Abstract
Bloom syndrome (BS) is a genetic disorder associated with dwarfism, immunodeficiency, reduced fertility, and an elevated risk of cancer. To investigate the mechanism of this disease, we isolated from human HeLa extracts three complexes containing the helicase defective in BS, BLM. Interestingly, one of the complexes, termed BRAFT, also contains five of the Fanconi anemia (FA) complementation group proteins (FA proteins). FA resembles BS in genomic instability and cancer predisposition, but most of its gene products have no known biochemical activity, and the molecular pathogenesis of the disease is poorly understood. BRAFT displays a DNA-unwinding activity, which requires the presence of BLM because complexes isolated from BLM-deficient cells lack such an activity. The complex also contains topoisomerase IIIα and replication protein A, proteins that are known to interact with BLM and could facilitate unwinding of DNA. We show that BLM complexes isolated from an FA cell line have a lower molecular mass. Our study provides the first biochemical characterization of a multiprotein FA complex and suggests a connection between the BLM and FA pathways of genomic maintenance. The findings that FA proteins are part of a DNA-unwinding complex imply that FA proteins may participate in DNA repair.
Humans and mice with mutations in either one or both copies of the BLM gene have a higher risk of developing cancer (8, 13, 14, 30). BLM belongs to the RecQ family of DNA helicases (8) and possesses a DNA-unwinding activity for several types of DNA substrates (3, 25, 26, 33, 39). Interestingly, two other members of the RecQ family are mutated in the Werner (51) and Rothmund-Thomson (27) syndromes, which feature both premature aging and genomic instability and predisposition to cancer (23, 32). Mutation in RecQ helicases in other species results in similar genome instability phenotypes. The fact that defects in three of five known human RecQ helicases cause genome instability diseases suggests that this family of proteins plays key roles in maintaining the integrity of the genome. Because the phenotypes of the three diseases are different, these helicases presumably function in distinct complexes and pathways.
In the case of BLM, several of its interacting proteins have been reported. These include topoisomerase IIIα (Τopo IIIα) (19, 22, 47), an enzyme that can stimulate helicase activity by relieving the torsional stress produced during unwinding of DNA; replication protein A (RPA) (2), a DNA-binding protein that plays essential roles in DNA replication and nucleotide excision repair; MLH1 (29, 38), a protein involved in mismatch repair and defective in colon cancer (1, 36); and p53, a tumor suppressor mutated in many cancers (45). In addition, BLM has been found in the BRCA1-associated genome surveillance complex, BASC (46). However, the endogenous BLM complexes have not been purified by unbiased biochemical approaches. Consequently, basic questions regarding BLM remain unanswered, including the number and composition of BLM complexes that exist in a given cell type.
Fanconi anemia (FA) is a genetic disease characterized by congenital defects, bone marrow failure, and cancer susceptibility (21). As with Bloom syndrome (BS), the cells derived from FA patients exhibit genomic instability. Eight complementation groups have been described for this disease, and their corresponding genes have been identified (18, 21, 41). Five FA proteins (A, C, E, F, and G) have been suggested to interact with each other to form a multiprotein nuclear complex, the “core complex” (7, 11, 31). Recent evidence suggests that FA proteins function in a DNA damage response pathway involving breast cancer susceptibility genes 1 and 2 (BRCA1 and BRCA2, respectively). For example, following DNA damage induced by mitomycin C (MMC), an FA protein, FANCD2, becomes monoubiquitinated and redistributes into nuclear foci, where it colocalizes with BRCA1 (12). In addition, another FA protein, FANCD1, has been identified as BRCA2 (18). BRCA2 can regulate the activity of Rad51 (6) and may participate in homologous repair of DNA damages as a DNA-binding protein (42, 50). However, the mechanism of this disease remains unclear because most FA proteins lack recognizable structure motifs, and none of them has been associated with any biochemical activity.
We have previously purified several ATP-dependent chromatin-remodeling complexes (44, 48, 49). They all contain an SWI2/SNF2-like ATPase or helicase. Often, one ATPase is present in several distinct complexes, each of which has a unique function. Thus, to understand the function of a particular ATPase, each complex containing the protein must be purified and analyzed. Because of the importance of RecQ helicases in maintaining genome stability, we sought to systematically purify each endogenous RecQ helicase complex and study their functions. We report here the purification and analysis of proteins in three distinct BLM-associated multiprotein complexes from human HeLa cells. Interestingly, one of these complexes includes five FA core complex proteins, which suggests a functional connection between the pathways disturbed in these genomic instability syndromes.
MATERIALS AND METHODS
Cell culture.
Three types of Epstein-Barr virus (EBV)-immortalized lymphoblasts—i.e., wild-type (ManEBV), FA-A (VU388), and BLM (2036) cell lines—were maintained in RPMI medium (Life Technology) supplemented with 10% heat-inactivated fetal calf serum and grown in a humidified 5% carbon dioxide (CO2)-containing atmosphere at 37°C. HeLa S3 cells were obtained from the National Cell Culture Center. For MMC-treated HeLa cells, the cells were treated with 40 ng of MMC/ml for 24 h.
Antibodies.
Monoclonal and polyclonal antibodies to FANCA, FANCF, FANCG, FANCC, and FANCE have been described elsewhere (7, 15, 35, 43). The rabbit BLM polyclonal antibody (69D) was raised against a chimeric protein containing a region of BLM (amino acids 230 to 330) fused to maltose-binding protein. This antibody was affinity purified and used for immunoaffinity purification of BRAFT. A goat polyclonal antibody (C-18) to BLM (Santa Cruz Biotechnology) was used for immunoblotting analysis. Antibodies to RPA34 (AB1) and RPA70 (AB2) were from Neomarker. Antibody to RPA14 was from Genetex. Monoclonal antibody to BRCA1(AB1) and polyclonal antibody to MLH1(AB2) were from Oncogene. Antibodies to human Topo IIIα were kind gifts of L. Guarente. A polyclonal FANCD2 antibody was generated by immunizing rabbit with recombinant protein containing maltose-binding protein and an N-terminal region of FANCD2 (Z. Yan, unpublished results).
Immunoprecipitation.
BLM complexes were isolated from HeLa S3 nuclear extract by using an immunoprecipitation protocol described previously (C. S. Lee et al., unpublished data). Briefly, 1 ml (8 mg/ml) of nuclear extract was diluted 10 times with IP buffer (20 mM HEPES [pH 7.9], 200 mM NaCl, 1 mM dithiothreitol [DTT], 0.2 mM phenylmethylsulfonyl fluoride [PMSF], 10% glycerol) and incubated with 5 μg of affinity-purified rabbit polyclonal antibody (69D) in the presence of 100 μl of protein A-beads (Amersham Pharmacia) for at least 12 h. The immunoprecipitate was washed four times with the IP buffer. The complex on the beads was either used directly for the helicase assay or was eluted from the beads by using 100 mM glycine-HCl buffer (pH 2.5). The eluted complex was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting analysis. Less than 10% of the input was loaded as controls for immunoblotting. For mass spectrometric analysis, the proteins were visualized by Coomassie blue staining, excised from the gel, and digested with trypsin. The peptides obtained were analyzed by matrix-assisted laser desorption ionization-time of flight and/or liquid chromatography-mass spectrometry analyses. The data are not shown but are available upon request. All other coimmunoprecipitations were carried out as described above or in the legends.
Nuclear extract fractionation.
The HeLa S3 nuclear extract (16 mg) was directly applied to a Superose-6 column (HR 16/50; Amersham) equilibrated with the column running buffer containing 20 mM HEPES (pH 7.9), 200 mM NaCl, 1 mM DTT, 0.1 mM PMSF, and 10% glycerol. Fractions were collected (1.5 ml each) and analyzed by SDS-8% PAGE and immunoblotting. The peak fraction was directly loaded onto antibody beads for immunoprecipitation as described above. For human lymphoblastoid cells, nuclear extracts were diluted twofold with buffer containing 20 mM HEPES (pH 7.9)-1 mM DTT-0.1 mM PMSF before they were loaded onto an HR 10/30 Superose-6 column (Amersham). Fractions were then collected (0.5 ml) and analyzed by immunoblotting.
DNA helicase assay.
A 5′-end-labeled oligonucleotide was annealed to an M13mp18 single-stranded DNA (New England Biolabs) to obtain a partial duplex DNA with a 3′ overhang region. Helicase assay reaction mixtures (40 μl) contained 200 cpm of partial duplex DNA substrate, 50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 2 mM ATP, 100 μg of bovine serum albumin/ml, 50 mM NaCl, and the indicated amounts of complexes isolated by using either BLM or FANCA antibody. Reactions were carried out at 37°C for 30 min. The reactions were stopped by the addition of 10 μl of 6× high-density gel loading dye (Novex) containing 50 mM EDTA and 0.8% SDS. The products of helicase reactions were resolved on 10% nondenaturing polyacrylamide gel and visualized by autoradiography.
RESULTS
BLM-associated polypeptides include Topo IIIα, RPA, MLH1, and five FA proteins.
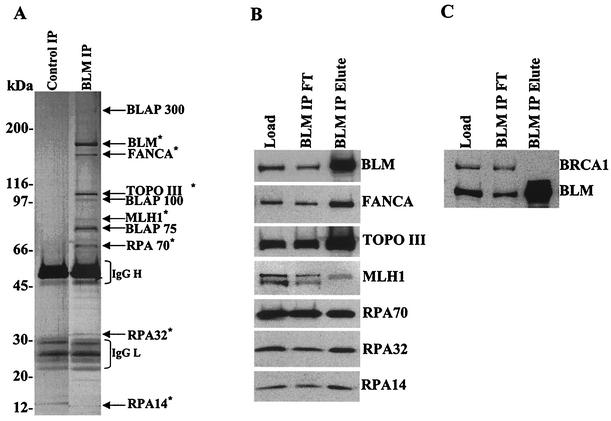
We isolated the endogenous BLM-associated complexes from human HeLa nuclear extract by direct immunoprecipitation with a BLM-specific antibody. BLM coimmunoprecipitated with about 10 major polypeptides (abbreviated as BLAPs for BLM-associated polypeptides) (Fig. 1A). These polypeptides were not immunoprecipitated with preimmune serum from the same rabbit, suggesting that they could represent components of BLM-associated complexes. The 170-kDa polypeptide was identified as BLM by both mass spectrometry and immunoblotting analysis (Fig. 1A and B). Five other BLAPs were similarly identified as Topo IIIα, three subunits of RPA, and MLH1. All of these proteins have been reported to interact with BLM (2, 19, 29, 38, 47). The fact that three known BLM-interacting proteins have been recovered validates the isolation protocol and suggests that the polypeptides obtained could all be components of one or several BLM-associated complexes.
FIG. 1.
Novel BS complexes contain Topo IIIα, RPA, and MLH1. (A) A silver-stained gel showing the novel BS complexes isolated by immunoprecipitation with a BLM antibody from a HeLa nuclear extract. The polypeptides identified by mass spectrometry analysis are indicated by an asterisk. The polypeptides that remain to be identified (termed BLAPs) are also marked. The presence of immunoglobulins (immunoglobulin G [IgG] H and IgG L) are indicated. A control immunoprecipitation was included with the preimmune serum of the same rabbit. (B and C) Autoradiographs illustrating the immunoblotting analysis of load, flowthrough (FT), and eluted (Elute) fractions from the immunoaffnity purification by the BLM antibody. The antibodies utilized for immunoblotting are shown.
BLAPs isolated in the present study do not contain BRCA1 and eight of the other known components of the previously described BLM-containing complex, BASC, which was also isolated from HeLa cells (Fig. 1C and data not shown) (46). MLH1 is the single component present in both the BASC and BLAPs. However, data below show that the BLM-MLH1 complex described here contains other components not reported to occur in the BASC. In addition, our complex is ∼600 kDa, which is considerably smaller than the reported mass of the BASC (∼2 MDa). Therefore, the BLM complexes reported here are different from the BASC.
The BLAPs described here also do not contain p53 (data not shown), which has been reported to interact with BLM (45). However, the present study used HeLa cells in which p53 was inactivated by association with HPV-E6 oncoprotein. Future studies with cell lines that contain wild-type p53 will be needed to isolate the BLM-p53 complex.
One of the remaining BLAPs was identified as an FA protein, FANCA, by mass spectrometry and immunoblotting (Fig. 1A and B). Five FA proteins (referred to as core complex FA proteins)—FANCA, FANCC, FANCE, FANCF, and FANCG—have previously been suggested to form a nuclear complex (7, 11, 31). Thus, the identification of FANCA as a BLAP hinted that the other four FA proteins could also be present in BLM complexes. This was confirmed by immunoblotting showing that all five FA proteins—FANCG, FANCC, FANCE, FANCF, and FANCA—all associate with BLM (Fig. 2B).
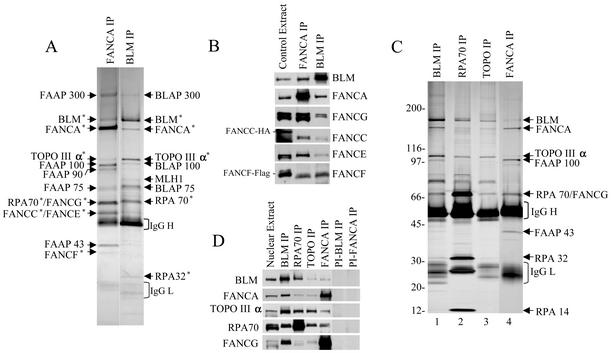
FIG. 2.
Five FA complementation group proteins are present in one complex with BLM, Topo IIIα, and RPA. (A) A silver-stained gel showing the polypeptides associated with endogenous FANCA (FAAPs) in comparison with BRAFT. Note that FANCA antibody was cross-linked to protein A-beads prior to immunoprecipitaion. The polypeptides identified by mass spectrometry analysis are marked with an asterisk. (B) Immunoblotting data showing the presence of multiple FA proteins in polypeptides isolated by using FANCA and BLM antibodies. As a positive control for FANCC and FANCF, whole-cell lysate from 293 cells transiently transfected with vectors expressing either epitope-tagged FANCC-HA or FANCF-Flag was used. HeLa nuclear extract was used as a positive control for BLM, FANCA, and FANCG. (C and D) A silver-stained gel (C) and Western blotting analysis (D) of the polypeptides associated with endogenous BLM, RPA70, Topo IIIα, and FANCA. The antibodies used for each immunoprecipitation are indicated on top of the figure. Mock immunoprecipitation with preimmune sera was used as the control. All immunoprecipitations were done with HeLa nuclear extract.
FANCA-associated polypeptides include five FA proteins, together with BLM, Topo IIIα, and RPA.
Although FANCA has been suggested to form a nuclear complex with four other FA proteins, this complex has not been purified and characterized. We immunoprecipitated FANCA-associated polypeptides (abbreviated as FAAPs for FANCA-associated polypeptides) by immunoprecipitation and found by both mass spectrometry and immunoblotting that they include all five core complex FA proteins (Fig. 2A and B). The finding that the FAAPs include all five FA proteins confirms and strengthens the earlier evidence for a nuclear FA complex that may play a critical role in FA. Speculatively, the remaining FAAPs may be components of the same FA complex.
Importantly, three of the remaining FAAPs were identified as BLM, Topo IIIα, and RPA70 by both mass spectrometry (Fig. 2A) and immunoblotting (Fig. 2D). Because BLM, core complex FA proteins, Topo IIIα, and RPA can be immunoprecipitated by both BLM and FANCA antibodies, they appear to be components of the same complex.
FANCA has been reported to associate with the human ATP-dependent chromatin-remodeling complex, SWI/SNF (34). However, we failed to detect any human SWI/SNF components in the FAAPs (data not shown).
RPA and Topo IIIα are components of the same complex containing BLM and core complex FA proteins.
RPA and Topo IIIα have been shown to interact independently with BLM (2, 19, 47) but have not previously been found together in a single complex. RPA and Topo IIIα-associated polypeptides were isolated by immunoprecitation and were found to exhibit similar mobilities on an SDS gel that not only resemble one another but also the mobilities of BLAPs and FAAPs (Fig. 2C). Immunoblotting showed that the two groups of proteins include not only RPA and Topo IIIα but also BLM, FANCA, and FANCG (Fig. 2D). Because these proteins can be coimmunoprecipitated by each respective antibody, they must be components of the same complex. We termed this complex BRAFT (for BLM, RPA, FA, and Topo IIIα).
The relative amounts of the BRAFT components isolated by different antibodies were compared based on their intensities on a silver-stained gel and their immunoblotting signals (Fig. 2C and D). The amounts of FANCA and several of its associated polypeptides (FAAP100, FANCG, and FAAP43) isolated with FANCA antibody are greater than those isolated by BLM and other antibodies, implying that significant amounts of core FA proteins may be present in complexes other than BRAFT (see also Fig. 4A). Likewise, the amounts of three RPA components immunoprecipitated with an RPA antibody are also significantly greater than the other subunits of BRAFT (Fig. 2C), suggesting that only a small percentage of RPA associates with BRAFT.
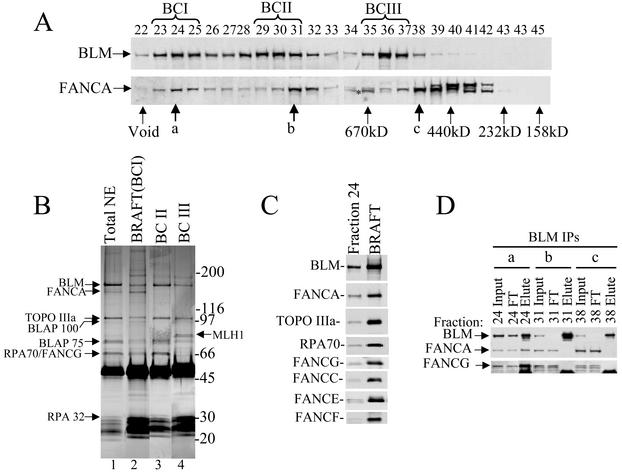
FIG. 4.
The BRAFT complex containing BLM and FA proteins is one of the three BLM complexes in the nuclear extract. (A) Immunoblotting analysis of different fractions from Superose-6 fractionation of HeLa nuclear extract. The fraction numbers are indicated on the top. The fractions corresponding to proteins of known molecular mass are denoted below the figure in kilodaltons. The void volume of the column is marked as “void.” The fractions in which BLM peaked are marked at the top as BCI, BCII, and BCIII (for BLM complexes I to III). The letters a, b, and c indicate the overlapping peak fractions between BLM and FANCA. A false-positive band derived from antibody cross-reactivity (which is also present in fractions from FANCA-deficient cells) is indicated by an asterisk. (B) Silver-stained gel showing the composition of three BLM complexes, which were immunoprecipitated with a BLM antibody (69D) from the three BLM peak fractions after Superose-6 fractionation (see panel A). The immunoprecipitations were carried out directly from each of the corresponding fractions (i.e., fractions 24, 30, and 36) without further dilution. BLM immunoprecipitate from total nuclear extract (NE) is shown for comparison. (C) Western blotting analysis showing the presence of multiple FANC proteins in the BRAFT complex. Fraction 24 of Superose-6, which corresponds to the 1.5- to 2-MDa fraction, was used as a positive control. (D) Immunoblotting of BLM immunoprecipitates (IPs) from three different Superose-6 fractions—a, b, and c—in which significant amounts of BLM and FANCA overlap. Notably, BLM coimmunoprecipitates with FANCA and FANCG only from fraction A, which corresponds to a molecular mass of 1.5 to 2 MDa (BRAFT).
The association between BLM and the core complex FA proteins is specific.
To rule out the possibility that BRAFT components may be isolated through nonspecific interactions with protein A-beads or antibodies, a control immunoprecipitation was performed with matched preimmune sera for both BLM and FANCA antisera. None of the BRAFT components was isolated, thus ruling out this possibility (Fig. 2D). To exclude another possibility that BLM and FANCA antibodies may cross-react, immunoprecipitation was done with nuclear extracts from human lymphoblastoid cell lines lacking either FANCA or BLM. These cell lines were derived from either FA or BS patients that carry mutated FANCA or BLM genes, respectively. It was noted that, in FANCA-deficient cells, FANCG was also absent (Fig. 3A), a finding consistent with earlier observations that these two proteins stabilize each other (10). BLM and FANCA antibodies immunoprecipitated each other and their associated proteins only from wild-type cells and not from their respective mutant cells, indicating that the antibodies do not cross-react (Fig. 3B). To eliminate a further possibility that BLM may associate with FA proteins through DNA, immunoprecipitation was performed in the presence of ethidium bromide (EtBr), a DNA-intercalating drug that can prevent proteins from binding to DNA and has been frequently used to identify DNA-independent protein associations (28). Both FANCA and FANCG were immunoprecipitated by BLM antibody in the presence of EtBr, and their levels are comparable to those immunoisolated in the absence of EtBr (Fig. 3C), indicating that the association between BLM and FA proteins is not through DNA.
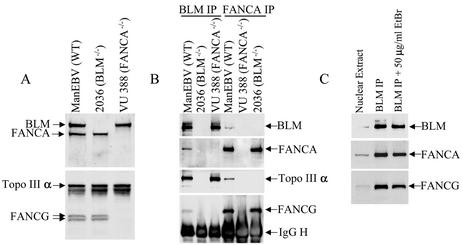
FIG. 3.
BLM and FANCA antibodies immunoprecipitate their associated complexes from wild-type but not their respective mutant cell lines. (A) Autoradiographs showing immunoblotting analysis of nuclear extracts prepared from wild-type (ManEBV), BLM−/− (2036), and FANCA−/− (VU388) human lymphoblastoid cell lines. After SDS-PAGE and Western transfer, the membrane was cut into two parts. The part corresponding to a molecular mass of >120 kDa was probed by using a mixture of BLM and FANCA antibodies (top panel). The other part was analyzed by using a mixture of Topo IIIα and FANCG antibodies (bottom panel). Note that FANCG was not detected in the FANCA-deficient cells. (B) Immunoblotting analysis of the polypeptides isolated by either BLM or FANCA antibodies from the three lymphoblastoid cell lines. The antibodies used for immunoprecipitation are indicated on the top, whereas those used for immunoblotting are listed on the right. (C) Immunoblotting analysis showing that the association between BLM and FA proteins remains intact in the presence of EtBr, a DNA-intercalating drug that can disassociate proteins from DNA. In this experiment, 50 μg of EtBr/ml was included in the immunoprecipitation mixture.
A proportion of BLM and core complex FA proteins can form independent complexes.
The amounts of BLM and Topo IIIα immunoisolated by BLM antibody from FANCA-deficient cells are similar to those obtained from wild-type cells (Fig. 3B), indicating that the interaction between BLM and Topo IIIα does not require FA proteins. The result also showed that BLM and Topo IIIα could form complexes that lack FA proteins, a finding consistent with our data below (Fig. 4). Likewise, the amounts of FANCA and FANCG immunoprecipitated by FANCA antibody from BLM-deficient cells are similar to those obtained from wild-type cells (Fig. 3B), indicating that the association between the core complex FA proteins is not dependent on BLM. Moreover, our results suggest that core complex FA proteins may form their own complexes without BLM. Interestingly, Topo IIIα was immuoprecipitated by FANCA antibody from wild-type cells but not from BLM-deficient cells, suggesting that the FA proteins may associate with Topo IIIα and other BRAFT subunits through BLM.
Only one of the three BLM complexes contains FA proteins.
We further characterized BLM complexes by fractionating HeLa nuclear extract through a Superose-6 sizing column. BLM fractionated in three separate complexes, termed BLM complexes I, II, and III, corresponding to masses of ca. 1.5 to 2.0 MDa, 1 MDa, and 600 kDa, respectively (Fig. 4A). No detectable BLM fractionated as the free protein at its calculated molecular mass (160 kDa), suggesting that essentially all BLM exists in high-molecular-mass complexes in cells. Each BLM complex was immunoprecipitated from its corresponding column fractions and displayed differential subunit composition by SDS-PAGE analysis (Fig. 4B). Only the 1.5- to 2-MDa complex contains the FANCA polypeptide (lane 2). The same complex also includes Topo IIIα, RPA, and other FA proteins (Fig. 4C), indicating that this complex is BRAFT. In comparison, BLM complexes II and III lack FANCA by silver-staining analysis (Fig. 4B), which was confirmed by immunoblotting analysis showing that they lack both FANCA and FANCG (data not shown; but see Fig. 4D). In complex I, the amounts of FANCA and BLM are more stoichiometric than those obtained by direct immunoprecipitation from nuclear extract (Fig. 4B, compare the relative intensities of FANCA and BLM bands in lanes 2 and 1). This is consistent with the greater purity of the BRAFT complex immunoisolated after Superose-6 fractionation, which has separated away the BLM complexes (II and III) that lack FA proteins. The 1-MDa BLM complex II contains Topo IIIα and RPA but no FA proteins (Fig. 4B). Because the only difference between complexes I and II appears to be the FA proteins, it is possible that complex II is a subcomplex of complex I. The 600-kDa BLM complex III contains Topo IIIα and MLH1 but no RPA or FA proteins.
Only a subset of FANCA associates with BLM.
Superose-6 fractionation also revealed that FANCA is in three different complexes corresponding to molecular masses of ca. 1.5 to 2.0 MDa, 900 kDa, and 500 kDa (Fig. 4A). Only the first one comigrates with BRAFT in a 1.5- to 2-MDa complex. Immunoprecipitation from the 1.5- to 2-MDa fraction by using FANCA antibody yielded BLM and other BRAFT components, a finding consistent with the results presented above indicating that BRAFT is a 1.5- to 2-MDa complex (data not shown). In addition, substantial amounts of BLM and FANCA overlap in the 900- and 500-kDa fractions. However, the possibility that these two proteins might also participate in smaller complexes was ruled out because BLM coimmunoprecipitated with FANCA only from the 1.5- to 2-MDa fraction but not from the two smaller fractions (Fig. 4D). Another FA core complex protein, FANCG, displayed a fractionation profile almost identical to that of FANCA by Superose-6 analysis (data not shown). We infer that these core complex FA proteins could occur in at least two complexes without BLM.
BLM complexes from a FANCA-deficient cell line have lower molecular masses.
We compared the molecular masses of BLM complexes from a FANCA-deficient cell line to those from a normal lymphoblastoid cell line by Superose-6 analysis. If FA proteins are required components of the BLM complex, the complexes isolated from a FANCA-deficient cell line should be smaller. This expectation was confirmed, since the majority of BLM from a FANCA-deficient cell line fractionated as a complex of 1 MDa (Fig. 5), corresponding to the BLM complex II of HeLa cells that lacks FA proteins. In contrast, most BLM from wild-type cells fractionated as the 1.5- to 2-MDa BRAFT complex. We infer that in the absence of FANCA, the BRAFT complex loses its FA components, becoming the smaller 1-MDa BLM complex.
FIG. 5.
The BLM complexes are present in smaller forms in an FA cell line. Immunoblotting analysis of different fractions from Superose-6 fractionation of the wild-type (ManEBV) and FANCA-deficient (VU388) lymphoblastoid cell lines. Notably, BLM peaks at fraction 21 in the FANCA-deficient cells, corresponding to a complex of ca. 1 MDa. This is smaller than the BLM from the wild-type cells, which peaks between fractions of 17 and 18, corresponding to a complex of 1.5 to 2 MDa (BRAFT). It was noted that BCII in this analysis is not that obvious, which may be due to its different levels in different cell lines.
It was noted that by Superose-6 analysis, the peak corresponding to BLM complex II of the normal lymphoblastoid cell line (Fig. 5, fractions 21 to 22) is not that obvious compared to that of HeLa cells (Fig. 4A). It is possible that the level of BLM complex II, which lacks FA proteins, is variable between cell lines.
BLM- and FANCA-associated complexes have a DNA-unwinding activity that requires the presence of BLM.
BLM complexes immunoisolated from HeLa or a normal lymphoblastoid cell line displayed a helicase activity that unwinds duplex DNA (Fig. 6A and B; see Fig. 3 for analyses of immunoprecipitates from various lymphoblastoid cell lines), a finding consistent with previous results that recombinant BLM is a DNA helicase (25). The observed activity should be derived from BLM, but not a contaminant, because a mock immunoprecipitation from a BLM-deficient cell line failed to isolate this activity. BLM immunoprecipitate from a FANCA-deficient cell line has a helicase activity indistinguishable from that isolated from the control cell line, suggesting that FA proteins do not affect the helicase activity of BLM, at least with this DNA substrate.
FIG. 6.
Complexes isolated by BLM and FANCA antibodies possess a DNA-unwinding activity that requires the presence of BLM. (A-D) Autoradiographs showing the results of a helicase assay for complexes isolated by antibodies to BLM or FANCA. The partial DNA duplex substrate and the displaced single-strand oligonucleotide (Oligo) are illustrated on the left of the figure. Also analyzed were fractions isolated by control (mock) immunoprecipitation (Mock IP) with preimmune serum of the BLM antibody or normal rabbit IgG for the FANCA antibody. The cell lines and antibodies used for immunoprecipitations are indicated on the top. For complexes isolated from HeLa cells, 0 to 200 ng of the BLM-associated complexes or 0 to 500 ng of FANCA-associated complexes are used for the helicase assay (A and C). For lymphoblastoid cells, 0 to 50 ng of BLM-associated complexes and 0 to 100 ng of FANCA-associated complexes were analyzed (B and D). Maximum amounts of complexes were assayed in those isolated from BLM-deficient cells. To ensure that comparable amounts of BLM were present in complexes isolated from wild-type (WT) and FANCA-deficient cells, each immunoprecipitate was analyzed by immunoblotting with a BLM antibody, and the results are shown as a small figure at the bottom of panel B.
Importantly, complexes isolated with the FANCA antibody from HeLa or a normal lymphoblastoid cell line displayed similar helicase activities (Fig. 6C and D), whereas mock immunoprecipitation from a FANCA-deficient cell line yielded no activity (Fig. 6D). Thus, this activity is apparently not due to a contaminant. FANCA immunoprecipitate from BLM-deficient cells also lacked helicase activity (Fig. 6D), suggesting that BLM is required for the observed activity. All of these data are consistent with a notion that BLM and FA proteins are components of one nuclear complex that can unwind DNA through the BLM helicase.
FA proteins and BLM do not function in a signaling pathway through monoubiquitination.
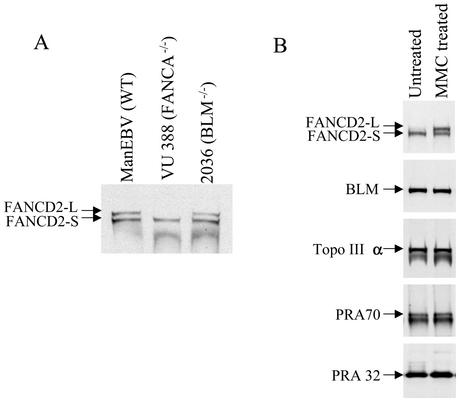
Recent evidence suggests that core complex FA proteins control the activation of FANCD2 by monoubiquitination in a DNA damage response pathway (12). The findings that BLM and core complex FA proteins are present in the same complex prompted us to explore the possibility that BLM may be involved in the same pathway. However, the level of FANCD2 monoubiquitination in BLM-deficient cells was found to be indistinguishable from that in normal lymphoblast cells (Fig. 7A). As a monoubiquitination was completely absent in the FANCA-deficient cells. The results suggest that BLM does not function upstream of the core complex FA proteins in this pathway. In addition, under DNA damage conditions in which a high level of FANCD2 monoubiquitination was induced, no ubiquitination was detected for BLM, Topo IIIα, and RPA (Fig. 7B), arguing against these proteins being downstream ubiquitination targets of the FA complex. Together, these data suggest that FA proteins and BLM do not act in a linear signaling pathway through monoubiquitination.
FIG. 7.
FA Proteins and BLM do not function in the linear signaling pathway through monoubiquitination. (A) Immunoblotting analysis to show that the monoubiquitination of FANCD2 is normal in BS cells. Nuclear extracts prepared from the indicated lymphoblast cells were probed with a FANCD2 antibody (FANCD2-L and FANCD2-S represent ubiquitinated and nonubiquitinated forms of this protein, respectively). (B) Immunoblotting analysis to show that monoubiquitination is not detectable for BLM, Topo IIIα, and RPA after HeLa cells are treated with MMC. As a control, monoubiquitination of FANCD2 is strongly induced under such conditions.
DISCUSSION
RecQ helicases play key roles in maintaining genome stability and preventing early onset of aging. However, their exact mechanisms of action remain unclear. One reason is that the functional units of these helicases in vivo—the endogenous complexes containing them—have not been purified and studied. We report here three human complexes containing the BLM helicase involved in BS and demonstrate that they have distinct subunit compositions. One complex contains five FA proteins, whereas another contains MLH1, a protein involved in mismatch repair and mutated in colon cancer. Identification of these distinct complexes permits further biochemical characterization of their roles in cellular pathways and in disease etiology.
The most important finding of the present study is the observation that five FA proteins are part of a BLM complex that possesses a DNA-unwinding activity. To our knowledge, this is the first biochemical activity for a complex containing the endogenous FA proteins. Our data imply that these FA proteins may have a function in DNA repair as part of this BLM-dependent DNA-unwinding complex. Thus far, the molecular function of the FA core complex proteins remains unknown. They have been proposed to function in at least two distinct pathways: DNA repair and detoxification of reactive oxygen species (4, 5). More-recent evidence suggests that these FA proteins control the activation of FANCD2 by monoubiquitination, a step required for DNA-damage-dependent colocalization with BRCA1 (12). Our Superose-6 analysis shows that FANCA is present in at least three nuclear complexes, only one of which contains BLM (Fig. 4). It will be important to determine the composition of the other two FANCA-containing complexes, as well as their function.
Although BS and FA cells have strikingly different phenotypes, with elevated sister chromatid exchanges in the former and cross-linker-induced quadriradials in the latter, a connection between the two diseases may be inferred from several lines of earlier evidence. First, both conditions feature genomic instability, predisposition to cancer, and reduced fertility. Second, the type of spontaneous chromosomal instability in FA was noted to be similar to that of BS (21), showing mainly chromatid breaks and interchanges. This is in contrast to other caretaker gene disorders that favor deletions, rearrangements, and variable chromosomal numbers. The similarity also extends to the chromosomal aberration observed in BRCA2-deficient cells, which was found to be similar to that in both BS and FA cells (37), long before BRCA2 was identified as an FA gene itself (18). Third, some FA cells have elevated homologous recombination activity (40) and a higher frequency of sister chromatid exchanges (9, 16, 24), which are reminiscent of BS cells. Fourth, certain BS cells exhibit increased sensitivity to MMC (17, 20), a hallmark of FA cells.
Our data link BS and FA by showing that BLM and five FA proteins are components of a single complex. The findings that this complex includes at least three DNA-binding and remodeling proteins (BLM helicase, Topo IIIα, and RPA) and possesses a DNA-unwinding activity suggest that it could participate in the removal of aberrant DNA structures formed during dynamic processes such as DNA replication and repair. In accord with this notion, BLM helicase is capable of unwinding several types of DNA structures (3, 26, 33, 39). Topo IIIα could facilitate unwinding by relieving the torsional stress generated by the helicase. RPA may play one or both roles in this process: it may participate in recognition of the DNA damage, just as it does for nucleotide excision repair, or it may facilitate the unwinding reaction by stabilizing single-stranded DNA generated by the BLM helicase. The single-stranded DNA thus generated could then serve as a critical intermediate in recombination or repair. Malfunction of proteins in this complex could result in the accumulation of aberrant DNA structures that lead to chromosomal aberrations observed in these diseases.
An important unanswered question is how BLM and FA proteins function together in the BRAFT complex. Currently, there is no biochemical activity described for the core complex FA proteins, which makes it difficult to establish a functional connection between BLM and FA proteins. Our ubiquitination analyses suggest that these proteins do not function in a linear signaling pathway through monoubiquitination (Fig. 7). However, our ongoing work has suggested a new possibility: that core complex FA proteins, or their associated FAAPs, may possess DNA-modifying activities that are complementary to those present in BLM, Topo IIIα, and RPA during unwinding of the damaged DNA. In other words, FA proteins and BLM may not regulate each other's activity but instead may modify DNA structures in concert in sequential steps during the DNA repair process. Indeed, we found that one of the uncharacterized FAAPs contains a DNA endonuclease domain highly homologous to that of a protein known to be involved in DNA damage repair (unpublished data). Speculatively, FA proteins, through this FAAP, may provide a DNA-cutting activity necessary for the subsequent unwinding step catalyzed by the BLM helicase. Future experiments will be needed to analyze whether BRAFT has multiple DNA-modifying activities that cooperate to unwind the damaged DNA.
Acknowledgments
We thank I. Hickson and N. Ellis for the BLM cDNAs, N. Neff and J. Qin for the BLM antibodies, K. J. Patel for the FANCE antibody, J. C. Wang for the Topo IIIα cDNA, B. Johnson and L. Guarente for the Topo IIIα antibodies, D. Smeets for the BLM-deficient cell line, J. Borowiec and K. Vasquez for the RPA, L. Li for DNA and advice, Z. Yan for the FANCD2 antibody, N. Sherman for the mass spectrometric analysis, and B. Field for technical assistance. We also thank the National Cell Culture Center for providing cells. We thank D. Schlessinger and J. Yin for comments on the manuscript.
W.W. is a scholar of the Ellison Medical Foundation.
S.S., M.W., and D.Y. contributed equally to this study.
REFERENCES
- 1.Bronner, C. E., S. M. Baker, P. T. Morrison, G. Warren, L. G. Smith, M. K. Lescoe, M. Kane, C. Earabino, J. Lipford, A. Lindblom, et al. 1994. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature 368:258-261. [DOI] [PubMed] [Google Scholar]
- 2.Brosh, R. M., Jr., J. L. Li, M. K. Kenny, J. K. Karow, M. P. Cooper, R. P. Kureekattil, I. D. Hickson, and V. A. Bohr. 2000. Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J. Biol. Chem. 275:23500-23508. [DOI] [PubMed] [Google Scholar]
- 3.Brosh, R. M., Jr., A. Majumdar, S. Desai, I. D. Hickson, V. A. Bohr, and M. M. Seidman. 2001. Unwinding of a DNA triple helix by the Werner and Bloom syndrome helicases. J. Biol. Chem. 276:3024-3030. [DOI] [PubMed] [Google Scholar]
- 4.Cumming, R. C., J. Lightfoot, K. Beard, H. Youssoufian, P. J. O'Brien, and M. Buchwald. 2001. Fanconi anemia group C protein prevents apoptosis in hematopoietic cells through redox regulation of GSTP1. Nat. Med. 7:814-820. [DOI] [PubMed] [Google Scholar]
- 5.D'Andrea, A. D. 2001. Cellular function of the Fanconi anemia pathway. Nat. Med. 7:1259-1260. [DOI] [PubMed] [Google Scholar]
- 6.Davies, A. A., J. Y. Masson, M. J. McIlwraith, A. Z. Stasiak, A. Stasiak, A. R. Venkitaraman, and S. C. West. 2001. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell 7:273-282. [DOI] [PubMed] [Google Scholar]
- 7.de Winter, J. P., L. van der Weel, J. de Groot, S. Stone, Q. Waisfisz, F. Arwert, R. J. Scheper, F. A. Kruyt, M. E. Hoatlin, and H. Joenje. 2000. The Fanconi anemia protein FANCF forms a nuclear complex with FANCA, FANCC, and FANCG. Hum. Mol. Genet. 9:2665-2674. [DOI] [PubMed] [Google Scholar]
- 8.Ellis, N. A., J. Groden, T. Z. Ye, J. Straughen, D. J. Lennon, S. Ciocci, M. Proytcheva, and J. German. 1995. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell 83:655-666. [DOI] [PubMed] [Google Scholar]
- 9.Fujiwara, Y., Y. Kano, and Y. Yamamoto. 1984. DNA interstrand cross-linking, repair, and SCE mechanism in human cells in special reference to Fanconi anemia. Basic Life Sci. 29:787-800. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Higuera, I., Y. Kuang, J. Denham, and A. D. D'Andrea. 2000. The fanconi anemia proteins FANCA and FANCG stabilize each other and promote the nuclear accumulation of the Fanconi anemia complex. Blood 96:3224-3230. [PubMed] [Google Scholar]
- 11.Garcia-Higuera, I., Y. Kuang, D. Naf, J. Wasik, and A. D. D'Andrea. 1999. Fanconi anemia proteins FANCA, FANCC, and FANCG/XRCC9 interact in a functional nuclear complex. Mol. Cell. Biol. 19:4866-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Higuera, I., T. Taniguchi, S. Ganesan, M. S. Meyn, C. Timmers, J. Hejna, M. Grompe, and A. D. D'Andrea. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7:249-262. [DOI] [PubMed] [Google Scholar]
- 13.Goss, K. H., M. A. Risinger, J. J. Kordich, M. M. Sanz, J. E. Straughen, L. E. Slovek, A. J. Capobianco, J. German, G. P. Boivin, and J. Groden. 2002. Enhanced tumor formation in mice heterozygous for Blm mutation. Science 297:2051-2053. [DOI] [PubMed] [Google Scholar]
- 14.Gruber, S. B., N. A. Ellis, G. Rennert, K. Offit, K. K. Scott, R. Almog, P. Kolachana, J. D. Bonner, T. Kirchhoff, L. P. Tomsho, K. Nafa, H. Pierce, M. Low, J. Satagopan, H. Rennert, H. Huang, J. K. Greenson, J. Groden, B. Rapaport, J. Shia, S. Johnson, P. K. Gregersen, C. C. Harris, and J. Boyd. 2002. BLM heterozygosity and the risk of colorectal cancer. Science 297:2013.. [DOI] [PubMed] [Google Scholar]
- 15.Heinrich, M. C., K. V. Silvey, S. Stone, A. J. Zigler, D. J. Griffith, M. Montalto, L. Chai, Y. Zhi, and M. E. Hoatlin. 2000. Posttranscriptional cell cycle-dependent regulation of human FANCC expression. Blood 95:3970-3977. [PubMed] [Google Scholar]
- 16.Hojo, E. T., P. C. van Diemen, F. Darroudi, and A. T. Natarajan. 1995. Spontaneous chromosomal aberrations in Fanconi anaemia, ataxia telangiectasia fibroblast and Bloom's syndrome lymphoblastoid cell lines as detected by conventional cytogenetic analysis and fluorescence in situ hybridisation (FISH) technique. Mutat. Res. 334:59-69. [DOI] [PubMed] [Google Scholar]
- 17.Hook, G. J., E. Kwok, and J. A. Heddle. 1984. Sensitivity of Bloom syndrome fibroblasts to mitomycin C. Mutat. Res. 131:223-230. [DOI] [PubMed] [Google Scholar]
- 18.Howlett, N. G., T. Taniguchi, S. Olson, B. Cox, Q. Waisfisz, C. De Die-Smulders, N. Persky, M. Grompe, H. Joenje, G. Pals, H. Ikeda, E. A. Fox, and A. D. D'Andrea. 2002. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 297:606-609. [DOI] [PubMed]
- 19.Hu, P., S. F. Beresten, A. J. van Brabant, T. Z. Ye, P. P. Pandolfi, F. B. Johnson, L. Guarente, and N. A. Ellis. 2001. Evidence for BLM and topoisomerase IIIα interaction in genomic stability. Hum. Mol. Genet. 10:1287-1298. [DOI] [PubMed] [Google Scholar]
- 20.Ishizaki, K., T. Yagi, M. Inoue, O. Nikaido, and H. Takebe. 1981. DNA repair in Bloom's syndrome fibroblasts after UV irradiation or treatment with mitomycin C. Mutat. Res. 80:213-219. [DOI] [PubMed] [Google Scholar]
- 21.Joenje, H., and K. J. Patel. 2001. The emerging genetic and molecular basis of Fanconi anaemia. Nat. Rev. Genet. 2:446-459. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, F. B., D. B. Lombard, N. F. Neff, M. A. Mastrangelo, W. Dewolf, N. A. Ellis, R. A. Marciniak, Y. Yin, R. Jaenisch, and L. Guarente. 2000. Association of the Bloom syndrome protein with topoisomerase IIIα in somatic and meiotic cells. Cancer Res. 60:1162-1167. [PubMed] [Google Scholar]
- 23.Johnson, F. B., D. A. Sinclair, and L. Guarente. 1999. Molecular biology of aging. Cell 96:291-302. [DOI] [PubMed] [Google Scholar]
- 24.Kano, Y., and Y. Fujiwara. 1981. Roles of DNA interstrand crosslinking and its repair in the induction of sister-chromatid exchange and a higher induction in Fanconi's anemia cells. Mutat. Res. 81:365-375. [DOI] [PubMed] [Google Scholar]
- 25.Karow, J. K., R. K. Chakraverty, and I. D. Hickson. 1997. The Bloom's syndrome gene product is a 3′-5′ DNA helicase. J. Biol. Chem. 272:30611-30614. [DOI] [PubMed] [Google Scholar]
- 26.Karow, J. K., A. Constantinou, J. L. Li, S. C. West, and I. D. Hickson. 2000. The Bloom's syndrome gene product promotes branch migration of holliday junctions. Proc. Natl. Acad. Sci. USA 97:6504-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitao, S., A. Shimamoto, M. Goto, R. W. Miller, W. A. Smithson, N. M. Lindor, and Y. Furuichi. 1999. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 22:82-84. [DOI] [PubMed] [Google Scholar]
- 28.Lai, J. S., and W. Herr. 1992. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. USA 89:6958-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langland, G., J. Kordich, J. Creaney, K. Heppner Goss, K. Lillard-Wetherell, K. Bebenek, T. A. Kunkel, and J. Groden. 2001. The BLM helicase interacts with hMLH1 but is not required for DNA mismatch repair. J. Biol. Chem. 276:30031-30035. [DOI] [PubMed] [Google Scholar]
- 30.Luo, G., I. M. Santoro, L. D. McDaniel, I. Nishijima, M. Mills, H. Youssoufian, H. Vogel, R. A. Schultz, and A. Bradley. 2000. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 26:424-429. [DOI] [PubMed] [Google Scholar]
- 31.Medhurst, A. L., P. A. Huber, Q. Waisfisz, J. P. de Winter, and C. G. Mathew. 2001. Direct interactions of the five known Fanconi anaemia proteins suggest a common functional pathway. Hum. Mol. Genet. 10:423-429. [DOI] [PubMed] [Google Scholar]
- 32.Mohaghegh, P., and I. D. Hickson. 2001. DNA helicase deficiencies associated with cancer predisposition and premature ageing disorders. Hum. Mol. Genet. 10:741-746. [DOI] [PubMed] [Google Scholar]
- 33.Mohaghegh, P., J. K. Karow, R. M. Brosh, Jr., V. A. Bohr, and I. D. Hickson. 2001. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 29:2843-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otsuki, T., Y. Furukawa, K. Ikeda, H. Endo, T. Yamashita, A. Shinohara, A. Iwamatsu, K. Ozawa, and J. M. Liu. 2001. Fanconi anemia protein, FANCA, associates with BRG1, a component of the human SWI/SNF complex. Hum. Mol. Genet. 10:2651-2660. [DOI] [PubMed] [Google Scholar]
- 35.Pace, P., M. Johnson, W. M. Tan, G. Mosedale, C. Sng, M. Hoatlin, J. de Winter, H. Joenje, F. Gergely, and K. J. Patel. 2002. FANCE: the link between Fanconi anaemia complex assembly and activity. EMBO J. 21:3414-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papadopoulos, N., N. C. Nicolaides, Y. F. Wei, S. M. Ruben, K. C. Carter, C. A. Rosen, W. A. Haseltine, R. D. Fleischmann, C. M. Fraser, M. D. Adams, et al. 1994. Mutation of a mutL homolog in hereditary colon cancer. Science 263:1625-1629. [DOI] [PubMed] [Google Scholar]
- 37.Patel, K. J., V. P. Yu, H. Lee, A. Corcoran, F. C. Thistlethwaite, M. J. Evans, W. H. Colledge, L. S. Friedman, B. A. Ponder, and A. R. Venkitaraman. 1998. Involvement of Brca2 in DNA repair. Mol. Cell 1:347-357. [DOI] [PubMed] [Google Scholar]
- 38.Pedrazzi, G., C. Perrera, H. Blaser, P. Kuster, G. Marra, S. L. Davies, G. H. Ryu, R. Freire, I. D. Hickson, J. Jiricny, and I. Stagljar. 2001. Direct association of Bloom's syndrome gene product with the human mismatch repair protein MLH1. Nucleic Acids Res. 29:4378-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun, H., J. K. Karow, I. D. Hickson, and N. Maizels. 1998. The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 273:27587-27592. [DOI] [PubMed] [Google Scholar]
- 40.Thyagarajan, B., and C. Campbell. 1997. Elevated homologous recombination activity in fanconi anemia fibroblasts. J. Biol. Chem. 272:23328-23333. [DOI] [PubMed] [Google Scholar]
- 41.Timmers, C., T. Taniguchi, J. Hejna, C. Reifsteck, L. Lucas, D. Bruun, M. Thayer, B. Cox, S. Olson, A. D. D'Andrea, R. Moses, and M. Grompe. 2001. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol. Cell 7:241-248. [DOI] [PubMed] [Google Scholar]
- 42.Venkitaraman, A. R. 2002. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108:171-182. [DOI] [PubMed] [Google Scholar]
- 43.Waisfisz, Q., J. P. de Winter, F. A. Kruyt, J. de Groot, L. van der Weel, L. M. Dijkmans, Y. Zhi, F. Arwert, R. J. Scheper, H. Youssoufian, M. E. Hoatlin, and H. Joenje. 1999. A physical complex of the Fanconi anemia proteins FANCG/XRCC9 and FANCA. Proc. Natl. Acad. Sci. USA 96:10320-10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, W., Y. Xue, S. Zhou, A. Kuo, B. R. Cairns, and G. R. Crabtree. 1996. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10:2117-2130. [DOI] [PubMed] [Google Scholar]
- 45.Wang, X. W., A. Tseng, N. A. Ellis, E. A. Spillare, S. P. Linke, A. I. Robles, H. Seker, Q. Yang, P. Hu, S. Beresten, N. A. Bemmels, S. Garfield, and C. C. Harris. 2001. Functional interaction of p53 and BLM DNA helicase in apoptosis. J. Biol. Chem. 276:32948-32955. [DOI] [PubMed] [Google Scholar]
- 46.Wang, Y., D. Cortez, P. Yazdi, N. Neff, S. J. Elledge, and J. Qin. 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14:927-939. [PMC free article] [PubMed] [Google Scholar]
- 47.Wu, L., S. L. Davies, P. S. North, H. Goulaouic, J. F. Riou, H. Turley, K. C. Gatter, and I. D. Hickson. 2000. The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 275:9636-9644. [DOI] [PubMed] [Google Scholar]
- 48.Xue, Y., J. C. Canman, C. S. Lee, Z. Nie, D. Yang, G. T. Moreno, M. K. Young, E. D. Salmon, and W. Wang. 2000. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl. Acad. Sci. USA 97:13015-13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xue, Y., J. Wong, G. T. Moreno, M. K. Young, J. Cote, and W. Wang. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2:851-861. [DOI] [PubMed] [Google Scholar]
- 50.Yang, H., P. D. Jeffrey, J. Miller, E. Kinnucan, Y. Sun, N. H. Thoma, N. Zheng, P. L. Chen, W. H. Lee, and N. P. Pavletich. 2002. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science 297:1837-1848. [DOI] [PubMed] [Google Scholar]
- 51.Yu, C. E., J. Oshima, Y. H. Fu, E. M. Wijsman, F. Hisama, R. Alisch, S. Matthews, J. Nakura, T. Miki, S. Ouais, G. M. Martin, J. Mulligan, and G. D. Schellenberg. 1996. Positional cloning of the Werner's syndrome gene. Science 272:258-262. [DOI] [PubMed] [Google Scholar]