Abstract
While roles in adhesion and morphogenesis have been documented for classical cadherins, the nonclassical cadherins are much less well understood. Here we have examined the functions of the giant protocadherin FAT by generating a transgenic mouse lacking mFAT1. These mice exhibit perinatal lethality, most probably caused by loss of the renal glomerular slit junctions and fusion of glomerular epithelial cell processes (podocytes). In addition, some mFAT1−/− mice show defects in forebrain development (holoprosencephaly) and failure of eye development (anophthalmia). In contrast to Drosophila, where FAT acts as a tumor suppressor gene, we found no evidence for abnormalities of proliferation in two tissues (skin and central nervous system [CNS]) containing stem and precursor cell populations and in which FAT is expressed strongly. Our results confirm a necessary role for FAT1 in the modified adhesion junctions of the renal glomerular epithelial cell and reveal hitherto unsuspected roles for FAT1 in CNS development.
Cell adhesion molecules are essential for the regulation of coordinated changes in cell shape and position that occur during morphogenesis and also for the maintenance of cell-cell interactions once development is complete. The best-studied families of cell adhesion molecules are the cadherins. These were originally identified as Ca2+-dependent cell-cell adhesion proteins characterized by five repeated cadherin-specific motifs in their extracellular domain. This domain is an approximately 110-amino-acid peptide that mediates homophilic interactions with other cadherin molecules, forming dimers which then interact with dimers on neighboring cells (41, 45, 46). The cytoplasmic domain of these so-called classical five-repeat cadherins is well conserved between different members of the family, containing motifs required for interaction with catenins that can then mediate linkage to the cytoskeleton (13, 17, 34, 42). In addition to the classical cadherins, at least five other classes of cadherins have been identified: nonclassical cadherins, in which the first extracellular repeat does not contain the HAV (histidine-alanine-valine) cell adhesion sequence present in all classical cadherins, and four additional families. Two of these, the desmosomal adhesion molecules desmocollins and desmogleins, all share the five extracellular cadherin repeat structure with the sole exception of desmocollin-1. Two other families, the flamingo cadherins and protocadherins, have variable numbers of cadherin repeats and additional motifs as well as differing cytoplasmic domain sequences (30, 44).
The functions of the protocadherin family, in particular, remain poorly understood. The presence of many different protocadherins, many encoded by three gene clusters comprising 52 genes in the human (47), could provide the basis for specificity of intercellular interactions based on differential adhesion. However, adhesive properties have been described only for Xenopus NF-protocadherin (the homologue of human pcdh7) (4), axial protocadherin (pcdh1) (21), and paraxial protocadherin (pcdh8) (19). In addition, the extracellular domain of pcdh2 shows homophilic adhesion properties when fused to the cytoplasmic domain of the classical cadherin (31). For the majority of protocadherins, therefore, no adhesive function has been documented, and the variability of structure within this class of molecules suggests that other functions may well be significant.
One other such function suggested for protocadherins has been growth suppression. This conclusion arose from studies of FAT, the largest protocadherin, with 34 extracellular cadherin repeats (30). In Drosophila, loss-of-function mutations cause hyperplasia of the pupal imaginal disks, suggesting a role as a tumor suppressor gene, while defects of epithelial morphogenesis are consistent with an adhesive function (25). This giant cadherin has been subsequently cloned in humans (10), rats (33), and mice (6). Structurally, these genes resemble the FAT gene of Drosophila, showing that FAT-like cadherins are conserved between vertebrates and invertebrates, and more recently two additional FAT genes (FAT2 and FAT3) have been identified in vertebrates (27, 29). The protein has a predicted size of 500 kDa, containing 5 epidermal growth factor (EGF) and 2 laminin-A-G domain repeats in addition to the 34 cadherin repeats in the extracellular domain. The cytoplasmic domain contains two potential β-catenin binding regions (FC1 and FC2), although these are only weakly conserved with classical cadherins (10). In addition, a putative PDZ domain binding sequence has been found at the C-terminal domain of the vertebrate homologues (33), although this is not present in Drosophila FAT.
Expression studies show FAT mRNA to be expressed widely in epithelial tissues, including skin, neuroepithelium, and renal glomerular epithelial cells (podocytes), during development (6, 33), with upregulation reported in human epithelial tumors (6). We previously reported that FAT expression within the central nervous system (CNS) persisted in the adult in areas in which proliferative neural stem cells remain throughout life (33). Given this localization of a putative growth suppressor molecule, we reasoned that FAT might contribute to regulation of proliferation in the CNS and other tissues. To test this and to examine other functions of this molecule, we have generated an mFAT1-deficient mouse. Our results provide no support for a role of mFAT1 in the regulation of proliferation, but they do reveal novel roles in adhesion and cell-cell signaling.
MATERIALS AND METHODS
Generation of Fat-deficient mice.
A mouse genomic PAC library from a female 129/SvevTacfBr mouse spleen was screened using probes from the 5′ and 3′ ends of rat Fat cDNA (33). Three different genomic clones were identified and isolated. Restriction analysis and further Southern blotting using as a probe the first 700 bp of the rat Fat cDNA (including the 5′ untranslated region and the transcription start codon) identified a 7-kb fragment containing the predicted ATG start codon for mFAT1. This fragment of 7 kb was then cloned in the Escherichia coli-yeast shuttle vector pRS414. Next, we modified a second plasmid (pRay-1) containing a cassette comprising the neomycin resistance gene under the control of the thymidine kinase promoter, the yeast selection marker URA3, and the lacZ reporter gene in front of these selection markers by inserting 400-bp recombinogenic arms on either side of the cassette. These arms were generated by PCR from the mouse DNA clone and were designed to allow homologous recombination in yeast so as to insert the lacZ gene in frame with the first ATG of FAT1, hence generating a targeting vector (43).
The entire cassette, now flanked by the two recombinogenic arms, was transferred into yeast cells together with the shuttle vector pRS414 containing the 7-kb mouse DNA clone. Recombinant yeast colonies in which homologous recombination had occurred were selected by plating on Trp/URA dropout plates, requiring the URA3 marker gene for survival, and plasmids from the selected colonies were transferred to E. coli (Top 10; Stratgene) by electroporation. Homologous recombination was confirmed using restriction analysis and sequencing. The final vector was linearized and electroporated into 129Sv embryonic stem (ES) cells. Electroporated cells were selected for 8 days in 250 μg of G418 (Gibco) per ml. Selected clones were expanded and analyzed by Southern blotting using an internal probe (lacZ) and an external probe that is adjacent to but not contained in the targeting construct (see Fig. 1A). Two of the three ES cell clones shown to have undergone correct homologous recombination, HB-5 and FB-2, were injected into C57BL/6 mouse blastocysts, and the blastocysts were then implanted into pseudo-pregnant females. No chimeric mice were born from HB-5-injected blastocysts. From FB-2-injected blastocysts, nine mice with chimerism varying from 40 to 100% were born. One of these animals gave germ line transmission of the mutant allele, and heterozygous progeny were intercrossed for more than five generations and also backcrossed onto C57BL/6 and 129Sv backgrounds. Genotyping of tail DNA was performed by either Southern blotting or PCR using two primer pairs flanking the insertion site. For these PCR amplifications, the two sense primers were 5′ CAGCAGTTTGGAGACA 3′ and 5′ TAGCTTGGCTGGACGT3′ and the antisense primer was 5′ ACGAGGGGTCTAAGATGT 3′, giving products of 150 nucleotides for the wild-type allele and 220 nucleotides for the mutant allele.
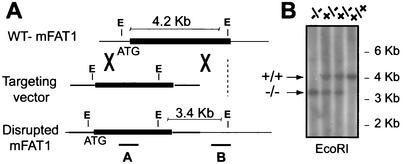
FIG. 1.
(A) Schematic representation of the protocol used to mutate the mFAT1 locus, showing the wild-type (WT) locus (top), mFAT1 targeting vector (middle), and expected mutant allele after homologous recombination (bottom). A lacZ/Neo cassette was inserted in frame with the first ATG of the mFAT1 gene as described in Materials and Methods. The positive ES clones undergoing correct homologous recombination were identified by Southern blot analysis using a 3′ external probe and an internal probe on the lacZ gene. (B) Southern blot analysis (using the 3′ flanking probe) of EcoRI-digested tail genomic DNA from progeny of mFAT1 heterozygous intercrosses, showing the three possible genotypes obtained.
Analysis of mFAT1 expression.
lacZ expression in embryos and sections (10 μm) from mFAT1 transgenic mice was detected by using 5-bromo-4-chloro-3-indolyl-β-d-galactopyronoside (X-Gal) staining solution containing 2 mM MgCl2 and 5 mM K4Fe(CN)6 in phosphate-buffered saline (PBS) plus 0.01% sodium deoxycholate, 0.02% NP-40, and 1 mg of X-Gal per ml. The sections were stained overnight at 37°C and then counterstained with Neutral Red before being mounted.
Western blotting.
Embryos (18.5 days postcoitum or E18.5, with the plug date being E0) were obtained by cesarean section following euthanasia of the pregnant female. The tail was removed for DNA extraction and genotyping. Skin was then removed, homogenized in lysis buffer (50 mM Tris-HCl, 5 mM EDTA, 150 mM NaCl, 1% NP-40, 1% sodium dodecyl sulfate, 2 mM phenylmethylsulfonyl fluoride, 1 μg of pepstatin A per ml, 2 mg of aprotinin per ml, 5 mg of leupeptin per ml) on ice for 30 min, and centrifuged at 15,000 rpm in an Eppendorf Microfuge for 15 min. The amount of total protein in the extracts was determined using the Bio-Rad detergent-compatible protein assay with bovine serum albumin as a standard. Equal amounts of protein from each animal were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (7% polyacrylamide) under reducing conditions and electroblotted onto a nitrocellulose membrane (Hybond-C; Pharmacia). The membranes were blocked for 1 h at room temperature in 5% nonfat dry milk in Tris-buffered saline containing 0.1% Tween 20 (TBS-T). The blots were incubated overnight at 4°C in TBS-T with a rabbit antiseum raised against a fusion protein comprising the cytoplasmic domain of rat FAT1 (to be described in detail elsewhere); this was followed by a 2-h incubation with an appropriate secondary peroxidase-conjugated antibody (Amersham) in TBS-T. The immunoreactive proteins were visualized using enhanced chemiluminescence (Amersham).
Proliferation assays.
To measure proliferation in stem and precursor cells derived from the CNS, neurospheres were grown from cells dissociated from E18.5 CNS tissue and plated on nonadherent substrates as previously described (15), using different concentrations (20, 2, and 0.2 ng/ml) of fibroblast growth factor 2 (FGF-2) or EGF. Proliferation was assessed by measuring bromodeoxyuridine (BrdU) incorporation with an immunofluorescence assay kit (Roche). BrdU was added to the cultures for 2 h after they had been grown for 5 days in the appropriate growth factor concentration. After the BrdU pulse, neurospheres were dissociated mechanically and cells were plated on poly-d-lysine-coated slides to ensure that all cells attached prior to labeling. BrdU-labeled cells were visualized using a mouse monoclonal anti-BrdU antibody followed by an anti-mouse fluorescein isothiocyanate-conjugated antibody. Before mounting, the slides were incubated for 10 min in propidium iodide (20 μg/ml) in PBS, and the percentage of BrdU+ cells was calculated. Three independent experiments were performed at each growth factor concentration, and in each case 500 cells from five distinct fields were counted. To measure proliferation in skin, pregnant mFAT1+/− females (previously crossed with mFAT1+/− males) were injected twice with BrdU (100 μg/g of body weight) at 1-h intervals. At 1 h after the second injection, the animals were sacrificed and the embryos were extracted and, after euthanasia, were frozen in isopentane cooled by liquid nitrogen (−30 to −40°C). Frozen sections were cut, thawed, and fixed in 95% ethanol-5% acetic acid for 20 min at −20°C, washed in PBS, and incubated for 1 h in blocking solution containing 10% normal goat serum (Sigma). For quantification of BrdU labeling, BrdU+ cells (detected as above) and propidium iodide-labeled cell nuclei were counted in five adjacent fields in the basal layer of the epidermis in skin of mutant embryos and their littermates, and the percentage of labeled cells was calculated for each genotype.
Electron microscopy.
Tissue was fixed by immersion for 4 h at 4°C in 4% glutaraldehyde in 0.1 M PIPES buffer containing 2.0 mM CaCl2 at pH 7.4. Immediately before use, H2O2 was added to the fixative to give a final concentration of 0.1%. The tissue was rinsed in PIPES buffer, incubated in 1% osmium ferricyanide for 1 h, rinsed in water, incubated in 1% uranyl acetate for 1 h, rinsed again in water, and then dehydrated in an ascending series of ethanol solutions followed by two rinses in propylene oxide before being embedded in Spurr's resin. Thin sections (50 nm) were cut with an Ultracut UCT microtome (Leica, Vienna, Austria), mounted on copper grids, stained with uranyl acetate and lead citrate, and viewed in a CM100 electron microscope (Philips, Eindhoven, The Netherlands) operated at 80 kV.
Immunofluorescence and TUNEL labeling in vivo.
Cryostat sections (10 μm) from mFAT1−/− mice were fixed for 20 min in 4% paraformaldehyde and then permeabilized in 0.1% Triton X-100 in 10% normal goat serum-4% bovine serum albumin for 1 h at room temperature. Desmosomal cadherins were visualized using polyclonal antibodies (a kind gift of T. Magee), used at 1/100, and a fluorescein isothiocyanate-conjugated secondary antibody. β-Catenin was identified using a mouse monoclonal antibody (Transduction Laboratories), used at 1/200, and a Texas red-conjugated secondary antibody. For TUNEL labeling, prefixed E13.5 mFAT1−/− embryos were sectioned as above and labeled using the terminal deoxyribonucleotide transferase-mediated dUTP nick-end labeling (TUNEL) method with a commercially available kit (ApoTag; Intergen).
RESULTS
To inactivate the mFAT1 gene in mice, we generated a targeting vector for homologous recombination by using the recombination efficiency of yeast (43) to insert a lacZ marker-selection cassette into a 7-kb genomic clone containing the first ATG of the mFAT1 gene as described in Materials and Methods. The targeting construct was then introduced into ES cells, and clones showing homologous recombination were identified by both genomic PCR and Southern blot analyses with an internal and an external 3′ probe. A total of 400 clones were screened, and the expected 3.4- and 4.2-kb EcoRI restriction fragments for the mutant and the wild-type allele, respectively (Fig. 1A), were obtained with three ES clones. Chimeras were generated from two of these clones and crossed with C57BL/6 mice, and the offspring were genotyped by Southern blotting (Fig. 1B) and PCR amplification (data not shown). One of these chimeric animals gave germ line transmission. Mice heterozygous for the mFAT1 mutant allele appeared normal and fertile and were indistinguishable from wild-type mice. Mice were then systematically backcrossed onto both C57BL/6 and 129Sv backgrounds.
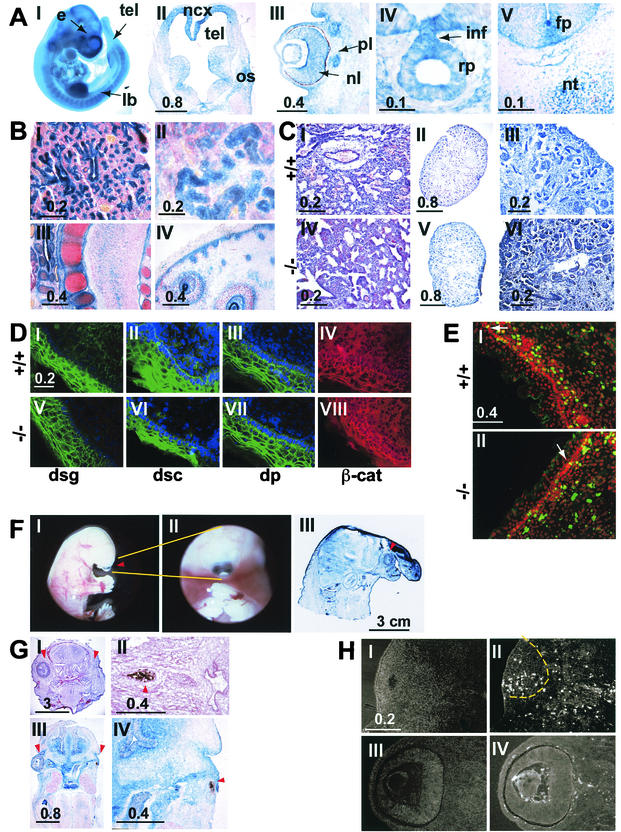
These heterozygous animals were first used for analysis of the pattern of expression of mFAT1, as determined by expression of the lacZ reporter gene, during embryonic development. The results obtained were in close agreement with results of previous in situ hybridization studies of mice (6) and rats (33), with no evidence for expression of the transgene in areas not identified by these studies. We found mFAT1 expression early in postimplantation development, with detectable X-Gal staining at E6.5 in the epiblast (data not shown). At mid-gestation (E12.5), mFAT1 expression was observed in the limb buds, telenchephalon, developing eye, Rathke's pouch, and spinal cord (Fig. 2A, I to IV). Within the spinal cord, expression was observed in the floor plate cells of the neural tube as well as in the notochord (Fig. 2A, V). At later embryonic stages, expression was also seen in the respiratory epithelium, renal epithelial cells, cartilage, and basal layer of the skin (Fig. 2B, I to IV).
FIG. 2.
(A) The lacZ reporter gene was used to analyze the pattern of expression of the FAT1 gene in the embryo. Micrographs I to V show X-Gal staining in an intact (I) or sectioned (II to V) E12.5 embryo. The staining is strong in the forebrain (tel in panel I), mainly in the neocortex around the telencephalic vesicles (top part of panel II; note the lower level of staining in the ganglionic eminences in the lower part of the section), in the limb buds (panel I), in the developing eye (panel III), and in Rathke's pouch (panel IV). Panel V shows lacZ expression in the spinal cord of an E12.5 embryo. Note the staining in the region of the floor plate. e, eye; fp, floor plate; inf, infundibulum; lb, limb bud; ncx, neocortex; nl, neural layer; nc, notochord; os, optic stalk; pl, pigment layer; rp, Rathcke's pouch; tel, telencephalon. Bars indicates sizes in millimeters. (B) lacZ staining in newborn mFAT1−/− pups. Epithelial cells in the lungs (I) and kidneys (II), cells surrounding developing cartilage (III), and the basal layer of the epidermis in the skin (IV) show a strong X-Gal staining. All sections were counter stained with Neutral Red. Bars indicate sizes in millimeters. (C) Hematoxylin-eosin staining of sections from lungs of wild-type and mFAT1−/− mice (panels I and IV) and kidneys (panels II and V; shown at higher power in panels III and VI, respectively). Light microscopy does not reveal any differences between the different genotypes. Bars indicate sizes in millimeters. (D) Distribution of adhesion-related proteins in skin from wild-type and mFAT−/− mice. No differences between the mutant and the wild-type animals were observed when using antibodies against the desmosomal proteins desmoglein (dsg), desmocollin (dsc), and desmoplakin (dp). The cellular distribution of β-catenin also does not shown any alteration between the wild-type and mFAT1−/− mice. (E) Anti-BrdU immunofluorescent labeling (green) and propidium iodide (red, to visualize nuclei) in the skin of wild-type and mutant mice after BrdU incorporation in vivo, as described in Materials and Methods. Labeled cells in the basal layer of the epidermis are indicated by white arrows, and no differences were observed in the labeling index in this cell layer. Note the lack of labeling in the more superficial epidermal cells. The labeled cells in the underlying dermis were not included in the analysis. (F) The holoprosencephaly phenotype seen in some mFAT1−/− mice is illustratedin micrographs I and II, which show lateral and ventral views of an intact embryo with cyclopia (an eye in the centre of the face [arrowhead in panel I]). Micrograph III shows a cross-section of a newborn mutant mouse with holoprosencephaly, showing a proboscis protruding above the eye (arrowhead). The section has been stained with X-Gal, showing the expression of lacZ from the transgene. (G) The eye phenotype observed in many of the FAT1−/− mice is shown. Micrographs I to IV are tissue sections from two different homozygous null embryos, one at E17.5 stained with hematoxylin and eosin (panels I and II) and one at E13.5 stained with X-Gal (panels III and IV). Note the complete loss of one eye in panels I and III (right arrowhead, with normal eye indicated by left arrowhead) and the presence of retinal pigment epithelium but not neural retina and lens in II and IV (arrowhead). Bars indicate sizes in millimeters. (H) TUNEL staining of an E13.5 embryo with an abnormal eye. The number of TUNEL-positive cells is dramatically increased in the abnormal eye (panels I and II, with panel I showing a 4′,6-diamidino-2-phenylindole (DAPI)-labeled section to visualize tissue and panel II showing TUNEL-positive cells, with the eye indicated by the dashed line) compared with the normal eye (panel III and IV). Bars indicate size in millimeters.
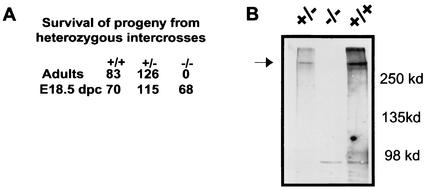
We next crossed heterozygous animals to determine the homozygous phenotype. Genotyping of E18.5 embryos obtained from these crosses showed no significant deviation from the expected Mendelian ratio (1:2:1) of wild-type, heterozygous, and homozygous animals. After birth, however, we found dead pups within 48 h of birth and none of the surviving pups were homozygous for the mutated mFAT1 allele (Fig. 3). Western blot analysis of skin from wild-type, heterozygous, and homozygous E18.5 mice, using a polyclonal affinity-purified antibody raised against a glutathione S-transferase fusion protein comprising the cytoplasmic domain of mFAT1, confirmed the complete loss of the mFAT1 protein in the homozygous animals (Fig. 3). We conclude, therefore, that mFAT1 is necessary for postnatal survival.
FIG. 3.
(A) Genotype distribution of progeny from mFAT1+/− intercrosses. Although the ratio of the three genotypes was normal at E18.5, no mFAT1−/− animals were found postnatally. (B) Western blot analysis of skin from wild-type, heterozygous, and homozygous mutant mice confirms the complete loss of the mFAT1 protein.
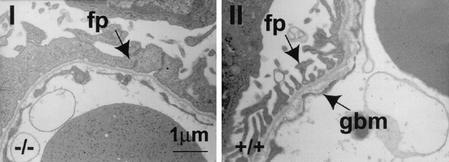
To explain the perinatal death of the mFAT1 null mice, we examined the lungs and kidneys by using light and electron microscopy. These organs were chosen because FAT1 is expressed in epithelial cells within both organs (6, 10, 14, 33) and their function is required only after birth, thus making dysfunction a possible cause of perinatal death. The lungs (Fig. 2C, I and IV) and kidneys (Fig. 2C, II, III, V, and VI) of mFAT1 null E18.5 mice were analyzed by light microscopy and showed no abnormalities compared with heterozygous and wild-type littermate controls. In particular, renal glomerular size was similar in the different genotypes. While the epithelium in the lungs was also normal by electron microscopy (data not shown), in the kidney the foot processes of the podocytes (the renal glomerular epithelial cells) were abnormal (Fig. 4). These processes are normally joined to each other adjacent to the glomerular basement membrane by slit junctions, specialized adhesion complexes that provide a gap of 30 to 45 nm between processes, sufficient to allow filtration from blood to renal tubules (36). In three null animals examined from two different litters, electron microscopy showed that these gaps were obliterated, with the processes fused as shown in Fig. 4 to generate a sheet of cell processes overlying the basement membrane. By contrast, slit junctions between podocyte foot processes were normal in two heterozygotes and one wild-type animal from the same litters.
FIG. 4.
Abnormalities in the kidneys of mFAT1−/− mice as seen by electron microscopy. The normal gaps (slit junctions [right panel]) between the podocyte (glomerular epithelial cell) foot processes (fp) adjacent to the glomerular basement membrane (gbm) have been lost, with flattening and fusion of the podocyte processes (left panel).
To determine the role of mFAT1 in other adhesion complexes, we examined skin, another tissue in which mFAT1 is expressed strongly (Fig 2B, IV), from E18.5 mFAT1 null mice. Epidermal cells in the skin are connected by both adherence junctions, in which classical cadherin cytoplasmic domains are associated with β-catenin (17), and desmosomes, in which the cytoplasmic domains of the cadherin family members desmocollin and desmoglein are associated with desmoplakin (11, 20). We therefore examined the distribution of these molecules by immunofluorescence to determine whether loss of mFAT1 altered the distribution of adhesion complex-related proteins. As shown in Fig. 2D, the distributions of desmoglein (I and IV), desmocollin (II and VI), desmopolakin (III and VII), and β-catenin (IV and VIII) were not altered in the mFAT1 null mice compared with wild-type littermates. In addition, electron microscopic studies of skin did not show any changes in cell morphology in null animals compared with wild-type and heterozygous littermates (not shown). We conclude, therefore, that mFAT1 is not essential for the formation and maintenance of intercellular adhesion complexes in the mouse skin.
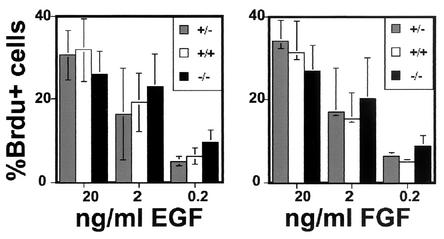
In light of the tumor suppressor role for FAT in Drosophila (25), we next asked whether loss of mFAT1 was associated with any changes in cell proliferation in two tissues: skin and the neuroepithelium of the CNS. These tissue were chosen because they both contain stem and transit-amplifying precursor cell populations (8, 16). Moreover, mFAT1 expression persists in postnatal animals in precisely those regions of these tissues where these cell populations are found, and cell proliferation with the generation of new differentiated cells continues throughout life (33). To measure proliferation in neuroepithelial cells, we used a cell culture technique in which stem and precursor cells taken from embryonic CNS were grown as neurospheres: three-dimensional aggregates of cells that can be grown in cell culture on nonadherent substrates (37, 38). These cells proliferate in response to either FGF-2 or EGF, and when neurospheres were grown from heterozygous or null CNS tissue, no significant differences in the responses to either growth factor (as measured by BrdU incorporation) were seen (Fig. 5). Proliferation in embryonic skin was measured in vivo following BrdU injections into the pregnant mother late in the pregnancy. As expected, BrdU incorporation within the epidermis was seen only in the basal layer of cells from which the postmitotic keratinocytes arise. Once again, there were no differences between heterozygous and null embryos (Fig. 2E), with the observed levels being 30 and 31% respectively.
FIG. 5.
BrdU proliferation assays on neural precursors grown in vitro. Neurospheres of the three different genotypes grown as described in Materials and Methods in different concentrations of the growth factors EGF and FGF-2 do not show any significant differences in the percentage of BrdU-positive cells.
Two unexpected phenotypes found in some of the mFAT1-deficient mice were holoprosencephaly and microphthalmia-anophthalmia. Holoprosencephaly was observed in 4 (6%) of 68 null embryos, with cyclopia (a single midline eye) and a midline proboscis being characteristic of this developmental abnormality of ventral forebrain induction (28) (Fig. 2F). Eye abnormalities were more common, with one or both eyes being either small or absent in 27 (40%) of 68 embryos (Fig. 2G, I and III). Histological analysis showed that the neural retinal layer and lens were absent in these animals and that the retinal pigment layer was misshapen and greatly reduced in size (Fig. 2G, II and IV). Interestingly, the phenotype of the eye in null embryos was variable even within a single embryo, since we often saw a severe eye phenotype on one side and normal eye development on the other side (Fig. 2G, I and III). Microphthalmia was also seen in one of three null animals (all of which exhibited perinatal lathality) generated within two litters of the transgenic line backcrossed five times onto a 129Sv background. To determine whether the abnormalities of eye development reflected increased levels of apoptosis, we used TUNEL assays on sections of E13.5 mFAT1 null embryos. Animals with one normal and one abnormal eye were chosen so as to have an internal control in the experiment. The normal eye showed very few apoptotic cells (Fig. 2H, III and IV), while the abnormal eye contained a large number of these cells (Fig. 2H, I and II), from which we conclude that the abnormal phenotype in mFAT1 null mice is associated with increased degeneration and apoptosis in the developing eye.
DISCUSSION
The abnormalities in the mFAT1 null mice described here confirm the essential role of this protocadherin for correct development in the mouse. This result contrasts with the normal viability and fertility reported for mice lacking PAPC (pcd8), the only other protocadherin molecule to have been knocked out so far (48). Our analysis of the mFAT1 null mice reveals renal glomerular abnormalities as the most likely cause of the perinatal death we observed and which was also noted in a survey of transgenic mice generated using a gene trap approach for secreted and membrane proteins (26). Our results also provide evidence for a novel and unexpected role for FAT1 in forebrain and eye development.
The expression of mFAT1 at the slit junctions, the intracellular junctions between podocytes in the glomerular filtration membrane, has been reported previously (14). Slit junctions are modified adhesion junctions, with the wide intercellular space allowing renal filtration. Based on the large size of mFAT1 and colocalization of the tight-junction protein ZO-1 with the FAT cytoplasmic domain, it has been suggested that mFAT1 acts as both an adhesion molecule and a spacer at the junction (14). As a consequence, mFAT1 generates the necessary intercellular adhesion between podocyte processes while at the same time maintaining an extracellular gap wider than is normally found in adhesion junctions. Our findings with the mFAT1 null mice support this hypothesis. In the absence of mFAT1, the ordered array of slit junctions disappears and the podocytes become closely apposed to one another, defining an essential role for mFAT1 as a slit junction adhesion molecule with a role in spacing the processes. These abnormalities in podocyte morphology are similar to those reported for mice lacking nephrin, another transmembrane molecule associated with the slit junction (35). In humans, nephrin mutations cause renal filtration abnormalities, as evidenced by a congenital nephrotic syndrome (18, 22). Together, these observations in mice and humans with nephrin mutations show that podocyte fusion, such as we observed in the mice lacking mFAT1, is associated with significant abnormalities of renal function. We conclude, therefore, that this most probably explains the perinatal death of the mFAT1 null mice.
Unlike the renal abnormalities, the defects in forebrain and eye development we observed in some of the mFAT1 null mice were completely unexpected on the basis of previous studies on the function of the protein. The partial or complete loss of the eye does not reflect a failure of the initial developmental events of optic vesicle formation from the forebrain, since remnants of pigment epithelium were present in the embryos. Since the development of the retinal layers requires a hierarchy of inductive interactions between optic vesicle and overlying ectoderm (40), these early inductive events must have occurred normally. Rather, the phenotype suggests a subsequent degeneration of the eye, as demonstrated by increased apoptosis as well as by the remnants of some retinal cell layers. The mechanisms linking mFAT1 to the maintenance of the embryonic eye are unknown. mFAT1 is expressed in both the neural retina and the underlying pigment epithelium and may therefore contribute to any adhesive interactions within and between these two cell layers necessary for development. The mFAT1 cytoplasmic domain contains both sequences that bind β-catenin in a two-hybrid assay (6) and a PDZ motif binding domain (33), thus allowing a number of potential intracellular signaling pathways to be regulated. Characterization of these pathways requires further experiments, but the interesting observations that the eye phenotype of the BMP-7 null mouse is very similar to the one we observed in our mFAT1 null mice (9) and that inhibition of BMP signaling in the chick causes microphthalmia and other abnormalities of optic cup development (1) suggest that FAT1 may interact with the BMP signaling mechanisms.
Holoprosencephaly results from a failure of ventral inductive signaling in forebrain development (12, 28), and the finding of this developmental abnormality in some of the mFAT1 null mice demonstrates another important role for FAT in neural development. A failure of sonic hedgehog (Shh) signaling is a well-recognized cause of holoprosencephaly in mice and humans (5, 39). However, we found no evidence for perturbation of Shh signaling in mFAT1−/− mice, since experiments on the spinal cord did not show any alterations in the expression pattern of genes known to be regulated by the concentration gradient of Shh derived from ventral midline sources such as the floorplate and notochord (L. Ciani, unpublished observations). Two other possible mechanisms for holoprosencepaly in the mice are suggested by previous work. First, in contrast to the spinal cord, where ventrally derived Shh is antagonized by dorsally derived BMP signaling (23, 24), in forebrain development Shh and BMP-7 are both produced ventrally by the prechordal mesoderm (or the prechordal plate) and signal cooperatively to induce ventral midline cells in the overlying neuroectoderm (7). Any alteration in BMP-7 signaling, as suggested by the eye phenotype, could also therefore increase the likelihood of holoprosencephaly. Second, an early event in forebrain development (before the formation of the prechordal plate) is the induction of the anterior neural ectoderm by signals transmitted in planar fashion through the developing ectoderm from Hensen's node (32). Studies of Drosophila have shown that FAT is involved in planar signaling in the eye (10a, 49), and a requirement for a similar role in neural ectoderm could explain abnormalities of forebrain development in mFAT1 null mice. Further studies of forebrain patterning in the mFAT1 null mice are clearly required, but these will be complicated by the low penetrance of the holoprosencephaly phenotype. However, at this stage our results clearly show that FAT1 mutations are a potential cause of holoprosencephaly in humans, with FAT1 being the first cell adhesion molecule to be implicated in this relatively common congenital abnormality (28).
Our studies of the skin and neuroepithelium, both of which contain a resident population of proliferating stem and precursor cells and in which mFAT1 is highly expressed, provide no support for the role for mFAT1 in growth suppression suggested by the Drosophila studies (25). The presence of two other FAT genes and the potential for compensation in null mice make it impossible to conclude that mFAT1 does not play such a role in normal development. However, expression of these other FAT genes appears to be restricted to the CNS (27), making a compensatory role in skin less likely. Generation of tissue- and stage-specific knockouts is required to examine this question of the role of FAT1 in proliferation more fully. However, our results do show that significant functional differences may be present between Drosophila FAT and vertebrate mFAT1, a point also emphasized by the observation that Drosophila FAT lacks the PDZ binding motif and may therefore interact with a different repertoire of downstream signaling molecules. Further genetic studies of FATs and other protocadherins will define the roles of this large family of molecules in vertebrates. They may also, as illustrated by our findings on holoprosencephaly and by the association between a human deafness syndrome and mutations in pcdh15 (2, 3), provide further novel insights into human diseases.
Acknowledgments
This work was funded by the Cancer Research Campaign (now Cancer Research UK) and the Wellcome Trust. C.F.-C. is a Wellcome Trust Research Leave fellow, and A.P. was funded by a studentship from Merck, Sharpe, and Dohme.
We are extremely grateful to Jeremy Skepper for help with the electron microscopy, to Tony Magee for the gift of antibodies and many helpful discussions throughout this work, and to Ian Horseman for care of the mice.
REFERENCES
- 1.Adler, R., and T. L. Belecky-Adams. 2002. The role of bone morphogenetic proteins in the differentiation of the ventral optic cup. Development 129:3161-3171. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed, Z. M., S. Riazuddin, S. L. Bernstein, Z. Ahmed, S. Khan, A. J. Griffith, R. J. Morell, T. B. Friedman, and E. R. Wilcox. 2001. Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am. J. Hum. Genet. 69:25-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alagramam, K. N., H. Yuan, M. H. Kuehn, C. L. Murcia, S. Wayne, C. R. Srisailpathy, R. B. Lowry, R. Knaus, L. Van Laer, F. P. Bernier, S. Schwartz, C. Lee, C. C. Morton, R. F. Mullins, A. Ramesh, G. Van Camp, G. S. Hageman, R. P. Woychik, R. J. Smith, and G. S. Hagemen. 2001. Mutations in the novel protocadherin PCDH15 cause Usher syndrome type 1F. Hum. Mol. Genet. 10:1709-1718. [DOI] [PubMed] [Google Scholar]
- 4.Bradley, R. S., A. Espeseth, and C. Kintner. 1998. NF-protocadherin, a novel member of the cadherin superfamily, is required for Xenopus ectodermal differentiation. Curr. Biol. 8:325-334. [DOI] [PubMed] [Google Scholar]
- 5.Chiang, C., Y. Litingtung, E. Lee, K. E. Young, J. L. Corden, H. Westphal, and P. A. Beachy. 1996. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383:407-413. [DOI] [PubMed] [Google Scholar]
- 6.Cox, B., A. K. Hadjantonakis, J. E. Collins, and A. I. Magee. 2000. Cloning and expression throughout mouse development of mfat1, a homologue of the Drosophila tumour suppressor gene fat. Dev. Dyn. 217:233-240. [DOI] [PubMed] [Google Scholar]
- 7.Dale, J. K., C. Vesque, T. J. Lints, T. K. Sampath, A. Furley, J. Dodd, and M. Placzek. 1997. Cooperation of BMP7 and SHH in the induction of forebrain ventral midline cells by prechordal mesoderm. Cell 90:257-269. [DOI] [PubMed] [Google Scholar]
- 8.Doetsch, F., I. Caille, D. A. Lim, J. M. Garcia-Verdugo, and A. Alvarez-Buylla. 1999. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 97:703-716. [DOI] [PubMed] [Google Scholar]
- 9.Dudley, A. T., K. M. Lyons, and E. J. Robertson. 1995. A requirement for bone morphogenetic protein-7 during development of the mammalian kidney and eye. Genes Dev. 9:2795-2807. [DOI] [PubMed] [Google Scholar]
- 10.Dunne, J., A. M. Hanby, R. Poulsom, T. A. Jones, D. Sheer, W. G. Chin, S. M. Da, Q. Zhao, P. C. Beverley, and M. J. Owen. 1995. Molecular cloning and tissue expression of FAT, the human homologue of the Drosophila fat gene that is located on chromosome 4q34-q35 and encodes a putative adhesion molecule. Genomics 30:207-223. [DOI] [PubMed] [Google Scholar]
- 10a.Fanto, M., L. Clayton, J. Meredith, K. Hardiman, B. Charroux, S. Kerridge, and H. McNeill. 2003. The tumour-suppressor and cell adhesion molecule Fat controls planar polarity via physical interactions with atrophin, a transcriptional co-repressor. Development 130:763-774. [DOI] [PubMed] [Google Scholar]
- 11.Garrod, D., M. Chidgey, and A. North. 1996. Desmosomes: differentiation, development, dynamics and disease. Curr. Opin. Cell Biol. 8:670-678. [DOI] [PubMed] [Google Scholar]
- 12.Golden, J. A. 1998. Holoprosencephaly: a defect in brain patterning. J. Neuropathol. Exp. Neurol. 57:991-999. [DOI] [PubMed] [Google Scholar]
- 13.Gumbiner, B. M. 2000. Regulation of cadherin adhesive activity. J. Cell Biol. 148:399-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue, T., E. Yaoita, H. Kurihara, F. Shimizu, T. Sakai, T. Kobayashi, K. Ohshiro, H. Kawachi, H. Okada, H. Suzuki, I. Kihara, and T. Yamamoto. 2001. FAT is a component of glomerular slit diaphragms. Kidney Int. 59:1003-1012. [DOI] [PubMed] [Google Scholar]
- 15.Jacques, T. S., J. B. Relvas, S. Nishimura, R. Pytela, G. M. Edwards, C. H. Streuli, and C. ffrench-Constant. 1998. Neural precursor cell chain migration and division are regulated through different beta1 integrins. Development 125:3167-3177. [DOI] [PubMed] [Google Scholar]
- 16.Jones, P. H., and F. M. Watt. 1993. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell 73:713-724. [DOI] [PubMed] [Google Scholar]
- 17.Kemler, R. 1993. From cadherins to catenins: cytoplasmic protein interactions and regulation of cell adhesion. Trends Genet. 9:317-321. [DOI] [PubMed] [Google Scholar]
- 18.Kestila, M., U. Lenkkeri, M. Mannikko, J. Lamerdin, P. McCready, H. Putaala, V. Ruotsalainen, T. Morita, M. Nissinen, R. Herva, C. E. Kashtan, L. Peltonen, C. Holmberg, A. Olsen, and K. Tryggvason. 1998. Positionally cloned gene for a novel glomerular protein—nephrin—is mutated in congenital nephrotic syndrome. Mol. Cell 1:575-582. [DOI] [PubMed] [Google Scholar]
- 19.Kim, S. H., A. Yamamoto, T. Bouwmeester, E. Agius, and E. M. Robertis. 1998. The role of paraxial protocadherin in selective adhesion and cell movements of the mesoderm during Xenopus gastrulation. Development 125:4681-4690. [DOI] [PubMed] [Google Scholar]
- 20.Koch, P. J., and W. W. Franke. 1994. Desmosomal cadherins: another growing multigene family of adhesion molecules. Curr. Opin. Cell Biol. 6:682-687. [DOI] [PubMed] [Google Scholar]
- 21.Kuroda, H., M. Inui, K. Sugimoto, T. Hayata, and M. Asashima. 2002. Axial protocadherin is a mediator of prenotochord cell sorting in Xenopus. Dev. Biol. 244:267-277. [DOI] [PubMed] [Google Scholar]
- 22.Lenkkeri, U., M. Mannikko, P. McCready, J. Lamerdin, O. Gribouval, P. M. Niaudet, C. K. Antignac, C. E. Kashtan, C. Homberg, A. Olsen, M. Kestila, and K. Tryggvason. 1999. Structure of the gene for congenital nephrotic syndrome of the finnish type (NPHS1) and characterization of mutations. Am. J. Hum. Genet. 64:51-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liem, K. F., Jr., T. M. Jessell, and J. Briscoe. 2000. Regulation of the neural patterning activity of sonic hedgehog by secreted BMP inhibitors expressed by notochord and somites. Development 127:4855-4866. [DOI] [PubMed] [Google Scholar]
- 24.Liem, K. F., Jr., G. Tremml, H. Roelink, and T. M. Jessell. 1995. Dorsal differentiation of neural plate cells induced by BMP-mediated signals from epidermal ectoderm. Cell 82:969-979. [DOI] [PubMed] [Google Scholar]
- 25.Mahoney, P. A., U. Weber, P. Onofrechuk, H. Biessmann, P. J. Bryant, and C. S. Goodman. 1991. The fat tumor suppressor gene in Drosophila encodes a novel member of the cadherin gene superfamily. Cell 67:853-868. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell, K. J., K. I. Pinson, O. G. Kelly, J. Brennan, J. Zupicich, P. Scherz, P. A. Leighton, L. V. Goodrich, X. Lu, B. J. Avery, P. Tate, K. Dill, E. Pangilinan, P. Wakenight, M. Tessier-Lavigne, and W. C. Skarnes. 2001. Functional analysis of secreted and transmembrane proteins critical to mouse development. Nat. Genet. 28:241-249. [DOI] [PubMed] [Google Scholar]
- 27.Mitsui, K., D. Nakajima, O. Ohara, and M. Nakayama. 2002. Mammalian fat3: a large protein that contains multiple cadherin and EGF-like motifs. Biochem. Biophys. Res. Commun. 290:1260-1266. [DOI] [PubMed] [Google Scholar]
- 28.Muenke, M., and P. A. Beachy. 2000. Genetics of ventral forebrain development and holoprosencephaly. Curr. Opin. Genet. Dev. 10:262-269. [DOI] [PubMed] [Google Scholar]
- 29.Nakayama, M., D. Nakajima, T. Nagase, N. Nomura, N. Seki, and O. Ohara. 1998. Identification of high-molecular-weight proteins with multiple EGF-like motifs by motif-trap screening. Genomics 51:27-34. [DOI] [PubMed] [Google Scholar]
- 30.Nollet, F., P. Kools, and F. van Roy. 2000. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J. Mol. Biol. 299:551-572. [DOI] [PubMed] [Google Scholar]
- 31.Obata, S., H. Sago, N. Mori, J. M. Rochelle, M. F. Seldin, M. Davidson, T. St. John, S. Taketani, and S. T. Suzuki. 1995. Protocadherin Pcdh2 shows properties similar to, but distinct from, those of classical cadherins. J. Cell Sci. 108:3765-3773. [DOI] [PubMed] [Google Scholar]
- 32.Pera, E. M., and M. Kessel. 1997. Patterning of the chick forebrain anlage by the prechordal plate. Development 124:4153-4162. [DOI] [PubMed] [Google Scholar]
- 33.Ponassi, M., T. S. Jacques, L. Ciani, and C. ffrench Constant. 1999. Expression of the rat homologue of the Drosophila fat tumour suppressor gene. Mech. Dev. 80:207-212. [DOI] [PubMed] [Google Scholar]
- 34.Provost, E., and D. L. Rimm. 1999. Controversies at the cytoplasmic face of the cadherin-based adhesion complex. Curr. Opin. Cell Biol. 11:567-572. [DOI] [PubMed] [Google Scholar]
- 35.Rantanen, M., T. Palmen, A. Patari, H. Ahola, S. Lehtonen, E. Astrom, T. Floss, F. Vauti, W. Wurst, P. Ruiz, D. Kerjaschki, and H. Holthofer. 2002. Nephrin TRAP mice lack slit diaphragms and show fibrotic glomeruli and cystic tubular lesions. J. Am. Soc. Nephrol. 13:1586-1594. [DOI] [PubMed] [Google Scholar]
- 36.Reiser, J., W. Kriz, M. Kretzler, and P. Mundel. 2000. The glomerular slit diaphragm is a modified adherens junction. J. Am. Soc. Nephrol. 11:1-8. [DOI] [PubMed] [Google Scholar]
- 37.Reynolds, B. A., and S. Weiss. 1996. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev. Biol. 175:1-13. [DOI] [PubMed] [Google Scholar]
- 38.Reynolds, B. A., and S. Weiss. 1992. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255:1707-1710. [DOI] [PubMed] [Google Scholar]
- 39.Roessler, E., E. Belloni, K. Gaudenz, P. Jay, P. Berta, S. W. Scherer, L. C. Tsui, and M. Muenke. 1996. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat. Genet. 14:357-360. [DOI] [PubMed] [Google Scholar]
- 40.Saha, M. S., M. Servetnick, and R. M. Grainger. 1992. Vertebrate eye development. Curr. Opin. Genet. Dev. 2:582-588. [DOI] [PubMed] [Google Scholar]
- 41.Shapiro, L., A. M. Fannon, P. D. Kwong, A. Thompson, M. S. Lehmann, G. Grubel, J. F. Legrand, J. Als-Nielsen, D. R. Colman, and W. A. Hendrickson. 1995. Structural basis of cell-cell adhesion by cadherins. Nature 374:327-337. [DOI] [PubMed] [Google Scholar]
- 42.Steinberg, M. S., and P. M. McNutt. 1999. Cadherins and their connections: adhesion junctions have broader functions. Curr. Opin. Cell Biol. 11:554-560. [DOI] [PubMed] [Google Scholar]
- 43.Storck, T., U. Kruth, R. Kolhekar, R. Sprengel, and P. H. Seeburg. 1996. Rapid construction in yeast of complex targeting vectors for gene manipulation in the mouse. Nucleic Acids Res. 24:4594-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki, S. T. 2000. Recent progress in protocadherin research. Exp. Cell Res. 261:13-18. [DOI] [PubMed] [Google Scholar]
- 45.Takeichi, M. 1995. Morphogenetic roles of classic cadherins. Curr. Opin. Cell Biol. 7:619-627. [DOI] [PubMed] [Google Scholar]
- 46.Tepass, U., K. Truong, D. Godt, M. Ikura, and M. Peifer. 2000. Cadherins in embryonic and neural morphogenesis. Nat. Rev. Mol. Cell. Biol. 1:91-100. [DOI] [PubMed] [Google Scholar]
- 47.Wu, Q., and T. Maniatis. 2000. Large exons encoding multiple ectodomains are a characteristic feature of protocadherin genes. Proc. Natl. Acad. Sci. USA 97:3124-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamoto, A., C. Kemp, D. Bachiller, D. Geissert, and E. M. De Robertis. 2000. Mouse paraxial protocadherin is expressed in trunk mesoderm and is not essential for mouse development. Genesis 27:49-57. [PubMed] [Google Scholar]
- 49.Yang, C. H., J. D. Axelrod, and M. A. Simon. 2002. Regulation of Frizzled by fat-like cadherins during planar polarity signaling in the Drosophila compound eye. Cell 108:675-688. [DOI] [PubMed] [Google Scholar]