndrome gene products in guarding genome integrity during DNA replication. J. Biochem. 129:501-507. [DOI] [PubMed] [Google Scholar]
- 6.Frei, C., and S. M. Gasser. 2000. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 14:81-96. [PMC free article] [PubMed] [Google Scholar]
- 7.Fricke, W. M., V. Kaliraman, and S. J. Brill. 2001. Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J. Biol. Chem. 276:8848-8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gangloff, S., J. P. McDonald, C. Bendixen, L. Arthur, and R. Rothstein. 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14:8391-8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gangloff, S., C. Soustelle, and F. Fabre. 2000. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 25:192-194. [DOI] [PubMed] [Google Scholar]
- 10.German, J. 1993. Bloom syndrome: a Mendelian prototype of somatic mutational disease. Medicine 72:393-406. [PubMed] [Google Scholar]
- 11.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 17:4713-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harmon, F. G., and S. C. Kowalczykowski. 1998. RecQ helicase, in concert with RecA and SSB proteins, initiates and disrupts DNA recombination. Genes Dev. 12:1134-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imamura, O., K. Fujita, C. Itoh, S. Takeda, Y. Furuichi, and T. Matsumoto. 2002. Werner and Bloom helicases are involved in DNA repair in a complementary fashion. Oncogene 21:954-963. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, F. B., D. B. Lombard, N. F. Neff, M. A. Mastrangelo, W. Dewolf, N. A. Ellis, R. A. Marciniak, Y. Yin, R. Jaenisch, and L. Guarente. 2000. Association of the Bloom syndrome protein with topoisomerase IIIα in somatic and meiotic cells. Cancer Res. 60:1162-1167. [PubMed] [Google Scholar]
- 15.Johnson, R. D., and M. Jasin. 2000. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19:3398-3407. [DOI] [PMC free article] [PubMed]
- 16.Kitao, S., I. Ohsugi, K. Ichikawa, M. Goto, Y. Furuichi, and A. Shimamoto. 1998. Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics 54:443-452. [DOI] [PubMed] [Google Scholar]
- 17.Kitao, S., A. Shimamoto, M. Goto, R. W. Miller, W. A. Smithson, N. M. Lindor, and Y. Furuichi. 1999. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 22:82-84. [DOI] [PubMed] [Google Scholar]
- 18.Kusano, K., M. E. Berres, and W. R. Engels. 1999. Evolution of the RECQ family of helicases: a Drosophila homolog, Dmblm, is similar to the human Bloom syndrome gene. Genetics 151:1027-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kusano, K., D. M. Johnson-Schlitz, and W. R. Engels. 2001. Sterility of Drosophila with mutations in the Bloom syndrome gene complementation by Ku70. Science 291:2600-2602. [DOI] [PubMed] [Google Scholar]
- 20.Li, W., and J. C. Wang. 1998. Mammalian DNA topoisomerase IIIα is essential in early embryogenesis. Proc. Natl. Acad. Sci. USA 95:1010-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindor, N. M., Y. Furuichi, S. Kitao, A. Shimamoto, C. Arndt, and S. Jalal. 2000. Rothmund-Thomson syndrome due to RECQ4 helicase mutations: report and clinical and molecular comparisons with Bloom syndrome and Werner syndrome. Am. J. Med. Genet. 90:223-228. [DOI] [PubMed] [Google Scholar]
- 22.Miyajima, A., M. Seki, F. Onoda, M. Shiratori, N. Odagiri, K. Ohta, Y. Kikuchi, Y. Ohno, and T. Enomoto. 2000. Sgs1 helicase activity is required for mitotic but apparently not for meiotic functions. Mol. Cell. Biol. 20:6399-6409. [DOI] [PMC free article] [PubMed]
- 23.Nakayama, K., N. Irino, and H. Nakayama. 1985. The recQ gene of Escherichia coli K-12: molecular cloning and isolation of insertion mutants. Mol. Gen. Genet. 200:266-271. [DOI] [PubMed] [Google Scholar]
- 24.Onoda, F., M. Seki, A. Miyajima, and T. Enomoto. 2000. Elevation of sister chromatid exchange in Saccharomyces cerevisiae sgs1 disruptants and the relevance of the disruptants as a system to evaluate mutations in Bloom's syndrome gene. Mutat. Res. 459:203-209. [DOI] [PubMed] [Google Scholar]
- 25.Onoda, F., M. Seki, A. Miyajima, and T. Enomoto. 2001. Involvement of SGS1 in DNA damage-induced heteroallelic recombination that requires RAD52 in Saccharomyces cerevisiae. Mol. Gen. Genet. 264:702-708. [DOI] [PubMed] [Google Scholar]
- 26.Puranam, K. L., and P. J. Blackshear. 1994. Cloning and characterization of RECQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J. Biol. Chem. 269:29838-29845. [PubMed] [Google Scholar]
- 27.Seki, M., H. Miyazawa, S. Tada, J. Yanagisawa, T. Yamaoka, S. Hoshino, K. Ozawa, T. Eki, M. Nogami, K. Okumura, et al. 1994. Molecular cloning of cDNA encoding human DNA helicase Q1 which has homology to Escherichia coli Rec Q helicase and localization of the gene at chromosome 12p12. Nucleic Acids Res. 22:4566-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seki, M., J. Yanagisawa, T. Kohda, T. Sonoyama, M. Ui, and T. Enomoto. 1994. Purification of two DNA-dependent adenosinetriphosphatases having DNA helicase activity from HeLa cells and comparison of the properties of the two enzymes. J. Biochem. 115:523-531. [DOI] [PubMed] [Google Scholar]
- 29.Shimamoto, A., K. Nishikawa, S. Kitao, and Y. Furuichi. 2000. Human RecQ5β, a large isomer of RecQ5 DNA helicase, localizes in the nucleoplasm and interacts with topoisomerases 3α and 3β. Nucleic Acids Res. 28:1647-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinclair, D. A., K. Mills, and L. Guarente. 1997. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science 277:1313-1316. [DOI] [PubMed] [Google Scholar]
- 31.Sonoda, E., M. S. Sasaki, C. Morrison, Y. Yamaguchi-Iwai, M. Takata, and S. Takeda. 1999. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol. 19:5166-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stewart, E., C. R. Chapman, F. Al-Khodairy, A. M. Carr, and T. Enoch. 1997. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 16:2682-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ui, A., Y. Satoh, F. Onoda, A. Miyajima, M. Seki, and T. Enomoto. 2001. The N-terminal region of Sgs1, which interacts with Top3, is required for complementation of MMS sensitivity and suppression of hyper-recombination in sgs1 disruptants. Mol. Genet. Genomics 265:837-850. [DOI] [PubMed] [Google Scholar]
- 34.Wang, W., M. Seki, Y. Narita, E. Sonoda, S. Takeda, K. Yamada, T. Masuko, T. Katada, and T. Enomoto. 2000. Possible association of BLM in decreasing DNA double strand breaks during DNA replication. EMBO J. 19:3428-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, W. S., M. Seki, T. Yamaoka, T. Seki, S. Tada, T. Katada, H. Fujimoto, and T. Enomoto. 1998. Cloning of two isoforms of mouse DNA helicase Q1/RecQL cDNA: the α form is expressed ubiquitously and β form specifically in the testis. Biochim. Biophys. Acta 1443:198-202. [DOI] [PubMed] [Google Scholar]
- 36.Watt, P. M., I. D. Hickson, R. H. Borts, and E. J. Louis. 1996. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144:935-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watt, P. M., E. J. Louis, R. H. Borts, and I. D. Hickson. 1995. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell 81:253-260. [DOI] [PubMed] [Google Scholar]
- 38.Yamagata, K., J. Kato, A. Shimamoto, M. Goto, Y. Furuichi, and H. Ikeda. 1998. Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc. Natl. Acad. Sci. USA 95:8733-8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yanagisawa, J., M. Seki, M. Ui, and T. Enomoto. 1992. Alteration of a DNA-dependent ATPase activity in xeroderma pigmentosum complementation group C cells. J. Biol. Chem. 267:3585-3588. [PubMed] [Google Scholar]
- 40.Yu, C. E., J. Oshima, Y. H. Fu, E. M. Wijsman, F. Hisama, R. Alisch, S. Matthews, J. Nakura, T. Miki, S. Ouais, G. M. Martin, J. Mulligan, and G. D. Schellenberg. 1996. Positional cloning of the Werner's syndrome gene. Science 272:258-262. [DOI] [PubMed] [Google Scholar]