Abstract
Rad53 of Saccharomyces cerevisiae is a checkpoint kinase whose structure and function are conserved among eukaryotes. When a cell detects damaged DNA, Rad53 activity is dramatically increased, which ultimately leads to changes in DNA replication, repair, and cell division. Despite its central role in checkpoint signaling, little is known about Rad53 substrates or substrate specificity. A number of proteins are implicated as Rad53 substrates; however, the evidence remains indirect. Previously, we have provided evidence that Swi6, a subunit of the Swi4/Swi6 late-G1-specific transcriptional activator, is a substrate of Rad53 in the G1/S DNA damage checkpoint. In the present study we identify Rad53 phosphorylation sites in Swi6 in vitro and demonstrate that at least one of them is targeted by Rad53 in vivo. Mutations in these phosphorylation sites in Swi6 shorten but do not eliminate the Rad53-dependent delay of the G1-to-S transition after DNA damage. We derive a consensus for Rad53 site preference at positions −2 and +2 (−2/+2) and identify its potential substrates in the yeast proteome. Finally, we present evidence that one of these candidates, the cohesin complex subunit Scc1 undergoes DNA damage-dependent phosphorylation, which is in part dependent on Rad53.
Checkpoints or surveillance mechanisms of eukaryotic cells ensure that critical stages of the cell cycle are initiated and completed in the proper order. These mechanisms become critical for survival when DNA is damaged and must be repaired before chromosome segregation lest genetic information is lost in cell division. A checkpoint is comprised of damage sensors and transducers, which convey a regulatory signal to the cell cycle and repair machinery (34, 64). In yeast cells, as in higher eukaryotes, there are three recognized DNA damage checkpoints (17). When DNA is damaged, cells can delay the G1-to-S transition, decelerate S phase progression, and pause before mitosis. Operation of the checkpoint involves activation and deactivation of the checkpoint kinases Mec1, Rad53, Chk1, and Dun1 (3, 42, 44, 45, 56). The key regulators, Rad53 and Mec1 kinases, are involved in all three of the checkpoints via partially overlapping circuits (17). While the phosphorylation preference of Mec1 is known (31), Rad53 specificity remains undetermined. Also, the targets of the Rad53 kinase in budding yeast are not well established. A number of proteins are reported to undergo phosphorylation in a Rad53- and DNA damage-dependent manner in vivo; however, it remains to be established whether these proteins are direct substrates of Rad53 and, if so, what specific residues are phosphorylated (64).
The sensitivity of the three DNA damage checkpoints to the levels of DNA damage appears to be quite different. For example, the same dose of gamma rays leads to a higher level of activation of Rad53 in G2 cells than in G1 cells (21). One double-strand break can hold cells at the G2/M boundary for many hours (46), whereas G1 cells do not activate the checkpoint in response to a single double-strand break (42).
G1 progression includes two waves of G1 cyclin (Cln) transcription, where CLN3 transcription peaks at the end of M and in early G1 and CLN1 and CLN2 mRNA levels peak in late G1 (37). Cln3/Cdk is an unstable activator of the Swi4/Swi6 transcription factor complex via a yet-unidentified mechanism (9, 55). When active, this complex brings about a rapid burst of transcription of dozens of genes (27), including CLN1 and CLN2, which enables budding and the G1-to-S transition (38).
Methyl methanesulfonate (MMS) treatment and UV and gamma irradiation during G1 can delay both budding and the onset of DNA replication. The treated cells remain sensitive to the mating pheromone, alpha factor (53, 54), and have low levels of CLN mRNA (51) and low Cln/Cdk activity (16). CLN1 and CLN2 transcription is downregulated upon addition of MMS (51). The rate of recovery of CLN1 and CLN2 mRNAs, which determines the timing of the G1-to-S transition, depends on at least one of the central checkpoint kinases, Rad53, and on the Swi4/Swi6 transcription factor complex, which activates CLN1 and CLN2 transcription. Moreover, Swi6 undergoes a DNA damage-inducible, Rad53-dependent phosphorylation in vivo and can be phosphorylated by Rad53 immunoprecipitates in vitro (51).
In the present study, we demonstrate that a short pulse of MMS administered to elutriated G1 cells delays the G1-to-S transition. MMS-treated G1 cells remain in G1 for a longer time and thus grow substantially larger than untreated cells before they transit into S phase. This delay of S phase is substantially reduced in rad53-11 checkpoint mutant cells. G1 cells lacking Swi6 also have a shorter delay of the G1-to-S transition after MMS treatment. Swi6 is phosphorylated by Rad53 in vitro, and we have identified five sites of this phosphorylation. At least one of these sites in Swi6 is phosphorylated by Rad53 in response to DNA damage in vivo. Alignment of the identified sites reveals conservation of flanking sequences, which we show to be critical for phosphorylation of Swi6 by Rad53 in vitro. This enables us to derive a consensus site for Rad53 phosphorylation and to implicate a number of other proteins as Rad53 targets. One of these new candidate proteins, the cohesin complex subunit Scc1, was tested and shown to undergo damage-induced phosphorylation in a Rad53-dependent manner. In cells with the Rad53 phosphorylation sites eliminated from Swi6, the checkpoint response to MMS in G1 is reduced but not abolished. This may be explained by the large number of other potential Rad53 substrates in G1 cells.
MATERIALS AND METHODS
Strains and plasmids.
The yeast strains BY2006 (MATa ura3 leu2 trp1 his3), BY2007 (MATa ura3 leu2 trp1 his3 rad53-11::URA3), and BY2390 (MATa ura3 leu2 trp1 his3 rad53-11) have been described elsewhere (50, 51). BY2917 and BY2981 are swi6::LEU2 and swi6::URA3 derivatives of BY2006. BY3256 and BY3257 are RAD53-HA-TRP1 and rad53-11-HA-TRP1 derivatives of BY2006 and BY2390, respectively, with Rad53 tagged with a hemagglutinin (HA) epitope tag (13). BY3258 is a swi6::LEU2 derivative of BY2390. The genotype of BY2918 is MATα ura3 leu2 trp1 his3 rad53-11::LEU2. SBY376 (MATa ura3 leu2 trp1::LacO his::pCUP1-GFP12-LacI12 can1 ade2 bar1 MCD1-HA3) (4) is a kind gift of S. Biggins. BY3242 and BY3243 are RAD+ and rad53-11 spores from the cross of strains SBY376 and BY2918. The genotype of BY479 is MATa dbf4-1 ura3 trp1 ade5.
The plasmids pBD1378 and pBD1265 are pRS316 and pZUC12 vectors, respectively, with the SWI6 gene promoter and open reading frame (ORF) (52). All mutations were introduced into the plasmid-borne SWI6 by using site-directed mutagenesis (32). The mutant constructs are listed in Table 1. Wild-type and alanine mutant constructs were expressed in swi6Δ strain BY2917 or BY2981. To generate glutathione S-transferase (GST)-Swi6 fusions, a 2.0-kb EcoRI fragment containing the SWI6 ORF encoding amino acids 1 to 573 was cloned into pGEX2T-1 (Pharmacia, Piscataway, N.J.), yielding pBD1998. A 1.0-kb EcoRI fragment containing SWI6 ORF codons for amino acids 573 to 803 was cloned into pGEX3X-1 (Pharmacia), yielding pBD2685. All mutant SWI6 genes were cloned into these plasmids, and the resulting constructs are listed in Table 1. Plasmid YQ118 encoding HA-tagged Dbf4 is a kind gift of R. Sclafani.
TABLE 1.
Mutations in SWI6
| Plasmid (pBD) | Background | Mutation(s)a |
|---|---|---|
| 1437 | pRS316 | S160D |
| 2659 | pRS316 | T169D, S170D, S547D |
| 2663 | pRS316 | T169A, S170A, S547A |
| 2664 | pRS316 | S149A, S152A, T169A, S170A, T267A, S268A, S547A |
| 2667 | pRS316 | L549A |
| 2668 | pRS316 | I545A |
| 2669 | pRS316 | T169A, S170A, T267A, S268A, S547A, S715A, T743A |
| 2673 | pZUC12 | S547A |
| 2742 | pZUC12 | T169D, S170D, S547D |
| 2675 | pGEX2T-1 | T267A, S268A |
| 2677 | pGEX2T-1 | I545A |
| 2676 | pGEX2T-1 | L549A |
| 2678 | pGEX2T-1 | S547A |
| 2681 | pGEX2T-1 | S547D |
| 2682 | pGEX2T-1 | T556A, T557A |
| 2680 | pGEX2T-1 | T169D, S170D |
| 2686 | pGEX3X-1 | S715E |
| 2687 | pGEX3X-1 | T743A |
Mutation designations consist of the wild-type amino acid, its position, and then the substituted amino acid.
Cell growth, synchronization, and [32P]orthophosphate labeling in vivo.
All rich (YEP) and minimal (YC) media, growth conditions, and alpha factor synchronizations were as described before (6). Cultures used for elutriation were grown in YC-Ura glucose media and then inoculated into YEPD media and grown to an optical density at 600 nm of 2.5 to 3. Elutriation and cell volume measurements were performed essentially as described earlier (50) in YEPD. For MMS treatments, elutriated cultures were split at time point 0, and MMS was added to one-half of these cultures. MMS was removed by filtration, and cultures were resuspended in YEPD and allowed to grow. Samples were taken every 10 or 15 min, and cell size and the percentage of budded cells were determined. The [32P]orthophosphate labeling conditions were as described before (52).
Protein extraction and purification.
Extraction of the labeled Swi6 protein from yeast was done as described previously (52). The full-length untagged recombinant Swi6 was described previously (51). GST fusions of Swi6 were purified out of 50-ml cultures of the BL21(DE3) Escherichia coli strain. Expression of the GST-Swi6 fusions was induced with 100 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 1 h. Bacteria were lysed by sonication in phosphate-buffered saline (PBS) supplemented with 0.1% Triton X-100, 10 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin/ml, and 1 μg of pepstatin A/ml. Precipitation with glutathione beads and elution of the precipitated proteins with PBS-glutathione solution were done according to the Pharmacia Biotech GST gene fusion system protocol. Fusions were eluted in a volume of 30 μl and adjusted to 1× HEPES kinase buffer (see below) and 10% glycerol by using 10× stock solutions. Then, 2 to 4 μl of these eluates was used in a kinase assay with Rad53.
To isolate HA-tagged Scc1, protein extracts from unlabeled or [32P]orthophosphate-labeled cells were made in the lysis buffer [50 mM Tris HCl (pH 8.0), 0.3 M (NH4)2SO4, 5% glycerol, 0.1% NP-40, 0.05% deoxycholate, 1 mM dithiothreitol, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 μg of pepstatin A/ml, 1 μg of leupeptin/ml, 20 mM β-glycerol phosphate, 20 mM NaF, 1 mM sodium orthovanadate]. These extracts were either loaded onto 7 or 8% (59:1 cross-link ratio) sodium dodecyl sulfate (SDS)-polyacrylamide gels or immunoprecipitated with 12CA5 antibodies. Immunoprecipitates were washed three times with lysis buffer, five times with the PBS containing 1% Triton X-100 and 10% glycerol (PBS-Triton-glycerol), and three times with PBS-Triton-glycerol containing 0.8 M NaCl. Immunoprecipitates were resolved on SDS-polyacrylamide gels and autoradiographed. Labeled Scc1 bands were excised and processed as described for Swi6 (52). Western blotting with 12CA5 antibodies was performed as described earlier for Swi6 (49) with horseradish peroxidase-conjugated goat anti-mouse secondary antibodies (Gibco-BRL, Rockville, Md.).
Rad53 kinase assay.
Immunoprecipitation of untagged or HA-tagged Rad53 out of yeast cell extracts was done as described previously (51) with rabbit polyclonal antibody to Rad53 (a gift of S. J. Elledge) or 12CA5 mouse monoclonal antibodies to the HA epitope. Baculovirus-expressed, HA-tagged Rad53 purified from insect cells was provided by S. J. Elledge. Kinase assays were performed with 50 to 100 ng of recombinant full-length (51) or GST-tagged and truncated Swi6 (see above) by using ca. 1 to 5 ng of baculovirus-expressed HA-Rad53 or Rad53 immunoprecipitated out of ca. 1 to 5 × 107 cells treated with either 4 μg of 4-nitroquinoline-N-oxide (4NQO)/ml or 0.1% MMS for 40 min. Reactions were carried out in HEPES kinase buffer (20 mM HEPES-NaOH [pH 7.5], 10 mM MgCl2, 10 mM MnCl2) for 20 to 30 min at 29°C. Products were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and autoradiographed. Phosphorylated Swi6 was then excised out of dried SDS-PAGE gels and processed as described below.
Phosphopeptide mapping and phosphoamino acid analysis.
Mapping and phosphoamino acid analysis were done as described previously (5) and as described for Swi6 (52). Chymotryptic digestion of Swi6 was conducted with 1 mg of chymotrypsin (Sigma, St. Louis, Mo.)/ml. Phosphopeptide maps of Scc1 digested with trypsin (Sigma) were run in the first dimension at pH 1.9 and in the second dimension in isobutyric buffer (5).
Searching of the yeast proteome for candidate Rad53 phosphorylation sites.
The searches were performed by using the Pattern Matching tool of the Saccharomyces cerevisiae database at Stanford (SGD). The output list of S. cerevisiae ORFs is available online (www.fhcrc.org/labs/breeden/Rad53). The most stringent consensus suggested by the analysis of Swi6 includes basic residues in the −3 position and L, M, V, or I at positions −2 and +2 and excludes proline from the +1 position. This site, [RK][LMVI]x[ST][noP][LMVI], is encountered at least once in 1,958 yeast ORFs, so we list only the 519 that contain two or more such sites.
RESULTS
Pulse treatment with MMS changes the size of a cell at budding.
We used elutriation to study cell cycle progression in small G1 cells after DNA damage. Typically, these elutriated cells grow at a specific rate and execute a G1-to-S transition when they reach the critical volume predetermined by medium conditions and cell's genotype (43). A pulse of MMS reduced the rate of volume enlargement of G1 cells as measured by plotting their mean volume increase over time (Fig. 1A). This growth retardation was dose dependent (data not shown), and it may result from a reduction of biosynthetic capacity, since transcription of many of the ribosomal protein genes is downregulated by this agent (28).
FIG. 1.
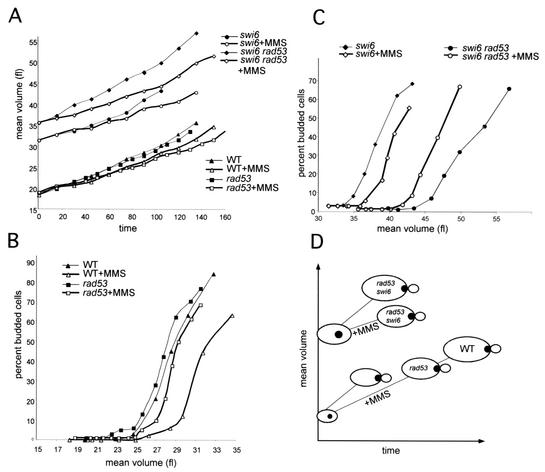
Pulse of MMS induces the delay of the G1-to-S transition in elutriated G1 cells. (A) Small G1 cells were harvested by centrifugal elutriation in fresh YEPD media. Cultures of elutriated G1 cells of wild type (BY2006), rad53-11 (BY2390), swi6Δ (BY2917), and swi6Δ rad53-11 (BY3258) were pulse treated with 0.1% MMS for 15 min as specified in Materials and Methods, and mean cell volume was measured as a function of time after harvesting. (B and C) Percent budded cells in the cultures of the untreated and MMS-treated wild-type (BY2006) and rad53-11 (BY2390) strains with starting mean volumes of 18.3 and 19 fl, respectively (B), and in the cultures of the untreated and MMS-treated swi6Δ (BY2917) and swi6Δ rad53-11 (BY3258) strains of the starting volume 31 and 35.5 fl, respectively, were plotted against their mean volume (C). Boldface lines represent MMS-treated cultures. (D) A schematic summarizing the effects of MMS on cell volume and budding.
As a consequence of the reduced rate of growth caused by MMS treatment, treated cells required more time to attain the same volume as untreated controls (Fig. 1A and D). However, MMS-treated wild-type cells also remained in G1 beyond the volume that was sufficient for budding in the untreated controls and grew up to 5 fl larger before they budded (Fig. 1B and 5C) and initiated DNA replication (data not shown). The rad53-11 mutation shortened the damage-induced delay of budding but did not eliminate it. rad53-11 cells treated with MMS initiated budding sooner than MMS-treated wild-type cells and did not grow as large as the wild type before budding (Fig. 1B and D), but they exhibited the same degree of growth retardation caused by MMS (Fig. 1A). In both types of cells, wild type and rad53-11, the response to MMS was the same whether MMS was added to small, early-G1 cells with the 18-fl mean volume (Fig. 1A) or to larger, mid-G1 cells with a 22-fl mean volume (data not shown).
FIG. 5.
S547 is the site of in vivo phosphorylation of Swi6 by Rad53. Wild-type (A) and S547A (B) Swi6 mutant cells were arrested in G1 with alpha factor and released into the media with [32P]orthophosphate and with or without 0.2% MMS for 40 min. Phosphopeptide maps of this in vivo-labeled Swi6 were generated as in Fig. 3. The arrows point to the two phosphopeptides that are MMS inducible and that are eliminated by S547A substitution. The insets in panels A and B are additional images of the S547-containing peptides derived from an independent experiment. An asterisk marks other damage-inducible peptide(s). (C) Elutriated G1 cells of swi6Δ (BY2917) carrying plasmids expressing the wild-type Swi6 or Swi6 mutants with alanine substitutions at the positions indicated were monitored in their progression through the cell cycle, with or without a 15-min pulse of 0.1% MMS, as described in Fig. 1. Budding and cell volume were measured, and the difference between mean volumes of the untreated and MMS-treated cultures at the point when these cells were 50% budded was plotted. The results of two to five measurements were averaged for each strain. The same data for the wild type (BY2006), the rad53-11 mutant (BY2390), and the swi6Δ mutant (BY2917) are shown for comparison. (D) Budding was measured for the swi6Δ BY2917 strain carrying plasmids expressing the wild-type Swi6 or the m5 mutant. The cells were arrested in G1 with alpha factor and either released from the arrest without genotoxic treatment or incubated with 0.1% MMS for 30 min prior to the release.
Since Swi6 is a potential target of Rad53 phosphorylation and is necessary for the prolonged repression of CLN1 and CLN2 transcription after MMS (51), we sought to determine whether the MMS-induced delay of the G1-to-S transition in elutriated cells required Swi6. The swi6Δ cells are large at birth compared to the wild type (30 to 35 fl versus 18 to 22 fl, Fig. 1A and C). However, they undergo the G1-to-S transition coordinately, as is evident from the relatively steep slope of the budding curves in Fig. 1C. Importantly, pulse treatment with MMS caused a reduced delay of the G1-to-S transition in swi6Δ cells. Similar to rad53-11 cells, the MMS-treated swi6Δ cells started to bud when they were only 2 to 3 fl larger than untreated controls (Fig. 1C; see also Fig. 5C).
Finally, the MMS-treated swi6Δ rad53-11 double mutant cells budded at the same or an even smaller volume than the untreated controls (Fig. 1C). The growth retardation caused by MMS in the double mutant was retained (Fig. 1A); however, there was no delay of budding. In fact, 50% budding was attained at a smaller volume and 10 min earlier in the treated swi6Δ rad53-11 culture than in the untreated control. This suggests that rad53-11 swi6Δ cells have lost the ability to delay in G1 in response to DNA damage. This synthetic phenotype of the swi6Δ rad53-11 strain may arise if the rad53-11 allele retains residual checkpoint activity, which is further compromised by the absence of Swi6 (e.g., RAD53 transcription may be Swi6 dependent [63]). It is also possible that there is a Rad53-independent branch of the checkpoint that operates through Swi6. Either way, these data indicate that both Rad53 and Swi6 contribute to the proper execution of the G1-to-S transition after DNA damage.
Rad53 phosphorylates Swi6 directly.
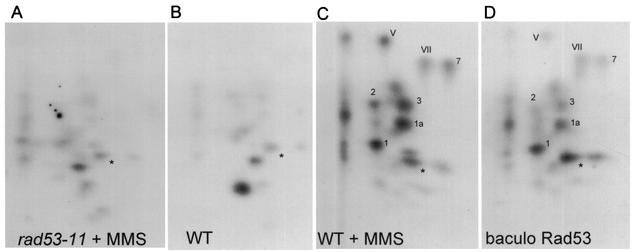
Swi6 undergoes DNA damage-inducible, Rad53-dependent phosphorylation in vivo (51). In addition, Swi6 can be phosphorylated in vitro with the immunoprecipitates of activated Rad53 isolated from cells which received DNA damage or with immunoprecipitates of Rad53 overproduced in undamaged yeast cells (51). To detemine whether Swi6 is a direct substrate of Rad53, we performed kinase reactions with purified recombinant Rad53 and Swi6 and compared phosphopeptide maps of Swi6 phosphorylated by recombinant Rad53 to the maps obtained by using immunoprecipitated wild-type and checkpoint-deficient Rad53.As seen in Fig. 2C, wild-type, damage-activated Rad53 from yeast cells phosphorylates Swi6 on a number of sites (1, 1a, 2, 3, 7, V, and VII). Neither mutant Rad53 from damaged cells (Fig. 2A), nor the wild-type Rad53 isolated from cells with no DNA damage (Fig. 2B) was capable of efficiently phosphorylating Swi6 on these sites. However, recombinant Rad53 provided in excess in vitro gave rise to a phosphorylation pattern on Swi6 that was qualitatively identical to the one generated by the damage-activated Rad53 from yeast cells (Fig. 2D), indicating that the damage-induced phosphorylations could be directly attributed to Rad53 activity. The phosphorylated peptides were subjected to phosphoamino acid analysis, and only phosphoserine and phosphothreonine residues were detected (data not shown).
FIG. 2.
Rad53 directly phosphorylates Swi6 in an MMS-inducible manner. Recombinant Swi6 purified from E. coli was incubated in the presence of [γ-32P]ATP and Rad53 immunoprecipitated from MMS-treated rad53-11 (BY2007) cells (A), untreated (B) or MMS-treated (C) wild-type (BY2006) cells, or with the baculovirus-expressed Rad53 purified from insect cells (D). Kinase-treated Swi6 was resolved on an SDS-polyacrylamide gel, excised, digested with trypsin, and resolved in two dimensions to generate phosphopeptide maps. Major Rad53-dependent phosphopeptides are numbered.
Swi6 is a 91-kDa protein with 104 serine and threonine residues. In order to map the sites of phosphorylation of Swi6 by Rad53, we used deletion derivatives of Swi6 purified as GST-tagged proteins. The tag did not affect the phosphorylation pattern (data not shown). Figure 3 shows a comparison of the phosphopeptide maps of the full-length untagged Swi6 and its two deletion derivatives, aa1-573 and aa573-803, that were phosphorylated by recombinant Rad53. The map positions of these Rad53-generated tryptic phosphopeptides were compared to the predicted positions of the tryptic phosphopeptides of Swi6 (5). Serine and/or threonine residues in the peptides that closely matched the observed migration patterns were mutated, and mutant substrates were purified and tested in a kinase reaction with the recombinant Rad53.
FIG. 3.
Identification of five major sites of Rad53 phosphorylation on Swi6. Tryptic phosphopeptide maps (as in Fig. 2) of the wild-type full-length Swi6 (E) or GST fusions aa1-573 (A to D) and aa573-803 (F to H) with amino acid substitutions as indicated. Arabic numerals correspond to the peptides mapping to the first 573 amino acids of Swi6, and roman numerals correspond to the C-terminal peptides mapping between amino acids 573 and 803. Rad53 used in these reactions was purified from baculovirus-infected insect cells.
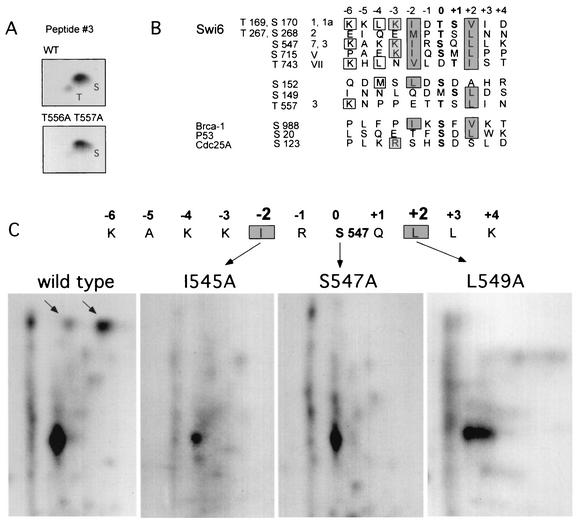
Figure 3 summarizes the results of this analysis. We identified five sites targeted by Rad53 in Swi6, which account for the major phophopeptides 1, 1a, 2, 3, 7, V, and VII. Peptides 1 and 1a were produced by the same sequence, [KKLKIDT(169)SVIDAE…] (Fig. 3B). Both serine and threonine residues can be phosphorylated in this sequence since the phosphoamino acid composition of this peptide is a mixture of both phosphoserine and phosphothreonine (data not shown). However, the mobility of these peptides suggests that they carry only one phosphate, either on T169 or on S170.
Another major phosphorylation site, [KAKKIRS(547)QLLKN…], accounts for at least two peptides, 3 and 7, where peptide 7 has a sequence of S(547)QLLK, and peptide 3 is most likely an incompletely digested peptide with the sequence S(547)QLLKNPPET(556)TSLINDVQNLLNS (Fig. 3C). Within this sequence, S547 remains a predominant site of phosphorylation and/or a primary recognition site by Rad53, since its substitution to aspartic acid eliminates both peptides 3 and 7 (Fig. 3C). However, in a fraction of molecules, the threonines 556 and 557 may be phosphorylated instead of S547, since peptide 3 contains both phosphothreonine and phosphoserine (Fig. 4A). Accordingly, substitution of threonines 556 and 557 to alanines does not abolish the phosphorylation of peptides 3 and 7, but it does eliminate threonine as a phosphoacceptor in peptide 3 (Fig. 4A).
FIG. 4.
Conserved hydrophobic aliphatic amino acid residues in the −2 and +2 positions of the Rad53 phosphorylation sites in Swi6 are important determinants of site specificity. (A) The identity of the phosphorylated residue was determined for phosphopeptide 3 from the wild type and the T556A T557A mutant. (B) Alignment of the Rad53 phosphorylation sites in Swi6 with conserved positions boxed and shaded. Coordinates of phosphorylated residues (in boldface) in five consensus sites and three “half” sites, as well as the numbers of the corresponding peptides in the phosphopeptide map, are shown on the left. The three identified in vivo sites of Chk2 phosphorylation are aligned below the Swi6 sites. (C) I545A or L549A mutations were introduced adjacent to S547, and the resulting mutants were phosphorylated in vitro by Rad53 immunoprecipitated out of 4NQO-treated yeast cells. Phosphorylated Swi6 fusions were digested with chymotrypsin and resolved in two dimensions as in Fig. 2 and 3. The arrows point to the peptides that correspond to S547 phosphorylation.
The site […QEMPT(267)SLNN…] maps to peptide 2, where mainly phosphothreonine is detected (Fig. 3D and data not shown). Peptide V has the sequence IQS(715)MLPPTVLLK, with S715 as the phosphorylatable residue (Fig. 3G), and peptide VII [HLTNVLDT(743)ISTK] has T743 as the phosphorylatable residue (Fig. 3H).
Alignment of these Rad53 phosphorylation sites reveals a strong preference for hydrophobic amino acids with long aliphatic side chains, I, L, V, or M at positions +2 and −2, where 0 and +1 are, respectively, the first and the second phosphorylatable residues (Fig. 4B). Also, a preference for a basic or hydrophilic residue at the −3 position is evident. We refer to this type of site as a −2/+2 consensus site. Inspection of the Swi6 protein sequence showed that in addition to the five identified −2/+2 sites there were five more sites of this type. However, based on the pattern of the Swi6 phosphopeptide maps, these other sites are unlikely to be phosphorylated by Rad53 to any significant level. Of these, only S160 is definitely located on the surface of the Swi6 protein and is accessible, since it is phosphorylated in vivo (52). However, it carries a proline residue in +1 position. At least in the context of short peptide libraries, an S/TP sequence was found to be a poor substrate for a human Rad53 homolog, Chk2 (41). It is therefore not surprising that S160 is not targeted by Rad53.
The weakly phosphorylated ET(556)TSLI site is a “half” site, with a hydrophobic aliphatic residue flanking the phosphorylatable residue only on one side (+2, Fig. 4B). We tested several half sites for phosphorylation by Rad53, and detected only weak reactivity at S149 and S152 of the sequence QDMS(149)LDS (data not shown), which contains two adjacent half sites (Fig. 4B).
The −2 and +2 positions are important determinants of the Rad53 site specificity.
As mentioned above, our analysis revealed conservation of the −2 and +2 positions in the Rad53 phosphorylation sites mapped on Swi6. Moreover, in two cases in which only one of these positions was occupied by the conserved residue I, L, M, or V, we observed only weak phosphorylation by Rad53. To address whether these hydrophobic aliphatic residues are important for the recognition of the site by Rad53, we replaced the −2 isoleucine or the +2 leucine around the S547 site with alanine, which is also nonpolar but less hydrophobic. The resulting mutants were phosphorylated in vitro by Rad53 immunoprecipitated out of 4NQO-treated yeast and digested with chymotrypsin. Chymotrypsin cleaves at least 10 amino acids away from I545 and L549 to release large peptides, and as such their mobilities will be less affected by the alanine substitutions. These digests show that the two major peptides that are generated due to S547 phosphorylation (compare the wild-type and S547A maps in Fig. 4C) disappear when either I545 or L549 are mutated to alanine (Fig. 4C). This result and the strong conservation we observed in the major Rad53 sites of Swi6 lead us to conclude that hydrophobic aliphatic residues at both the −2 and +2 positions are critical determinants of the Rad53 phosphorylation specificity.
S547 is the site of in vivo damage-dependent phosphorylation of Swi6.
Only a subset of in vitro Rad53 phosphorylation sites can be readily observed on an in vivo phosphopeptide map of Swi6. The in vitro peptides 3 and 7 are the closest in their phosphopeptide map positions to the DNA damage-inducible peptides previously detected in vivo (51). These in vivo peptides are also Rad53 dependent, as we have shown previously (see above). We therefore asked whether S547 of Swi6, which maps to the peptides 3 and 7, is the site of a detectable phosphorylation by Rad53 in vivo. As shown before (51), the wild-type Swi6 isolated from MMS-treated RAD+ cells exhibits two damage-inducible phosphopeptides that are well separated from the bulk of the peptides on the two-dimensional map (Fig. 5A). When S547 is substituted for A, these phosphopeptides are no longer observed upon MMS treatment (Fig. 5B). We conclude that S547 is a site of in vivo phosphorylation by Rad53 under conditions of DNA damage. The maps in Fig. 5 also show other changes in the in vivo map upon MMS treatment. This suggests that additional, yet-unidentified sites on Swi6 may be targets of damage-inducible phosphorylation. However, we cannot attribute them to Rad53 at this time (Fig. 2) (51).
Rad53 phosphorylation site mutants of Swi6 interfere with but do not eliminate the G1-to-S transition delay.
The MMS-induced G1/S checkpoint activity of wild-type cells was compared to that of cells carrying alanine substitutions in serine and threonine residues at sites of Rad53 phosphorylation in Swi6 (Fig. 5C). G1 cells were collected by elutriation, and the G1-to-S transition was monitored by counting budding over time with or without MMS treatment, as in Fig. 1. All mutants tested behaved as wild type in driving the G1-to-S transition when no DNA damage was applied. That is, they transited to the S phase at the same critical volume as the wild type and thus were not measurably defective under normal growth conditions (data not shown). When the single mutants, S547A or I545A, were compared to the wild type under DNA damage conditions, there was a slight acceleration of budding (Fig. 5C and data not shown). Further alanine substitutions of serines and threonines in up to five mapped Rad53-phosphorylatable sites had a similarly modest effect, and none of these mutants reduced the damage-induced delay as much as rad53-11 or swi6Δ mutants (Fig. 5C). To follow this up, we used alpha factor as means of synchronizing the cells in G1. Seven and six independent measurements were done on strains with the wild-type Swi6 and a mutant Swi6 with alanines substituted for serines and threonines in all five Rad53 sites (m5, see example in Fig. 5D). These measurements showed that, on average, the wild type achieved 50% budding in 84.8 ± 2.53 min after a 30-min pulse of 0.1% MMS during alpha factor arrest, whereas the mutant had 50% buds after 73.4 ± 3.2 min. Therefore, in the absence of Rad53 phosphorylation sites in Swi6, cells were somewhat less capable of delaying the G1-to-S transition after DNA damage. In rad53-11 cells, eliminating multiple Rad53 phosphorylation sites in Swi6 had no additional effect on the G1-to-S transition delay compared to rad53-11 cells alone (data not shown). Overall, these data are consistent with the notion that phosphorylation of Swi6 by Rad53 contributes to the mechanism through which Rad53 delays S phase in response to DNA damage but is not its only component.
The residues T169 and S170 are adjacent to the Swi6 nuclear localization sequence, which is inactivated by phosphorylation of S160 in late G1 (52), or by substituting S160 with aspartic acid. We thus asked whether analogous substitutions of T169 and S170 affected nuclear localization of Swi6 in G1. We found that Swi6 remained nuclear throughout the MMS-induced delay of the G1-to-S transition, whether T169 or S170 were mutated or not (data not shown). In addition, the S160D mutant of Swi6, which is not restricted to the nucleus in G1 (52), was still found in the cytoplasm in MMS and had no effect on the MMS-induced delay of the G1-to-S transition (data not shown). We conclude that changes in the subcellular localization of Swi6 do not have a role in the MMS-induced pausing of the cell cycle in G1.
Search for Rad53 phosphorylation site matches in yeast ORFs reveals a number of prospective Rad53 targets.
The most stringent consensus suggested by the analysis of Swi6 includes basic residues (lysine or arginine) in the −3 position, excludes proline from +1 position, and requires L, M, V, or I at positions −2 and +2. Using the version of this consensus −2/+2 site, we sought to identify new candidate Rad53 targets. A total of 519 proteins were identified that carried two or more such sites (see Materials and Methods). The search returned all of the mismatch repair Msh proteins (Msh1 to Msh6) and some Mlh proteins (Mlh1 to Mlh3) (35). −2/+2 sites were found in the three proteins critical for telomere tethering to the nuclear periphery, Nup145, Mlp1, and Mlp2 (18). Both Swi6 partners, the Mbp1 and Swi4 proteins, contain −2/+2 sites, and four of these in Swi4 are located within the Swi6-binding domain of Swi4. Finally, we noted that all SMC (for structural maintenance of chromosomes) proteins and the SMC-like Rad50 contained Rad53 sites. The six SMC proteins form the cohesin (SMC1 and -3), condensin (SMC2 and -4), and the SMC5-SMC6 complex that is involved in DNA repair (22). Cohesin and condensin complexes have additional subunits (Scc1/Mcd1, Scc3 and Ycs4, Ycs5, and Brn1, respectively) (22). Remarkably, all of these subunits also carry one or more stringent −2/+2 sites.
Scc1 is a cohesin subunit whose proteolysis at the G2/M boundary allows sister chromatid separation (40). G2/M-specific phosphorylation of Scc1 by the Polo-like kinase Cdc5 promotes this proteolysis (2). There are three perfect −2/+2 sites in Scc1: S112, S273, and S367. Interestingly, two of these −2/+2 sites in Scc1 are adjacent to or overlap the mapped Cdc5 phosphorylation sites (2) (Fig. 6A). In SDS-PAGE, Scc1 from untreated cells migrates predominantly as a single band, with trace amounts of a slower-migrating form (Fig. 6B). This latter form can be attributed to Cdc5 phosphorylation. A 20- to 30-min pulse of MMS or 4NQO triggers the appearance of a more prominent slowly migrating form of Scc1 (Fig. 6B and C). In the rad53-11 cells treated with the same doses of 4NQO, the appearance of this slowest-migrating, damage-inducible form of Scc1 was virtually eliminated and only a band of intermediate mobility was detected (Fig. 6C). The cell cycle distribution of both wild-type and rad53-11 cells was comparable before and after this short 4NQO treatment (data not shown). Thus, the observed differences in the modification pattern of Scc1 cannot be attributed to the damage-induced accumulation of cells at the G2/M border in the wild-type cells and not in the rad53 cells. Rather, it is likely that at least a subset of DNA damage-induced modifications on Scc1 in vivo are occurring in a Rad53-dependent manner.
FIG. 6.
Scc1 undergoes DNA damage-induced phosphorylation. (A) Schematic of Scc1 phosphorylation sites. Vertical lines correspond to the positions of the phosphorylatable residues. The black circles above the diagram indicate Cdc5 phosphorylation sites mapped in vitro (2). The open circles below represent −2/+2 sites, which may be the targets of Rad53 phosphorylation. Black bars mark the two Esp1 cleavage sites (58). (B) Steady-state cultures of the cells carrying the HA-tagged Scc1 (SBY376) were split, and one-half of the culture was treated for 30 min with 0.1% MMS or 4 μg of 4NQO/ml. Extracts were prepared from these cells and loaded on SDS-polyacrylamide gels. Scc1 was visualized on Western blots by using 12CA5 antibodies to the tag. Center lane is a negative control expressing HA-tagged Dbf4 and no HA-Scc1 (plasmid YQ118 from R. Sclafani). (C) Wild-type (BY3242) and rad53-11 (BY3243) strains expressing HA-Scc1 were treated with 0, 0.25, 0.5, and 1 μg of 4NQO/ml for 20 min. Cell extracts of these cultures and of the untreated controls were made, and HA-Scc1 was visualized on Western blots as described above. A line marks the position of the hypomodified, fast-mobility form of Scc1. An arrow on the left shows the damage-specific, slow-mobility form of the protein. An arrow on the right marks the position of the intermediate mobility form observed in rad53-11 mutant cells.
To further test this, we generated in vivo phosphopeptide maps of HA-tagged Scc1 immunoprecipitated from wild-type and rad53-11 cells treated with 0.5 μg of 4NQO/ml. The arrow in Fig. 7 denotes one peptide within Scc1 that is more prominently phosphorylated after 4NQO treatment in RAD+ but not in rad53-11 cells (Fig. 7A and B). This finding is consistent with the Rad53 and damage-specific mobility shift observed in Scc1 in SDS-PAGE and suggests that Scc1 undergoes DNA damage-induced phosphorylation, a part of which is attributable to Rad53.
FIG. 7.
Scc1 is phosphorylated in a DNA damage-, Rad53-dependent manner in vivo. Steady-state cultures of the wild-type (BY3242 [A and B]) and rad53-11 (BY3243 [C and D]) strains with HA-tagged Scc1 were incubated with [32P]orthophosphate for 30 min in the presence (B and D) or absence (A and C) of 0.5 μg of 4NQO/ml. Scc1 was isolated by immunoprecipitation with 12CA5 antibodies, and phosphopeptide maps of Scc1 were generated. An arrow marks the peptide, which is both DNA damage and Rad53 dependent. Asterisks mark the positions of 32P-labeled inorganic phosphate.
DISCUSSION
We have identified five sites of Rad53-mediated phosphorylation on Swi6 by using both recombinant Rad53 and the kinase immunoprecipitated out of yeast cells undergoing DNA damage. Importantly, Rad53 from both of these sources produces nearly identical profiles of phosphorylation on Swi6. This suggests that Rad53 alone and not a copurifying yeast kinase is responsible for Swi6 phosphorylation in our assays. Moreover, it indicates that DNA damage does not alter the site specificity of Rad53 but only increases kinase activity. This result is consistent with our previous observation that Rad53 overproduced in undamaged yeast cells can also phosphorylate Swi6 in vitro.
At least one of the identified sites, S547, is phosphorylated in vivo in an MMS-inducible, Rad53-dependent manner, suggesting that Rad53 recognizes Swi6 as one of its substrates in a cell undergoing DNA damage. Phosphorylation of this site is substoichiometric, suggesting that only a subpopulation of Swi6 molecules can be phosphorylated. For example, only DNA-bound or Swi4-bound fractions of Swi6 could be targeted by Rad53. We did not detect in vivo phosphorylation of the other sites mapped in vitro on Swi6. These sites may not be accessible, or they may be phosphorylated in a even more transient or substochiometric manner. Alternatively, there may be inducible phosphopeptides that are obscured by overlapping constitutive phosphopeptides on the peptide map.
Alignment of the five Rad53 phosphorylation sites of Swi6 reveals a remarkable degree of conservation, particularly at −2 and +2 positions (Fig. 4B). Both of these positions are occupied only by hydrophobic aliphatic amino acids, and conservative substitution of these residues to alanine prevents phosphorylation by Rad53. Position −3 shows a preference for basic or hydrophilic residues, and proline may be unfavorable at +1. There may also be a bias toward basic residues at the −6 position. Finally, it appears that either position 0 or +1 can be phosphorylated within a given site, if occupied by serine or threonine. Interestingly, comparison of the Swi6 sites to the in vivo phosphorylation site recognized by Chk2, the mammalian homolog of Rad53 (36), on human BRCA1 (S988) (33) shows exactly the same sequence preference at −2 and +2, as the one identified in the present study (Fig. 4A). Another in vivo Chk2-recognized site, S20 of p53 (8, 23, 48), a reportedly less-than-optimal site, has a leucine only in the +2 position. This is consistent with our finding that “half” sites can be phosphorylated by Rad53 in vitro, albeit weakly. However, the other reported in vivo Chk2 phosphorylation site, S123 of CDC25A (15), has no obvious similarity to the −2/+2 consensus (Fig. 4A).
The specificities of the Chk1 and Chk2 kinases have been studied by using peptide libraries (41). The peptide substrate preferences derived for human Chk2 are similar to our consensus in that they include hydrophobic residues at positions following the phosphorylatable residue, and a basic residue, arginine, at position −3. However, there is no preference seen for the −2 position in the peptide study. These differences may be attributed to the use of peptide versus protein substrates or to the necessarily limited array of sequences present in any peptide library. It is also possible that human Chk2 and yeast Rad53 have somewhat different substrate preferences or that the identified Swi6 sites are not the highest affinity sites for Rad53 phosphorylation, just as S988 in BRCA1 is not the best Chk2 substrate (41). The in vivo targets of checkpoint kinases may carry multiple suboptimal sites as a way of transforming their phosphorylation into an on-off switch actuated by a substantial increase of the kinase activity, as is the case for Sic1 phosphorylation (39).
The −2/+2 consensus allows us to derive insights into Rad53 activity and regulation. All of the proteins implicated as targets or binding partners of Rad53 (Crt1, Dbf4, Dun1, Cdc5, Rad55, Bfa1, Asf1, Rad9, and Mrc1 [1, 13, 14, 25, 26, 57, 59, 64]) carry one to nine matches to the −2/+2 consensus. In Rad53 itself there are eight potential −2/+2 sites, most of which are within 30 amino acids of S/TQ sites, which are potential sites for phosphorylation by the upstream checkpoint kinases Mec1 and Tel1. The association of potential Rad53 and Mec1/Tel1 sites suggests that both transcatalytic (via Mec1 and Tel1) and autocatalytic pathways may activate Rad53 in response to damage (20).
In addition, three −2/+2 sites are located between the β-sheets of the FHA2 domain of Rad53 (12). FHA1 and FHA2 are protein domains that bind to phosphorylated threonine residues embedded within the FHA1- or FHA2-binding motifs (TxxD or TxxL/I, Fig. 8 [11]). Autophosphorylation of Rad53 on these sites could change the properties of the FHA2 domain. For example, the phosphorylated FHA2 domain of Rad53 may no longer associate with Rad9. This could provide a mechanistic explanation for the observed release of Rad53 from the complex with Rad9 after autophosphorylation (20).
FIG. 8.
(A) −2/+2 sites can overlap FHA-binding motifs. FHA1- and FHA2-binding motifs identified by using peptide libraries (12) were aligned with the known FHA1-binding motifs of p53 (10) and Rad9 (62), the known FHA2-binding motifs of Rad9 (7, 47), and the putative FHA2-binding motifs adjacent to the mapped −2/+2 sites of Swi6. Boxed residues are similar or identical to the ones identified by using peptide libraries in (12). Black circles over serine or threonine residues mark the known Chk2 phosphorylation site in p53 (S20), and the known Rad53 phosphorylation sites in Swi6. In Rad9, gray circles show serine or threonine residues that lie within matches to the −2/+2 consensus and could be phosphorylated by Rad53.
Interestingly, four of five Swi6 −2/+2 sites overlap or adjoin sequences that match the FHA2-binding motif (Fig. 8). This is analogous to the in vivo Chk2 phosphorylation site S20 in p53, which overlaps the FHA1-binding motif that binds the FHA1 domain of Rad53 (10). In Rad9, there are three −2/+2 sites within the cluster of the FHA-binding motifs that were shown to be necessary for Rad53 binding and activation in vivo (47). Among these, two −2/+2 sites completely overlay both the putative Mec1/Tel1 phosphorylation sites (S/TQ) and the FHA-binding motifs. The third −2/+2 site in Rad9 adjoins the FHA-binding motif (Fig. 8). It is tempting to speculate that Rad53 phosphorylation adjacent to FHA-binding motifs affects their ability to bind to FHA domains.
Our search for consensus sites for Rad53 phosphorylation in other proteins revealed a number of potentially interesting new candidate Rad53 targets among cell cycle and DNA metabolism proteins, in particular, the cohesin complex. This complex is known to be targeted by the DNA damage checkpoint in G2 both in yeast (60) and in mammals (30, 61). We have shown that one of the cohesin complex subunits, Scc1, undergoes DNA damage-induced phosphorylation in vivo. We also established that at least one of the damage-inducible phosphorylation events on Scc1 is largely Rad53 dependent. Although an in vitro kinase assay is needed to prove that Scc1 is indeed a direct Rad53 target, our data are certainly consistent with this possibility.
Given the fact that the DNA damage checkpoint pathways in budding yeast (19, 26, 44, 60) consist of multiple parallel branches, it is not surprising that elimination of the Rad53 phosphorylation sites in Swi6 has only a minor effect on the length of the checkpoint-mediated delay in the G1-to-S transition after damage. It is possible that Rad53 acts upon other components of the late-G1 transcription machinery, e.g., Swi4 and Mbp1, since both of these proteins carry multiple Rad53 consensus sites. Other kinases, such as Dun1, Chk1, and Mec1, are also activated and could phosphorylate Swi6 and additional G1 targets. This is consistent with our findings that SWI6-null mutants have a G1 checkpoint defect and that the rad53-11 swi6Δ double mutant has a more extreme G1 checkpoint-deficient phenotype than either mutation alone.
It is unclear at present what biochemical activities of Swi6 are affected by Rad53 phosphorylation and DNA damage in general. MMS-induced damage does not abolish Swi6 nuclear localization in G1. Nor does it prevent Swi6 from binding to the CLN1 promoter in vivo in RAD+ or rad53-11 cells (J. M. Sidorova, unpublished data). Loss of Sin3 repressor, or of Stb1, which may connect Swi6 to the Sin3 repressor complex (24, 29), does not give rise to a G1/S checkpoint defect (Sidorova, unpublished). Perhaps other, yet-unidentified interacting factors may confer the effects of Swi6 phosphorylation by Rad53 on the G1-to-S transition.
Checkpoints are likely to include multiple layers of control imposed on key events, with dozens of substrates targeted by more than one kinase and phosphorylated either in an additive or competitive fashion. A network thus organized would be highly responsive to changing conditions and failsafe. Understanding of the Rad53 phosphorylation site preferences provides a new tool for dissecting the complexity of checkpoint controls in budding yeast.
Acknowledgments
We gratefully acknowledge members of the Breeden lab for support and numerous discussions. We particularly thank Bernard Mai for generating pBD1998; Steve Elledge for helpful discussions and for providing baculovirus-expressed Rad53 and anti-Rad53 serum; and Andrew Emili, Robert Sclafani, and Sue Biggins for strains and constructs.
This work was supported by the NIH grant GM41073 to L.L.B. and by a Leukemia and Lymphoma Society senior fellowship to J.M.S.
REFERENCES
- 1.Alcasabas, A. A., A. J. Osborn, J. Bachant, F. Hu, P. J. Werler, K. Bousset, K. Furuya, J. F. Diffley, A. M. Carr, and S. J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3:958-965. [DOI] [PubMed] [Google Scholar]
- 2.Alexandru, G., F. Uhlmann, K. Mechtler, M. A. Poupart, and K. Nasmyth. 2001. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell 105:459-472. [DOI] [PubMed] [Google Scholar]
- 3.Allen, J. B., Z. Zhou, W. Siede, E. C. Friedberg, and S. J. Elledge. 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8:2401-2415. [DOI] [PubMed] [Google Scholar]
- 4.Biggins, S., F. F. Severin, N. Bhalla, I. Sassoon, A. A. Hyman, and A. W. Murray. 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13:532-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyle, W. J., P. Van Der Geer, and T. Hunter. 1991. Phosphopeptide mapping and phosphoamino acid anaylsis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201:110-149. [DOI] [PubMed] [Google Scholar]
- 6.Breeden, L., and G. Mikesell. 1991. Cell cycle-specific expression of the SWI4 transcription factor is required for the cell cycle regulation of HO transcription. Genes Dev. 5:1183-1190. [DOI] [PubMed] [Google Scholar]
- 7.Byeon, I. J., S. Yongkiettrakul, and M. D. Tsai. 2001. Solution structure of the yeast Rad53 FHA2 complexed with a phosphothreonine peptide pTXXL: comparison with the structures of FHA2-pYXXL and FHA1-pTXXD complexes. J. Mol. Biol. 314:577-588. [DOI] [PubMed] [Google Scholar]
- 8.Chehab, N. H., A. Malikzay, M. Appel, and T. D. Halazonetis. 2000. Chk2/hCds1 functions as a DNA damage checkpoint in G1 by stabilizing p53. Genes Dev. 14:278-288. [PMC free article] [PubMed] [Google Scholar]
- 9.Dirick, L., T. Bohm, and K. Nasmyth. 1995. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 14:4803-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durocher, D., J. Henckel, A. R. Fersht, and S. P. Jackson. 1999. The FHA domain is a modular phosphopeptide recognition motif. Mol. Cell 4:387-394. [DOI] [PubMed] [Google Scholar]
- 11.Durocher, D., and S. P. Jackson. 2002. The FHA domain. FEBS Lett. 513:58-66. [DOI] [PubMed] [Google Scholar]
- 12.Durocher, D., I. A. Taylor, D. Sarbassova, L. F. Haire, S. L. Westcott, S. P. Jackson, S. J. Smerdon, and M. B. Yaffe. 2000. The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol. Cell 6:1169-1182. [DOI] [PubMed] [Google Scholar]
- 13.Emili, A. 1998. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol. Cell 2:183-189. [DOI] [PubMed] [Google Scholar]
- 14.Emili, A., D. M. Schieltz, J. R. Yates III, and L. Hartwell. 2001. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell 7:13-20. [DOI] [PubMed] [Google Scholar]
- 15.Falck, J., N. Mailand, R. G. Syljuasen, J. Bartek, and J. Lukas. 2001. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410:842-847. [DOI] [PubMed] [Google Scholar]
- 16.FitzGerald, J. N., J. M. Benjamin, and S. J. Kron. 2002. Robust G1 checkpoint arrest in budding yeast: dependence on DNA damage signaling and repair. J. Cell Sci. 115:1749-1757. [DOI] [PubMed] [Google Scholar]
- 17.Foiani, M., A. Pellicioli, M. Lopes, C. Lucca, M. Ferrari, G. Liberi, M. Muzi Falconi, and P. Plevani. 2000. DNA damage checkpoints and DNA replication controls in Saccharomyces cerevisiae. Mutat. Res. 451:187-196. [DOI] [PubMed] [Google Scholar]
- 18.Galy, V., J. C. Olivo-Marin, H. Scherthan, V. Doye, N. Rascalou, and U. Nehrbass. 2000. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature 403:108-112. [DOI] [PubMed] [Google Scholar]
- 19.Gardner, R., C. W. Putnam, and T. Weinert. 1999. RAD53, DUN1 and PDS1 define two parallel G2/M checkpoint pathways in budding yeast. EMBO J. 18:3173-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilbert, C. S., C. M. Green, and N. F. Lowndes. 2001. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8:129-136. [DOI] [PubMed] [Google Scholar]
- 21.Grenon, M., C. Gilbert, and N. F. Lowndes. 2001. Checkpoint activation in response to double-strand break requires the Mre11/Rad50/Xrs2 complex. Nat. Cell Biol. 3:844-847. [DOI] [PubMed] [Google Scholar]
- 22.Hirano, T. 2002. The ABCs of SMC proteins: two-armed ATPases for chromosome condensation, cohesion, and repair. Genes Dev. 16:399-414. [DOI] [PubMed] [Google Scholar]
- 23.Hirao, A., Y. Y. Kong, S. Matsuoka, A. Wakeham, J. Ruland, H. Yoshida, D. Liu, S. J. Elledge, and T. W. Mak. 2000. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287:1824-1827. [DOI] [PubMed] [Google Scholar]
- 24.Ho, Y., M. Costanzo, L. Moore, R. Kobayashi, and B. J. Andrews. 1999. Regulation of transcription at the Saccharomyces cerevisiae start transition by Stb1, a Swi6-binding protein. Mol. Cell. Biol. 19:5267-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu, F., A. A. Alcasabas, and S. J. Elledge. 2001. Asf1 links Rad53 to control of chromatin assembly. Genes Dev. 15:1061-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu, F., Y. Wang, D. Liu, Y. Li, J. Qin, and S. J. Elledge. 2001. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell 107:655-665. [DOI] [PubMed] [Google Scholar]
- 27.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409:533-538. [DOI] [PubMed] [Google Scholar]
- 28.Jelinsky, S. A., and L. D. Samson. 1999. Global Response of Saccharomyces cerevisiae to an alkylating agent. Proc. Natl. Acad. Sci. USA 96:1486-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasten, M. M., and D. J. Stillman. 1997. Identification of the Saccharomyces cerevisiae genes STB1-STB5 encoding Sin3p binding proteins. Mol. Gen. Genet. 256:376-386. [DOI] [PubMed] [Google Scholar]
- 30.Kim, S. T., and M. B. Kastan. 2002. Involvement of the cohesin protein, Smc1, in ATM-dependent and independent responses to DNA damage. Genes Dev. 16:560-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, S. T., D. S. Lim, C. E. Canman, and M. B. Kastan. 1999. Substrate specificities and identification of putative substrates of ATM kinase family members. J. Biol. Chem. 274:37538-37543. [DOI] [PubMed] [Google Scholar]
- 32.Kunkel, T. A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82:488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, J. S., K. M. Collins, A. L. Brown, C. H. Lee, and J. H. Chung. 2000. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature 404:201-204. [DOI] [PubMed] [Google Scholar]
- 34.Lowndes, N. F., and J. R. Murguia. 2000. Sensing and responding to DNA damage. Curr. Opin. Genet. Dev. 10:17-25. [DOI] [PubMed] [Google Scholar]
- 35.Marti, T. M., C. Kunz, and O. Fleck. 2002. DNA mismatch repair and mutation avoidance pathways. J. Cell Physiol. 191:28-41. [DOI] [PubMed] [Google Scholar]
- 36.Matsuoka, S., M. Huang, and S. J. Elledge. 1998. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science 282:1893-1897. [DOI] [PubMed] [Google Scholar]
- 37.McInerny, C. J., J. F. Partridge, G. E. Mikesell, D. P. Creemer, and L. L. Breeden. 1997. A novel Mcm1-dependent promoter element in the SWI4, CLN3, CDC6, and CDC47 promoters activates M/G1-specific transcription. Genes Dev. 11:1277-1288. [DOI] [PubMed] [Google Scholar]
- 38.Mendenhall, M., and A. Hodge. 1998. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1191-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nash, P., X. Tang, S. Orlicky, Q. Chen, F. B. Gertler, M. D. Mendenhall, F. Sicheri, T. Pawson, and M. Tyers. 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414:514-521. [DOI] [PubMed] [Google Scholar]
- 40.Nasmyth, K. 2001. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35:673-745. [DOI] [PubMed] [Google Scholar]
- 41.O'Neill, T., L. Giarratani, P. Chen, L. Iyer, C. H. Lee, M. Bobiak, F. Kanai, B. B. Zhou, J. H. Chung, and G. A. Rathbun. 2002. Determination of substrate motifs for human Chk1 and hCds1/Chk2 by the oriented peptide library approach. J. Biol. Chem. 277:16102-16115. [DOI] [PubMed] [Google Scholar]
- 42.Pellicioli, A., C. H. Lee, C. Lucca, M. Foiani, and J. Haber. 2001. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol. Cell 7:293-300. [DOI] [PubMed] [Google Scholar]
- 43.Rupes, I. 2002. Checking cell size in yeast. Trends Genet. 18:479-485. [DOI] [PubMed] [Google Scholar]
- 44.Sanchez, Y., J. Bachant, H. Wang, F. Hu, D. Liu, M. Tetzlaff, and S. J. Elledge. 1999. Control of the DNA damage checkpoint by Chk1 and Rad53 protein kinases through distinct mechanisms. Science 286:1166-1171. [DOI] [PubMed] [Google Scholar]
- 45.Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang, and S. J. Elledge. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271:357-360. [DOI] [PubMed] [Google Scholar]
- 46.Sandell, L. L., and V. A. Zakian. 1993. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell 75:729-739. [DOI] [PubMed] [Google Scholar]
- 47.Schwartz, M. F., J. K. Duong, Z. Sun, J. S. Morrow, D. Pradhan, and D. F. Stern. 2002. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell 9:1055-1065. [DOI] [PubMed] [Google Scholar]
- 48.Shieh, S. Y., J. Ahn, K. Tamai, Y. Taya, and C. Prives. 2000. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 14:289-300. [PMC free article] [PubMed] [Google Scholar]
- 49.Sidorova, J., and L. Breeden. 1993. Analysis of the SWI4/SWI6 protein complex, which directs G1/S-specific transcription in Saccharomyces cerevisiae Mol. Cell. Biol. 13:1069-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sidorova, J., and L. L. Breeden. 2002. Precocious S-phase entry in budding yeast prolongs replicative state and increases dependence upon Rad53 for viability. Genetics 160:123-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sidorova, J., and L. L. Breeden. 1997. Rad53-dependent phosphorylation of Swi6 and downregulation of CLN1 and CLN2 transcription occur in response to DNA damage in Saccharomyces cerevisiae. Genes Dev. 11:3032-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sidorova, J., G. Mikesell, and L. Breeden. 1995. Cell cycle regulated phosphorylation of Swi6 controls its nuclear localization. Mol. Biol. Cell 6:1641-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siede, W., A. S. Friedberg, I. Dianova, and E. C. Friedberg. 1994. Characterization of G1 checkpoint control in the yeast Saccharomyces cerevisiae following exposure to DNA-damaging agents. Genetics 138:271-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siede, W., A. S. Friedberg, and E. C. Friedberg. 1993. RAD9-dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90:7985-7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stuart, D., and C. Wittenberg. 1996. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 9:2780-2794. [DOI] [PubMed] [Google Scholar]
- 56.Sun, Z., D. S. Fay, F. Marini, M. Foiani, and D. F. Stern. 1996. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 10:395-406. [DOI] [PubMed] [Google Scholar]
- 57.Sun, Z., J. Hsiao, D. S. Fay, and D. F. Stern. 1998. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281:272-274. [DOI] [PubMed] [Google Scholar]
- 58.Uhlmann, F., F. Lottspeich, and K. Nasmyth. 1999. Sister chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400:37-42. [DOI] [PubMed] [Google Scholar]
- 59.Vialard, J. E., C. S. Gilbert, C. M. Green, and N. F. Lowndes. 1998. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 17:5679-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, H., D. Liu, Y. Wang, J. Qin, and S. J. Elledge. 2001. Pds1 phosphorylation in response to DNA damage is essential for its DNA damage checkpoint function. Genes Dev. 15:1361-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yazdi, P. T., Y. Wang, S. Zhao, N. Patel, E. Y. Lee, and J. Qin. 2002. SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev. 16:571-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan, C., S. Yongkiettrakul, I. J. Byeon, S. Zhou, and M. D. Tsai. 2001. Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53. J. Mol. Biol. 314:563-575. [DOI] [PubMed] [Google Scholar]
- 63.Zheng, P., D. S. Fay, J. Burton, H. Xiao, J. L. Pinkham, and D. F. Stern. 1993. SPK1 is an essential S-phase specific gene of Saccharomyces cerevisiae that encodes a nuclear serine/threonine/tyrosine kinase. Mol. Cell. Biol. 13:5829-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]