Abstract
Robust transcription of human T-cell leukemia virus type 1 (HTLV-1) genome requires the viral transactivator Tax. Although Tax has been previously shown to interact with the KIX domain of CBP/p300 in vitro, the precise functional relevance of this interaction remains unclear. Using two distinct approaches to interrupt the physical interaction between Tax and KIX, we find that Tax transactivation from chromatin templates is strongly dependent on CBP/p300 recruitment via the KIX domain. Additionally, we find that the primary functional contribution of CBP/p300 to Tax transactivation resides in the intrinsic acetyltransferase activity of the coactivators. These studies unexpectedly uncover a specific requirement for CBP/p300 acetyltransferase activity on chromatin templates assembled with nucleosomes lacking their amino-terminal tails. Together, these data indicate that the KIX domain of CBP/p300 is essential for targeting the acetyltransferase activity of the coactivator to the Tax-CREB (Tax/CREB) complex. Significantly, these observations reveal the presence of one or more CBP/p300 acetyltransferase targets that function specifically on chromatin templates, are independent of the histone tails, and are critical to Tax transactivation.
The human T-cell leukemia virus type 1 (HTLV-1)-encoded Tax protein is required for high-level transcription of the viral genome (for review, see reference 66). Tax stimulates HTLV-1 transcription through interaction with three conserved 21-bp repeat DNA elements, called viral CREs, located in the transcriptional control region of the virus. These sequences serve as binding sites for Tax in complex with the cellular transcription factor CREB (or other members of the ATF/CREB family of transcription factors) (1, 16, 22, 67). Tax binds to the viral CRE sequences through protein-DNA interactions with GC-rich minor groove sequences (34, 43, 44, 48) and protein-protein interactions with CRE-bound CREB (1, 22). The formation of this promoter-bound Tax-CREB complex (Tax/CREB) is critical for the recruitment of the multifunctional cellular coactivators CBP and p300 (19, 26, 32, 39, 65).
CBP and p300 are very large paralogous proteins that coordinate highly regulated gene expression in metazoans. Several conserved domains in CBP and p300 serve as binding sites for a wide variety of structurally unrelated cellular and viral transcription factors. These domains in CBP/p300 include two cysteine-histidine-rich domains (CH1 and CH3), the SRC-interacting domain, and the KIX domain (for recent reviews, see references 11 and 21). The transcription factor-coactivator interactions mediated by these domains facilitate recruitment of the coactivators to target promoters. Subsequent to recruitment by the transcription factors, CBP and p300 appear to stimulate transcription through multiple, distinct mechanisms. For example, there is evidence that CBP/p300 may directly stabilize components of the general transcription machinery at target promoters (13, 40). Additionally, CBP and p300 have intrinsic acetyltransferase activity that targets multiple lysine residues present on the four core histone tails (8, 51). A significant body of evidence suggests that histone tail acetylation weakens internucleosomal interactions, which may lead to a more relaxed chromatin structure, providing greater transcription factor or RNA polymerase access at target promoters (5, 17, 24). Additionally, combinations of differentially acetylated histone tails may serve as recognition sites for the assembly of factors that facilitate transitions from transcriptionally silent to transcriptionally active chromatin (30, 60). Although the evidence for histone acetylation by CBP/p300 is strong, the precise mechanistic events that couple transcriptional activation with acetylation of promoter-proximal nucleosomes are unknown.
The HTLV-1 Tax protein has previously been shown to bind at three of the four major transcription factor interaction domains in CBP/p300: CH1, KIX, and the carboxy-terminal SRC-interacting domain (19, 26, 39, 42, 58, 65). The interaction between Tax and KIX has been studied in the greatest detail and is the only Tax-CBP/p300 interaction that has been reported to occur with the Tax-containing, promoter-bound ternary complex (Tax/CREB/viral CRE DNA) (19, 26, 39, 65). This observation suggests that the ternary complex interaction with KIX may play a major role in CBP/p300 recruitment to the HTLV-1 promoter. However, little is known about the functional significance of this protein-protein interaction.
In this study, we explore the transcriptional relevance of the Tax-KIX interaction in Tax transactivation from chromatin-assembled templates in vitro. Using polypeptides specifically designed to inhibit CBP/p300 association with the Tax/CREB/viral CRE complex, we demonstrate that the KIX domain of CBP/p300 is critical for mediating coactivator function at the HTLV-1 promoter. The inhibition is specific to Tax transactivation on chromatin templates and, unexpectedly, is independent of the four core histone amino-terminal tails. We extend these studies to examine the contribution of the acetyltransferase function of the coactivators on Tax transactivation from the HTLV-1 promoter. We find that the CBP/p300-selective acetyltransferase inhibitor Lys-coenzyme A (CoA) inhibits Tax transactivation from chromatin templates to the same levels observed with the polypeptide inhibitors. These observations indicate that CBP and p300 play a prominent role in Tax transactivation from chromatin templates and that the acetyltransferase activity of the coactivators provides the major functional contribution to transcriptional activation in vitro. Interestingly, a strong CBP/p300-specific acetyltransferase requirementwas also observed on chromatin templates prepared from tailless histones. We identify multiple nonhistone acetylation targets, a subset of which are dependent upon Tax and CREB. Together, these data indicate that CBP and p300 participate in critical chromatin-specific, tail-independent acetylation events during transcriptional activation by Tax.
MATERIALS AND METHODS
Purification of recombinant proteins.
The four Xenopus laevis core histones, both as wild-type and amino-terminal deletion mutant proteins, were individually expressed in Escherichia coli and purified to homogeneity as previously described (46, 47). The amino acid coordinates of the histone deletion mutants are: H2A (Δ1-12 and Δ119-128), H2B (Δ1-23), H3 (Δ1-26), and H4 (Δ1-19). Drosophila NAP-1 (28), ISWI, and Acf1 (29) were expressed and purified as previously described (18). Full-length, recombinant CREB (16) and Tax (68) were expressed and purified as previously described (19). Six-histidine (His6)-tagged p300 was expressed and purified as previously described (37). Recombinant glutathione S-transferase (GST) and the GST fusion proteins GST-KIX (CBP amino acids [aa] 588 to 683) (19), GST-KIXmut (CBP aa 597 to 719) (65), and GST-SREBP-1a (SREBP-1 aa 1 to 50) (50) were expressed and purified as previously described.
Chromatin assembly and in vitro transcription.
Chromatin templates were assembled as previously described at core histone/DNA ratios that were empirically determined to give complete chromatin assembly (18). The 3.2-kbp p4TxRE/G-less plasmid DNA used in the transcription assays carried four tandem copies of the HTLV-1 promoter-proximal viral CRE cloned upstream of the HTLV-1 core promoter and drove the synthesis of a 380-nucleotide (nt) transcript (4). The p-52/G-less plasmid DNA carried positions −52 to +1 (relative to the transcription start site) of the natural HTLV-1 core promoter and drove the synthesis of a 190-nt transcript (4). This plasmid contained a TATA box but no Tax-responsive viral CRE sequences. Following chromatin assembly, 75 fmol of the chromatin-assembled template was incubated in the absence or presence of 80 nM purified recombinant Tax, 80 nM CREB, 3 nM p300, and 40 μg of CEM (HTLV-1-negative human T-lymphocyte) nuclear extract, as previously described (18, 43). For experiments using unassembled or naked DNA templates, samples were prepared as described above, but without the chromatin assembly step. All reaction mixtures contained 100 μM acetyl-CoA unless otherwise indicated. Molecular weight markers (radiolabeled HpaII-digested pBR322) were used to estimate the size of the RNA products.
Biotin-streptavidin DNA pull-down assay.
Double-stranded DNA fragments containing a single viral CRE (and biotinylated at the 5′-end of one strand, [Integrated DNA Technologies]) were annealed and bound to streptavidin-agarose (Novagen) in TM buffer (50 mM Tris, 100 mM KCl, 12.5 mM MgCl2, 1 mM EDTA, 20% glycerol, 0.1% Tween 20, 2 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF]). The HTLV-1 promoter-proximal viral CRE sequence used in this assay was modified to contain a consensus CRE site to enhance CREB binding. The sequence of the DNA fragment is 5′-GAAGATCTCTCAGGCGTTGACGTCAACCCCTCACCAGATCTT-3′. The core CRE region is indicated in boldface, and the conserved GC-rich flanks are underlined. The immobilized viral CRE DNA (2 pmol) was incubated with Tax (10 pmol), protein kinase A-phosphorylated CREB (10 pmol), p300 (10 pmol), GST-KIX or GST-KIXmut (5 or 10 pmol), and GST-SREBP-1a or GST (10 or 50 pmol) as indicated in the relevant figures. The reaction mixtures were incubated in binding buffer (25 mM Tris, 50 mM KCl, 6.25 mM MgCl2, 0.5 mM EDTA, 10% glycerol, 0.05% Tween 20, 50 mM NaCl, 2 mM DTT, 1 mM PMSF) for 1 h at 4°C, washed three times in binding buffer, resuspended in sodium dodecyl sulfate (SDS) sample dyes, boiled, and analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (5 to 20% polyacrylamide). Bound proteins were detected by Western blot analysis with a mixture of anti-GST (to detect GST-KIX, GST-KIXmut, GST, and GST-SREBP-1a; Sigma), anti-His6 (to detect Tax and p300; Santa Cruz Biotechnology), and anti-CREB (Santa Cruz Biotechnology) antibodies.
GST pull-down assay.
The GST pull-down assay was performed by incubating 10 pmol of GST or GST-SREBP-1a with 15 μl of swollen glutathione-agarose (Sigma) in 400 μl of 0.5× Superdex buffer (12.5 mM HEPES [pH 7.9], 6.25 mM MgCl2, 75 mM KCl, 5 μM ZnSO4, 0.5 mM EDTA, 10% glycerol, 0.05% NP-40) at 4°C for 2 h. The beads were then washed twice with 0.5× Superdex buffer and incubated with 2 pmol of p300 (in a total volume of 400 μl of 0.5× Superdex buffer) at 4°C for 18 h. After being washed twice with 0.5× Superdex, the beads were resuspended in SDS sample dyes, boiled, and analyzed by SDS-PAGE (5 to 20% polyacrylamide). Bound protein was detected by Western blot analysis with an antibody against p300 (N-15; Santa Cruz Biotechnology).
In vitro acetylation assay.
The p4TxRE/G-less plasmid template was assembled into chromatin by using wild-type Xenopus histones (2 μg), or tailless Xenopus histones (6 μg), NAP-1, and ACF as previously described (18). Chromatin assembly was performed at empirically determined histone/DNA mass ratios; for samples containing tailless chromatin, this resulted in a 30% larger amount of template than that in the wild-type chromatin samples. However, the amounts of recombinant proteins and nuclear extract that were subsequently added remained constant for all samples. p300 (12 nM) and [14C]acetyl-CoA (0.09 mM; 57 mCi/mmol) were added following chromatin assembly, together with 400 nM Tax and CREB where applicable, in a solution containing 50 mM Tris (pH 8), 10% glycerol, 10 mM sodium butyrate, 1 mM DTT, and 1 mM PMSF. Samples were incubated at 30°C for 60 min, and protein was precipitated by methanol-chloroform extraction and analyzed by SDS-PAGE (18% polyacrylamide). For the acetylation assays using purified chromatin templates, chromatin assembly was carried out as described above, and templates were preincubated for 60 min at 30°C with 12 nM p300, 400 nM Tax, 400 nM CREB, and 400 μg of CEM nuclear extract, where indicated. Following preincubation, samples were fractionated by Sephacryl 200 chromatography, and the void volume eluate, which contained the chromatin template (and associated proteins), was collected. The samples were then incubated with [14C]acetyl-CoA (0.09 mM; 57 mCi/mmol) as described above. Proteins were precipitated by methanol-chloroform extraction and analyzed by SDS-PAGE (5 to 20% polyacrylamide).
RESULTS
The KIX domain of CBP/p300 mediates coactivator function in Tax transactivation from chromatin templates.
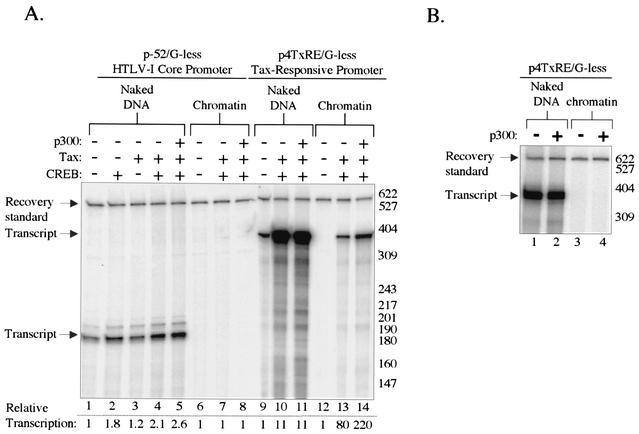
High-level transcription of the HTLV-1 genome requires the binding of the virally encoded Tax protein, in complex with the cellular transcription factor CREB, to the viral CRE promoter elements. This promoter-bound complex is believed to recruit the cellular coactivators CBP and p300 via interactions with the KIX domain. The physical interaction between the KIX domains of both CBP and p300 and the Tax/CREB complex has been well characterized (9, 19, 26, 39, 65). However, the precise functional relevance of this interaction in Tax transactivation remains unclear. We utilized a chromatin-based transcription system that includes recombinant core histones (2, 18) to characterize the mechanisms of Tax-activated transcription. Transcription reactions were performed in the presence of acetyl-CoA and nuclear extract prepared from CEM cells (HTLV-1-negative human T lymphocyte). We used the p4TxRE/G-less template, which carries four copies of the Tax-responsive viral CRE driving synthesis of a 380-nt guanine-less transcript (4). This template was chosen because the four viral CREs are the only known activator binding sites in the promoter region of the plasmid. Therefore, the transcriptional readout should be fully dependent upon the addition of Tax/CREB. This template was responsive to Tax/CREB, both as free DNA and assembled into recombinant chromatin (Fig. 1A, lanes 9, 10, 12, and 13). Exogenous p300 further activated Tax/CREB transcription from the p4TxRE/G-less template, in a chromatin-specific manner (Fig. 1A, compare lanes 10 and 11 with lanes 13 and 14). In contrast, a template containing only the HTLV-1 core promoter sequence (−52 to +1 relative to the transcription start site; designated p-52/G-less) and lacking the viral CREs was minimally responsive to Tax/CREB and Tax/CREB/p300 (Fig. 1A, lanes 1 to 5). Additionally, this core promoter template was unresponsive to Tax/CREB and Tax/CREB/p300 when assembled into chromatin (Fig. 1A, lanes 6 to 8). Importantly, p300 coactivator function required Tax/CREB, suggesting that p300 cannot function independently of promoter-bound activators in this system (Fig. 1B).
FIG. 1.
p300 coactivator function requires the viral CREs and Tax/CREB for HTLV-1 transcription. (A) The Tax-responsive viral CRE enhancer elements are required for transcriptional activation. The p-52/G-less and p4TxRE/G-less cassette templates were assayed both as free DNA and as DNA assembled into chromatin by using recombinant Xenopus histones. Transcription reaction mixtures contained CEM nuclear extract and purified recombinant Tax, CREB, Tax/CREB, or Tax/CREB/p300 as indicated. The recovery standard, size markers, and the positions of the full-length transcript are indicated. Relative transcription was calculated separately for each set of templates, with basal transcription set equal to 1. (B) p300 coactivator function requires Tax/CREB for HTLV-1 transcriptional activation. The p4TxRE/G-less cassette template was assayed as free DNA and as DNA assembled into chromatin by using recombinant Xenopus histones. Transcription reaction mixtures contained CEM nuclear extract and purified recombinant p300 as indicated.
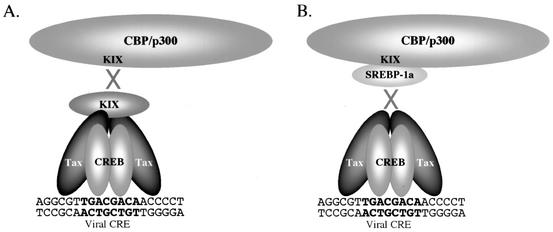
In this study, we were interested in evaluating the role of CBP/p300, and specifically the contribution of the KIX domain, in Tax-mediated transcriptional activation from chromatin templates in vitro. To biochemically characterize the role of KIX in CBP/p300 recruitment by the Tax/CREB complex, we expressed and purified two polypeptides designed to specifically block the physical interaction between promoter-bound Tax and CBP/p300. A schematic showing the relevant polypeptide interactions predicted to block coactivator recruitment is shown in Fig. 2. The first polypeptide, KIX, carries CBP aa 588 to 683. (CBP and p300 share 93% homology within the KIX domain.) This represents the minimal region of KIX required for high-affinity binding to Tax (65). We hypothesized that the binding of KIX to the Tax/CREB/viral CRE complex would block CBP/p300 recruitment to the HTLV-1 promoter (Fig. 2A). The second polypeptide, SREBP-1a, carries the first 50 aa of SREBP-1 (sterol regulatory element binding protein 1) and has previously been shown to bind to the KIX domain of CBP (50). We hypothesized that SREBP-1a would bind to the KIX domain of CBP/p300, thus preventing association of CBP/p300 with the Tax/CREB/viral CRE complex (Fig. 2B).
FIG. 2.
Strategies for the disruption of CBP/p300 recruitment to the Tax-responsive HTLV-1 promoter. (A) KIX binding to the promoter-bound Tax/CREB complex blocks the recruitment of full-length CBP/p300. (B) SREBP-1a binding to the KIX domain of full-length CBP/p300 prevents binding of the coactivators to the Tax/CREB/viral CRE DNA complex.
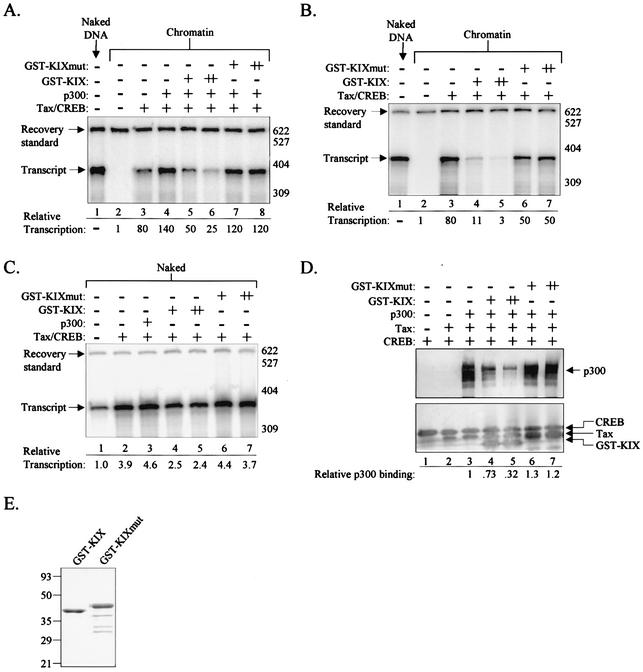
We first asked whether addition of the KIX domain to in vitro chromatin transcription reactions would inhibit Tax/CREB/p300-mediated transcriptional activation. Figure 3A shows that the addition of recombinant, purified Tax/CREB and Tax/CREB/p300 strongly activated HTLV-1 transcription (lanes 2 to 4). As hypothesized, the addition of purified GST-KIX to the transcription reaction mixtures significantly diminished Tax transactivation in a dose-dependent manner (Fig. 3A, lanes 4 to 6). As a negative control, we tested a KIX mutant (GST-KIXmut), carrying CBP aa 597 to 719. We have previously shown that deletion of aa 588 to 597 abolishes KIX interaction with Tax (65). The addition of GST-KIXmut to the transcription reaction mixtures had only a modest effect on Tax/CREB/p300 transcriptional activation (Fig. 3A, lanes 7 and 8).
FIG. 3.
The KIX peptide inhibits Tax-activated transcription on chromatin templates. (A) KIX inhibits Tax/CREB/p300-mediated transcription. The p4TxRE/G-less cassette template was assembled into chromatin by using recombinant Xenopus histones. Transcription reaction mixtures contained CEM nuclear extract and purified recombinant Tax/CREB or Tax/CREB/p300, as indicated. GST-KIX (aa 588 to 683) and GST-KIXmut (aa 597 to 719) were added to the transcription reaction mixtures at equimolar concentrations (+) or fivefold molar excess (++) relative to the Tax/CREB complex (see Materials and Methods). Recovery standard, size markers, and the position of the full-length transcript are indicated. (B) KIX inhibits Tax/CREB transcriptional activation in the absence of exogenous p300. Transcription reactions were performed as described in panel A, except in the absence of exogenous p300. (C) KIX does not affect Tax/CREB transcriptional activation on naked DNA templates. In vitro transcription reactions were performed on the unassembled p4TxRE/G-less template. Transcription reaction mixtures contained CEM nuclear extract nd purified Tax/CREB, Tax/CREB/p300, GST-KIX, or GST-KIXmut, as indicated. (D) KIX disrupts p300 binding to the Tax/CREB/viral CRE DNA complex. Binding reaction mixtures contained a DNA fragment carrying a single viral CRE linked to a streptavidin-agarose bead. CREB, Tax, and p300 were added to the immobilized viral CRE DNA as indicated. GST-KIX or GST-KIXmut was added at 0.5× molar (+) or equimolar (++) amounts relative to the Tax/CREB complex. Reactions were analyzed by Western blotting. For this experiment, Ser133-phosphorylated CREB was used. (E) Analysis of purified proteins used in the transcription and DNA binding reactions. Purified recombinant GST-KIX and GST-KIXmut were analyzed by SDS-PAGE and staining with Coomassie brilliant blue. The sizes of molecular mass markers (in kilodaltons) are indicated.
Because the addition of KIX reduced activated transcription to levels below that observed with Tax/CREB alone, we were interested in testing the effect of KIX on Tax/CREB-activated transcription in the absence of exogenous p300. Figure 3B shows that GST-KIX strongly inhibited transcription in the absence of the recombinant coactivator, suggesting that endogenous CBP/p300 present in the CEM nuclear extract participates in Tax-activated transcription from the chromatin template. Interestingly, the addition of GST-KIX or GST-KIXmut to non-chromatin, or naked, p4TxRE/G-less templates had minimal effect on Tax/CREB-activated transcription (Fig. 3C). These data are consistent with previous studies showing that p300 does not activate transcription on naked DNA (6, 18, 36, 38, 45) (Fig. 3C, lane 3).
To directly test whether KIX blocks p300 recruitment to Tax/CREB/viral CRE complexes, we performed a biotin-streptavidin-agarose DNA pull-down assay using an immobilized DNA fragment carrying a single viral CRE binding site for the Tax/CREB complex. We used a naked DNA fragment in this assay, since p300 has previously been shown to interact directly with chromatin (49). As shown in Fig. 3D, full-length p300 forms a quaternary complex with Tax and CREB on the viral CRE DNA (lane 3). Addition of GST-KIX to the binding reaction displaced p300 from the complex in a dose-dependent fashion (Fig. 3D, lanes 3 to 5). Displacement of p300 from the Tax/CREB/DNA complex likely occurred as a consequence of GST-KIX binding, because GST-KIX association with the DNA-bound fraction correlated directly with p300 dissociation (Fig. 3D, lanes 3 to 5). As expected, GST-KIXmut was unable to compete with p300 for binding to the Tax/CREB/DNA complex (Fig. 3D, lanes 6 and 7). The results of SDS-PAGE showing purified GST-KIX and GST-KIXmut used in these assays are presented in Fig. 3E. Together, these data support a model in which KIX inhibits Tax/CREB/p300-mediated transcription via disruption of CBP/p300 binding to the Tax/CREB/viral CRE DNA complex (Fig. 2A).
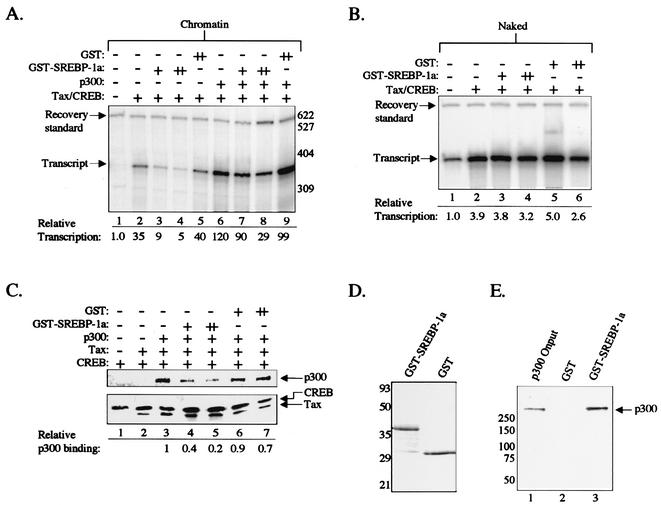
Although the KIX peptide repression data shown above are consistent with inhibition of CBP/p300 recruitment, it is also possible that the tight binding of KIX to the Tax/CREB complex simply blocks access of another coactivator or ancillary factor to the HTLV-1 promoter. To more specifically define a role for CBP/p300 in Tax transactivation, we next tested whether the addition of purified GST-SREBP-1a could similarly inhibit activated transcription from the chromatin-assembled HTLV-1 promoter. Since SREBP-1a binds directly to the CBP/p300 KIX domain, it is a highly specific inhibitor of KIX function (50). Figure 4A shows that addition of GST-SREBP-1a to the chromatin-based transcription reactions inhibited both Tax/CREB-activated transcription (lanes 2 to 4) and Tax/CREB/p300-activated transcription (lanes 6 to 8). GST-SREBP-1a-mediated transcriptional inhibition was weaker in the presence of exogenously added p300 (Fig. 4A, lanes 6 to 8), consistent with the idea that saturation of CBP/p300 KIX domain is required. Since SREBP-1a was expressed as a GST fusion, we used GST alone as a negative control. GST had no effect on Tax/CREB- or Tax/CREB/p300-activated transcription (Fig. 4A, lanes 5 and 9). Similar to our observation with the GST-KIX polypeptide, addition of GST-SREBP-1a or GST alone had minimal effect on Tax/CREB-activated transcription on naked templates (Fig. 4B). Finally, the addition of GST-SREBP-1a to immobilized biotin-streptavidin DNA pull-down reaction mixtures containing Tax, CREB, p300, and the viral CRE DNA inhibited p300 association with the Tax/CREB/DNA complex in a dose-dependent fashion (Fig. 4C). Unlike KIX, SREBP-1a did not replace p300 in the Tax/CREB/viral CRE DNA complex (data not shown), consistent with our model for SREBP-1a inhibition (Fig. 2B). The purified GST-SREBP-1a and GST used in these experiments are shown in Fig. 4D. The SREBP-1a used in these experiments was functional for binding to full-length p300, as shown by a GST pull-down assay (Fig. 4E).
FIG. 4.
The SREBP-1a peptide inhibits Tax-activated transcription on chromatin templates. (A) SREBP-1a inhibits Tax/CREB/p300-mediated transcription. Transcription reaction mixtures contained the chromatin-assembled p4TxRE-G-less template, CEM nuclear extract, and Tax/CREB or Tax/CREB/p300, as indicated. GST-SREBP-1a and GST alone were added to the transcription reactions at equimolar concentrations (+) or fivefold molar excess (++) relative to the Tax/CREB complex, as indicated. The recovery standard, size markers, and position of the full-length transcript are indicated. (B) SREBP-1a does not affect Tax/CREB transcriptional activation on naked DNA templates. Transcription reaction mixtures contained the unassembled p4TxRE-G-less template, CEM nuclear extract, Tax/CREB, SREBP-1a, or GST, as indicated. (C) SREBP inhibits p300 binding to the Tax/CREB/viral CRE DNA complex. Streptavidin-agarose DNA pull-down reactions were performed as described in Fig. 2D. GST-SREBP-1a or GST was added at equimolar concentrations (+) or fivefold molar excess (++) relative to the Tax/CREB complex, as indicated. Reactions were analyzed by Western blotting. (D) Analysis of purified proteins used in the transcription and DNA binding reactions. Purified recombinant GST-SREBP-1a and GST were analyzed by SDS-PAGE and staining with Coomassie brilliant blue. The sizes of molecular mass markers (in kilodaltons) are indicated. (E) GST-SREBP-1a interacts with p300. The GST pull-down assay contained purified recombinant p300 and GST or GST-SREBP-1a bound to glutathione-agarose, as indicated. p300 input (10%) is shown in lane 1. Reactions were analyzed by Western blotting.
Together, these data suggest that KIX and SREBP-1a similarly inhibit Tax transactivation by physically blocking coactivator access to the Tax/CREB complex bound to the HTLV-1 promoter. Furthermore, the strong transcriptional inhibition produced by KIX and SREBP-1a in the absence of exogenous p300 strongly suggests that Tax-activated transcription in a chromatin context in vitro is highly dependent upon CBP/p300 provided by the nuclear extract. Western blot analysis of the CEM nuclear extracts reveals the presence of approximately 0.7 pmol of CBP/p300 in each transcription reaction (data not shown). This level of CBP/p300 in the nuclear extract is likely sufficient for the strong Tax transactivation observed in these assays and may account for the relatively modest stimulation observed in the presence of exogenous p300.
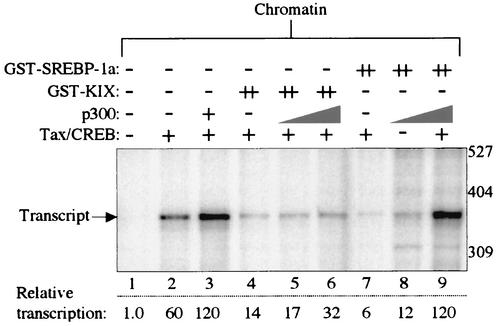
We next examined the specificity of KIX and SREBP-1a function by performing a p300 “add-back” experiment. We reasoned that if KIX and SREBP-1a inhibited transcription by blocking Tax/CREB access to the coactivator, then addition of exogenous p300 would overcome the observed effect. Figure 5 shows that the GST-KIX inhibition was modestly relieved by the addition of recombinant p300 (lanes 5 and 6). However, GST-SREBP-1a inhibition was fully reversed in the presence of the largest amount of added coactivator (Fig. 5, lane 9). This difference in the degree of transcriptional response to exogenous p300 in the presence of KIX and SREBP-1a is not unexpected, because the two polypeptides mediate transcriptional inhibition via distinct mechanisms (Fig. 2). It is possible that a slow off-rate of GST-KIX from the Tax/CREB complex inhibits access to the exogenous p300. Alternatively, SREBP-1a transcriptional inhibition likely occurs via direct stoichiometric binding to endogenous CBP/p300; therefore, the excess p300 should be available for recruitment to the Tax/CREB/viral CRE complex.
FIG. 5.
Exogenous p300 relieves KIX and SREBP-1a-mediated inhibition of Tax-activated transcription. Transcription reaction mixtures contained the chromatin-assembled p4TxRE-G-less template, CEM nuclear extract, and Tax/CREB, as indicated. GST-KIX and GST-SREBP-1a were added to the indicated transcription reaction mixtures at a fivefold molar excess (++) relative to the Tax/CREB complex. Recombinant p300 was added at 0.05 and 0.1 pmol. Size markers and the position of the full-length G-less transcript are indicated.
The CBP/p300-selective acetyltransferase inhibitor Lys-CoA inhibits Tax transactivation on chromatin templates.
The experiments presented above support a prominent role for CBP/p300 in Tax/CREB-activated transcription from Tax-responsive chromatin templates. Since these experiments reveal a significant contribution of endogenous CBP/p300 present in our transcription system, we were interested in selectively testing whether the acetyltransferase function of endogenous CBP/p300 was responsible for Tax/CREB transcriptional activation. We have previously observed that activation of Tax transcription by exogenous p300 required intact acetyltransferase activity, because a p300 acetyltransferase mutant was inactive for transcriptional stimulation relative to wild-type p300 (18). However, these experiments did not address the contribution of endogenous CBP/p300 acetyltransferases in mediating Tax function.
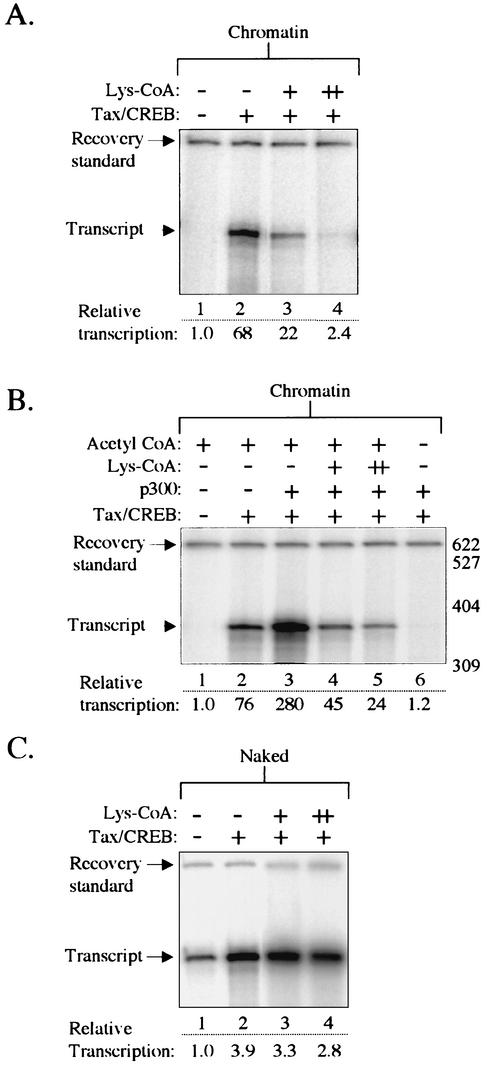
To specifically evaluate the contribution of endogenous acetyltransferase activity in our transcription system, we tested the effect of the selective CBP/p300 inhibitor Lys-CoA on Tax/CREB-activated transcription. Lys-CoA has previously been shown to inhibit p300 (and by extension CBP) both in vitro and in vivo, with a >100-fold selectivity for p300 relative to P/CAF (6, 38, 41, 45, 54). Figure 6A shows that the addition of 50 μM Lys-CoA to Tax-activated chromatin transcription reaction mixtures containing 10 μM acetyl-CoA reduced RNA synthesis by >90% (lane 4), consistent with a previous observation (46). The concentrations of Lys-CoA used in these experiments have previously been shown to selectively inhibit p300 acetyltransferase activity by >90% (41). We next tested Lys-CoA inhibition in the presence of Tax/CREB and exogenous p300 and found that Lys-CoA inhibited transcription to a degree similar to that observed with Tax/CREB alone (Fig. 6B, lanes 4 and 5). As expected, Tax/CREB/p300-activated transcription was essentially abolished in the absence of acetyl-CoA (Fig. 6B, lane 6). Lys-CoA had minimal effect on Tax/CREB-activated transcription from naked DNA templates (Fig. 6C).
FIG. 6.
The CBP/p300-selective HAT inhibitor Lys-CoA inhibits Tax-activated transcription on chromatin templates. (A) Transcription reaction mixtures contained the chromatin-assembled p4TxRE-G-less template, CEM nuclear extract, and Tax/CREB, as indicated. Lys-CoA was added to the transcription reaction mixtures at final concentrations of 10 μM (+) and 50 μM (++), as indicated. Acetyl-CoA was added to a final concentration of 10 μM in all samples. The recovery standard and the position of the full-length G-less transcript are indicated. (B) Lys-CoA inhibits Tax-activated transcription in the presence of exogenous p300. Transcription reactions were performed as described in panel A, except in the presence of exogenous p300, as indicated. Lane 6 shows Tax/CREB-activated transcription in the absence of acetyl-CoA. (C) Lys-CoA does not inhibit Tax-activated transcription on naked DNA templates. Transcription reactions were performed on the unassembled p4TxRE-G-less template. Transcriptional activation was analyzed in the presence of Tax/CREB and 10 μM (+) or 50 μM (++) Lys-CoA, as indicated.
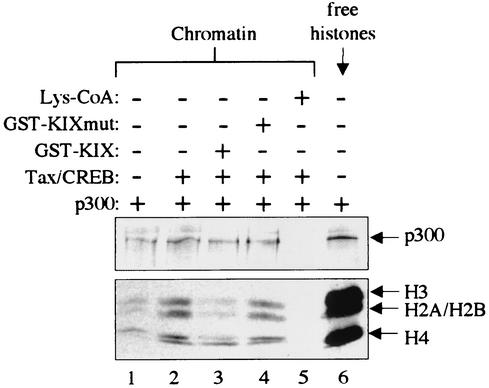
The Lys-CoA inhibition data strongly suggest that Tax/CREB transcriptional activation associated with CBP/p300 recruitment results primarily from the intrinsic acetyltransferase activity associated with the coactivator(s). If this is correct, then it would follow that the transcriptional inhibition observed in the presence of the KIX and SREBP-1a polypeptides results from an inhibition of targeted p300 histone acetyltransferase (HAT) activity. To directly test this idea, we performed an in vitro HAT assay in the presence of Tax/CREB and KIX. Figure 7 shows that, as expected, Tax/CREB addition increased the p300-dependent acetylation of histones assembled on the Tax-responsive p4TxRE/G-less template (lanes 1 and 2). However, Tax/CREB did not increase p300-dependent acetylation of histones assembled on the p-52/G-less plasmid template lacking the Tax-responsive viral CRE enhancer elements (data not shown). The addition of GST-KIX reduced histone acetylation to levels observed in the absence of added activators, presumably through inhibition of p300 recruitment (Fig. 7, lane 3). This inhibition was specific to chromatin, because free histone acetylation and p300 autoacetylation were unaffected by GST-KIX (Fig. 7, lane 3, and data not shown). GST-KIXmut, which is defective for Tax/CREB binding, had minimal effect on p300 HAT activity (Fig. 7, lane 4). As expected, 50 μM Lys-CoA completely inhibited p300 acetyltransferase activity (Fig. 7, lane 5). Together, these data indicate that the primary mechanism of CBP/p300 transcriptional coactivation on the HTLV-1 promoter is through the acetyltransferase activity of the coactivators.
FIG. 7.
KIX disrupts the recruitment of p300 HAT activity to chromatin templates in a Tax-dependent manner. Both chromatin-assembled templates and free histones were acetylated in vitro in the presence of p300 and [14C]acetyl-CoA. Tax/CREB, GST-KIX, and GST-KIXmut were added to the indicated acetylation reaction mixtures. The CBP/p300-specific HAT inhibitor Lys-CoA was added to a final concentration of 50 μM, as indicated. GST-KIX and GST-KIXmut were each added at a fivefold molar excess relative to the Tax/CREB complex.
Histone tail deletion reveals an unknown target for CBP/p300 acetyltransferase function in Tax transactivation.
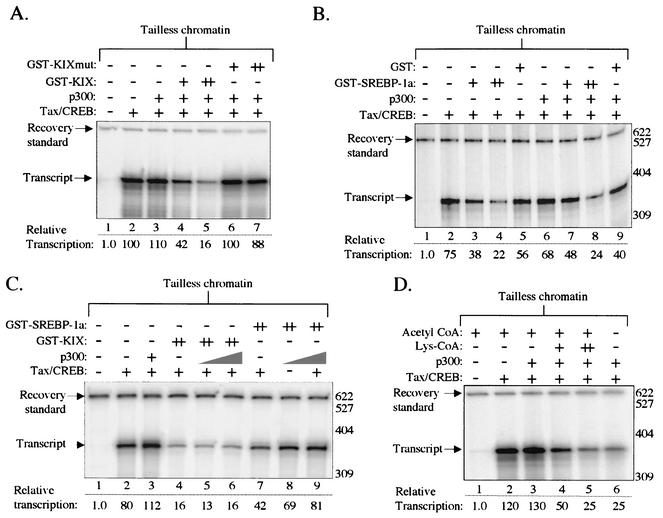
The data presented thus far support a role for the KIX domain in Tax/CREB recruitment of CBP/p300 to the HTLV-1 promoter. We also show data indicating that CBP and p300 cooperate with Tax to activate transcription of the p4TxRE/G-less chromatin template via their intrinsic acetyltransferase activity. Since the histone tails are established targets of coactivator acetylation during transcriptional activation, we reasoned that deletion of the histone tails would render the transcription system insensitive to CBP/p300 inhibition. To test this hypothesis, we used “tailless” histones carrying amino-terminal deletions (H2A Δ1-12 and Δ119-128, H2B Δ1-23, H3 Δ1-26, and H4 Δ1-19) to assemble chromatin on the p4TxRE/G-less template as previously described (18). All of the previously identified targets of CBP/p300 acetylation are deleted in our tailless histones (57). We tested whether chromatin composed of tailless histones was sensitive to KIX/SREBP-1a inhibition of Tax/CREB-mediated transcriptional activation. As shown before, nucleosomal templates formed with tailless histones were strongly activated by Tax/CREB and insensitive to exogenous p300 (Fig. 8A, lanes 1 to 3) (18). Unexpectedly, the addition of GST-KIX, but not GST-KIXmut, inhibited Tax/CREB transcriptional activation over 80% on the tailless templates (Fig. 8A, lanes 4 to 7). Similar transcriptional inhibition was observed with GST-SREBP-1a, in both the presence and absence of exogenous p300 (Fig. 8B). These results were surprising, because the tailless histones carry deletions of the regions encompassing the previously characterized sites of acetylation. We confirmed the specificity of the effects of KIX and SREBP-1a on tailless chromatin by performing a p300 “add-back” experiment (Fig. 8C). Consistent with the results for wild-type chromatin templates (Fig. 5), we found that inhibition by GST-KIX is highly stable and is not responsive to exogenous p300, while inhibition by GST-SREBP-1a was reversed by exogenous p300. To determine if KIX- and SREBP-1a-mediated transcriptional inhibition was associated with disrupted recruitment of the acetyltransferase activity of CBP/p300, we tested the effect of Lys-CoA on Tax-activated transcription from the tailless chromatin templates. Figure 8D shows that 50 μM Lys-CoA inhibited transcription by up to 80%. Interestingly, this effect was indistinguishable from the level of inhibition observed in the absence of acetyl-CoA, suggesting that acetyl-CoA is utilized exclusively by CBP/p300 on these templates. Together, these data strongly support a role for CBP/p300 acetyltransferase activity in Tax/CREB-dependent activation of transcription from tailless nucleosomal templates.
FIG. 8.
KIX, SREBP-1a, and Lys-CoA inhibit Tax-activated transcription on chromatin templates lacking histone amino-terminal tails. Transcription reaction mixtures contained the p4TxRE-G-less template assembled into chromatin by using tailless histones, CEM nuclear extract, and Tax/CREB, as indicated. The recovery standard and full-length G-less transcripts are indicated. (A) The KIX polypeptide specifically inhibits Tax/CREB transcription on tailless chromatin. GST-KIX or GST-KIXmut was added at an equimolar concentration (+) or fivefold molar excess (++) relative to the Tax/CREB complex, as indicated. (B) SREBP-1a specifically inhibits Tax/CREB transcription on tailless chromatin. GST-SREBP-1a and GST were added at equimolar concentration (+) or fivefold molar excess (++) relative to the Tax/CREB complex, as indicated. (C) Exogenous p300 relieves KIX and SREBP-1a-mediated inhibition of Tax/CREB transcription on tailless chromatin. GST-KIX and GST-SREBP-1a were added to the indicated transcription reaction mixtures at fivefold molar excess (++) relative to the Tax/CREB complex. Recombinant p300 was added at 0.05 and 0.1 pmol. (D) Lys-CoA inhibits Tax/CREB transcription on tailless chromatin. For this experiment, acetyl-CoA was added to a final concentration of 10 μM in all samples. Lys-CoA was added to samples at final concentrations of 10 μM (+) and 50 μM (++).
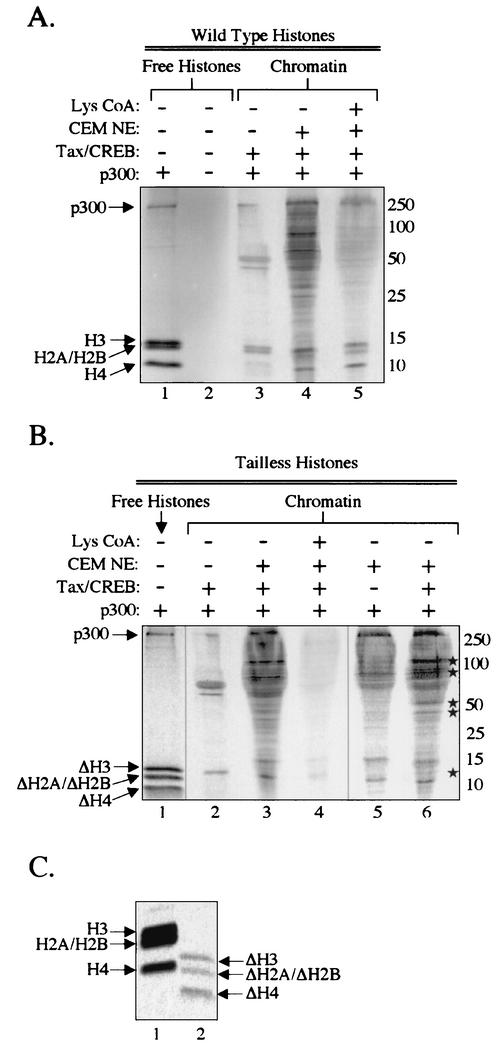
The CBP/p300 acetyltransferase inhibitor Lys-CoA strongly inhibited transcription from both wild-type and tailless chromatin templates, but had a minimal effect on transcription from naked DNA templates. These data suggest that chromatin-related acetylation substrates, distinct from the histone tails, are targeted by CBP/p300 to promote transcriptional activation from the HTLV-1 promoter in vitro. Many previous studies have identified a wide variety of non-histone proteins that are acetylated by CBP/p300 (for review, see reference 59). To determine what proteins might be acetylated during Tax-activated transcription, we performed in vitro acetylation assays on isolated transcription complexes formed on the p4TxRE/G-less chromatin template. First, we prepared chromatin templates with full-length histones and incubated them with Tax, CREB, p300, and CEM nuclear extract. These template-bound complexes were then separated from unbound proteins by gel filtration chromatography. Following incubation with [14C]acetyl-CoA, we examined the acetylation state of the proteins associated with the chromatin-assembled templates. Figure 9A shows that in addition to the four core histones, many proteins present in the CEM nuclear extract used in our transcriptional studies were acetylated in the presence of p300 (lane 4). Acetylation of these proteins was highly dependent on the acetyltransferase activity of CBP/p300, because the addition of 50 μM Lys-CoA strongly reduced the appearance of labeled bands (Fig. 9A, lane 5). This is the same concentration of Lys-CoA that strongly inhibited Tax transactivation from chromatin templates (Fig. 6B). Interestingly, core histone acetylation was only modestly affected by the addition of Lys-CoA, suggesting a role for HATs other than CBP/p300. These data correlate the acetylation of numerous proteins by CBP/p300 with the transcriptional output of the HTLV-1 promoter. As a control for acetylation events that occur in the absence of nuclear extract, we performed p300 acetylation assays on chromatin-assembled templates that were incubated with only Tax and CREB. Figure 9A reveals the presence of several prominently labeled bands that appear to correspond to the core histones H3 and H2A/2B, Tax, CREB, p300, and the chromatin assembly protein NAP-1 (lane 3). The acetylation of NAP-1 by p300 in vitro has been reported previously (7). As an additional control, we show that free full-length histones are acetylated in a p300-dependent manner (Fig. 9A, lanes 1 and 2). These data suggest that in addition to many unidentified proteins in the nuclear extract, Tax and/or CREB may also be targets for CBP/p300 acetylation.
FIG. 9.
Histone tail-independent acetylation of nuclear extract proteins by CBP/p300. Chromatin templates were assembled with NAP-1, ACF, and wild-type or tailless histones, as indicated. These templates were preincubated with Tax, CREB, p300, and CEM nuclear extract (as indicated), and preinitiation complexes were isolated by gel filtration chromatography. The samples were incubated with [14C]acetyl-CoA, and analyzed by SDS-PAGE (5 to 20% polyacrylamide). The sizes of molecular mass markers (in kilodaltons) are indicated. (A) CBP/p300-dependent acetylation of proteins associated with wild-type chromatin templates. Free histones (0.5 μg) were assayed in the absence and presence of p300 as a positive control for acetylation (lanes 1 and 2). Chromatin-assembled templates were assayed in the absence or presence of CEM nuclear extract, as indicated (lanes 3 to 5). Lys-CoA was added to a final concentration of 50 μM (lane 5). (B) CBP/p300-dependent acetylation of proteins associated with tailless chromatin templates. Free histones (2 μg) were assayed as a positive control for acetylation (lane 1). Chromatin-assembled templates were assayed in the absence or presence of CEM nuclear extract and/or Tax/CREB, as indicated (lanes 2 to 6). Lys-CoA was added to a final concentration of 50 μM (lane 4). The Tax/CREB-dependent acetylated protein bands are indicated (*). For this experiment, we used a longer exposure of the image to provide band intensities comparable to those in panel A. (C) Tailless histones are weakly acetylated by p300. Equivalent amounts (2 μg) of wild-type and tailless free histones were incubated in the presence of p300.
We next examined the acetylation state of the proteins present in transcription complexes formed on chromatin templates assembled with the tailless histones. These templates were prepared and analyzed as described above for Fig. 9A. Similar to our observations using templates assembled with full-length histones, we found that many proteins present in preinitiation complexes formed on the tailless chromatin templates were also acetylated and that their acetylation was CBP/p300 dependent (Fig. 9B, lanes 3, 4, and 6). Although we observed acetylation of tailless histone H3 on chromatin templates prepared in the absence of nuclear extract (Fig. 9B, lane 2), it was unclear whether tailless H3 or any of the other tailless histones were acetylated in the presence of the extract (Fig. 9B, lanes 3, 5, and 6). This result was not unexpected, because all four tailless histones were only weakly acetylated in a p300-dependent manner (Fig. 9C and data not shown), in agreement with previous observations (3, 18). We were next interested in determining whether the pattern of CBP/p300 acetylation was Tax/CREB dependent. Figure 9B shows that a subset of CEM nuclear extract proteins appeared to exhibit Tax/CREB-dependent acetylation (lanes 5 and 6). It is not known whether this was due to enhanced acetylation via Tax/CREB tethering of p300 to the template or to enhanced recruitment of specific factors to the template following Tax/CREB binding. Together, the observations presented above support the idea that CBP/p300 acetyltransferase activity is directed at multiple targets, in addition to the histone amino-terminal tails. Furthermore, these data correlate specific non-histone acetylation events with transcriptional activation by Tax.
DISCUSSION
Several previous studies have characterized a physical interaction between the KIX domain of CBP/p300 and the viral transcriptional activator protein Tax (19, 26, 39, 65). Here, we provide the first direct evidence that KIX-mediated Tax recruitment of CBP/p300 promotes strong Tax transactivation from chromatin templates. Using specific polypeptide inhibitors of the Tax-KIX interaction (GST-KIX and GST-SREBP-1a), we find that Tax recruitment of CBP/p300 present in the nuclear extract is a critical step in mediating Tax transactivation in a chromatin context, accounting for >80% of the observed Tax stimulation in this system. The SREBP-1a inhibition is counteracted by the addition of recombinant p300 to the transcription reactions, providing strong evidence that CBP and p300 are the primary endogenous coactivators that mediate Tax transactivation. The effect of the coactivators is dependent on chromatin, because Tax transactivation from unassembled (naked) templates is refractory to the stimulatory effects of acetyl-CoA and p300 and is not inhibited by the polypeptides.
We also investigated the specific role of the intrinsic acetyltransferase activity of CBP/p300 in Tax transactivation. As previously shown, we find that Tax enhances nucleosomal histone acetylation by recruiting p300 to chromatin templates (18, 45). We find that the addition of KIX peptide dramatically inhibits this effect, directly coupling Tax recruitment of CBP/p300, via the KIX domain, with nucleosome acetylation and transcriptional activation. We find that the p300-selective inhibitor Lys-CoA significantly blocks Tax-activated transcription, reducing transcription to the same levels observed with the KIX-specific polypeptide inhibitors. This observation is consistent with a prominent role for a CBP/p300 acetyltransferase activity in transcriptional initiation, as previously suggested (36, 38). Additionally, a recent study has shown that a p300 acetyltransferase mutant was unable to activate transcription from the integrated HTLV-1 promoter in vivo (52). These data provide further evidence that CBP and p300 are the prominent coactivators mediating Tax transcription function and, importantly, that the acetyltransferase activity intrinsic to CBP/p300 provides the dominant transcriptional stimulatory activity associated with coactivator recruitment to the Tax-responsive promoter.
Although we observe residual Tax transactivation in the presence of Lys-CoA, transcription is abolished in the absence of acetyl-CoA. While this may represent incomplete Lys-CoA inhibition of endogenous CBP/p300, it is also possible that acetyltransferases unrelated to CBP/p300 participate in Tax transactivation from chromatin templates. This observation is not surprising, because a broad range of coactivators and ancillary factors utilize acetyl-CoA to activate transcription (59). These additional coactivators may include proteins that acetylate lysine residues on the core histone amino-terminal tails that are distinct from those lysines targeted by CBP/p300. These proteins may also include acetyltransferases that cooperate with CBP/p300. For example, lysine 14 of histone H3 is acetylated not only by CBP/p300, but also by GCN5, P/CAF, and TAFII250 (59). Of these acetyltransferases, P/CAF has been shown to interact with Tax in both vitro and in vivo and to activate HTLV-1 transcription in transient transfection assays (25, 31). It is possible that P/CAF cooperates with CBP/p300 in histone tail acetylation and thus participates in Tax transactivation of HTLV-1 transcription.
We previously reported that the amino-terminal histone tails are required for Tax transactivation in response to exogenously added p300 and that tail deletion rendered the chromatin templates insensitive to recombinant p300 addition, while remaining moderately sensitive to acetyl-CoA (18). In the previous study, however, the role of endogenous CBP/p300 in Tax transactivation was not addressed. In the present study, we make the unexpected observation that the acetyltransferase activity required for Tax transactivation on the tailless chromatin templates is contributed by CBP/p300 present in the nuclear extract. We observe potent inhibition of Tax-activated transcription from the tailless chromatin templates with both Lys-CoA and the polypeptide inhibitors. Interestingly, Lys-CoA inhibits transcription to the same level as that observed in the absence of acetyl-CoA. These data indicate that CBP/p300 play a prominent and perhaps exclusive role in Tax transactivation from tailless chromatin templates and that the acetyltransferase activity provides the coactivator function in this context.
It seems paradoxical that tailless chromatin templates require CBP/p300 acetyltransferase activity and yet are unresponsive to exogenous p300. However, responsiveness to exogenous p300 may be a function of the number of potential acetylation sites present in the transcription system. For example, previous characterization of CBP/p300 suggests that wild-type chromatin templates contain up to 22 CBP/p300 acetylation targets per nucleosome, in addition to other potential CBP/p300 acetyltransferase targets present in the nuclear extract (57). Although we have found a substantial concentration of endogenous CBP/p300 in our CEM nuclear extract, it appears that the endogenous CBP/p300 may be limiting. Therefore, Tax transactivation from wild-type chromatin templates is enhanced by exogenous p300. However, in the absence of histone tails, the number of potential CBP/p300 acetylation substrates present in the transcription system is dramatically reduced. Therefore, endogenous CBP/p300 acetyltransferase activity may be saturating for strong Tax transactivation from tailless chromatin templates; consequently, exogenous p300 has no effect. Consistent with the idea that a reduced number of acetylation substrates leads to a reduced requirement for acetyltransferase enzyme, we have previously observed that tailless chromatin templates have a reduced requirement for acetyl-CoA in Tax transactivation compared with that of wild-type chromatin templates (18; data herein).
In vitro acetylation assays of transcription complexes formed on tailless chromatin templates revealed the presence of CBP/p300-specific acetylation targets that are histone tail independent. These unidentified acetylation targets may be components of the general transcription machinery and/or chromatin remodeling factors that are acetylated to facilitate negotiation of chromatin templates during transcriptional activation. In recent years, the collective work of many groups has produced an impressive list of non-histone CBP/p300 substrates. These include HMG proteins, the general transcription factors TFIIE and TFIIF, various transcriptional activators, and proteins involved in nuclear import (reviewed in reference 59). Our data also suggest that Tax, and/or CREB, may be acetylated by CBP/p300. Whether these transcriptional activators are bona fide acetylation substrates for CBP/p300 and whether acetylation has any effect on their function is not known. The role of these known and unknown CBP/p300 acetylation substrates in Tax transactivation remains to be elucidated.
Another intriguing possibility is that the nucleosomal core is itself acetylated by CBP/p300. Recent studies indicate that histones can be modified at lysine residues in regions other than the well-characterized amino-terminal tails (14, 56, 61, 63). In the experiments presented herein, we used a histone H3 deletion mutant that carried a lysine residue at the amino-terminal end of the protein (corresponding to aa 27). Although lysine 27 may be acetylated in yeast (33), it is not one of the previously identified CBP/p300 acetylation targets (57). It is a formal possibility that this lysine residue is acetylated by CBP/p300, because we observe tailless H3 acetylation on chromatin templates. However, when these templates were incubated with nuclear extract, the acetylated H3 band was not readily discernible. It is therefore equivocal whether acetylation within the globular domain of histone H3 plays a role in transcriptional activation.
While our observation of strong transcriptional activation from chromatin templates assembled with tailless histones is in contrast to a recently published study (2), it is consistent with a number of previous studies showing that tailless chromatin templates are more accessible to binding by transcriptional activators (20, 53, 64) and are generally more transcriptionally active than intact chromatin templates (12, 27, 55, 64). This may reflect the ability of tailless chromatin to adopt a less-condensed structure than intact chromatin (10, 15, 35, 62; reviewed in reference 23). The fact that both our wild-type and tailless chromatin templates respond similarly to Tax transactivation suggests that histone acetylation and histone tail deletion are functionally equivalent at the HTLV-1 promoter.
In summary, the data presented in this study demonstrate a prominent and perhaps exclusive role for the KIX domain in CBP/p300 recruitment by the HTLV-1 Tax protein. We find that Tax transactivation from chromatin templates is largely dependent upon CBP/p300 present in the nuclear extract and that the acetyltransferase activity is the prominent functional property of the coactivators responsible for mediating transcriptional activation. By specifically inhibiting CBP/p300 enzymatic activity in transcription from tailless chromatin templates, we have discovered a mode of CBP/p300 transcriptional regulatory function that is acetyl-CoA dependent and histone tail independent. We find that CBP/p300 acetyltransferase activity may not be directed at the histone tails, but instead at undefined targets whose acetylation is critical to the activation of viral gene expression in a chromatin context. Our data provide insight into a novel mechanism of HTLV-1 transcriptional regulation by CBP/p300 that may have broad implications for CBP/p300 function.
Acknowledgments
We thank Anders Näär and Robert Tjian for the gift of SREBP-1a, Jes Kuruvilla and Raji Edayathumangalam for histone octamers, and Jeanne Mick for purified Tax and CREB. We especially thank W. Lee Kraus for very helpful discussions that led to the initiation of this project.
This study was supported by National Institutes of Health grants CA87540 to J.K.N. and P.J.L. and GM62437 to P.A.C.
REFERENCES
- 1.Adya, N., and C.-Z. Giam. 1995. Distinct regions in human T-cell lymphotropic virus type I Tax mediate interactions with activator protein CREB and basal transcription factors. J. Virol. 69:1834-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, W., V. B. Palhan, M. A. Karymov, S. H. Leuba, and R. G. Roeder. 2002. Selective requirements for histone H3 and H4 N termini in p300-dependent transcriptional activation from chromatin. Mol. Cell 9:811-821. [DOI] [PubMed] [Google Scholar]
- 3.An, W., and R. G. Roeder. 2003. Direct association of p300 with unmodified H3 and H4 N termini modulates p300-dependent acetylation and transcription of nucleosomal templates. J. Biol. Chem. 278:1504-1510. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, M. G., K. E. S. Scoggin, C. M. Simbulan-Rosenthal, and J. A. Steadman. 2000. Identification of poly(ADP-ribose) polymerase as a transcriptional coactivator of the human T-cell leukemia virus type 1 Tax protein. J. Virol. 74:2169-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Annunziato, A. T., and J. C. Hansen. 2000. Role of histone acetylation in the assembly and modulation of chromatin structures. Gene Expr. 9:37-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asahara, H., B. Santoso, E. Guzman, K. Du, P. A. Cole, I. Davidson, and M. Montminy. 2001. Chromatin-dependent cooperativity between constitutive and inducible activation domains in CREB. Mol. Cell. Biol. 21:7892-7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asahara, H., S. Tartare-Deckert, T. Nakagawa, T. Ikehara, F. Hirose, T. Hunter, T. Ito, and M. Montminy. 2002. Dual roles of p300 in chromatin assembly and transcriptional activation in cooperation with nucleosome assembly protein 1 in vitro. Mol. Cell. Biol. 22:2974-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641-643. [DOI] [PubMed] [Google Scholar]
- 9.Bex, F., M.-J. Yin, A. Burny, and R. B. Gaynor. 1998. Differential transcriptional activation by human T-cell leukemia virus type 1 Tax mutants is mediated by distinct interactions with CREB binding protein and p300. Mol. Cell. Biol. 18:2392-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carruthers, L. M., and J. C. Hansen. 2000. The core histone N termini function independently of linker histones during chromatin condensation. J. Biol. Chem. 275:37285-37290. [DOI] [PubMed] [Google Scholar]
- 11.Chan, H. M., and N. B. La Thangue. 2001. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114:2363-2373. [DOI] [PubMed] [Google Scholar]
- 12.Chirinos, M., F. Hernandez, and E. Palacian. 1998. Repressive effect on oligonucleosome transcription of the core histone tail domains. Biochemistry 37:7251-7259. [DOI] [PubMed] [Google Scholar]
- 13.Dallas, P. B., P. Yaciuk, and E. Moran. 1997. Characterization of monoclonal antibodies raised against p300: both p300 and CBP are present in intracellular TBP complexes. J. Virol. 71:1726-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dover, J., J. Schneider, M. A. Boateng, A. Wood, K. Dean, M. Johnston, and A. Shilatifard. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by RAD6. J. Biol. Chem. 17:28368-28371. [DOI] [PubMed] [Google Scholar]
- 15.Fletcher, T. M., and J. C. Hansen. 1995. Core histone tail domains mediate oligonucleosome folding and nucleosomal DNA organization through distinct molecular mechanisms. J. Biol. Chem. 270:25359-25362. [DOI] [PubMed] [Google Scholar]
- 16.Franklin, A. A., M. F. Kubik, M. N. Uittenbogaard, A. Brauweiler, P. Utaisincharoen, M. A. Matthews, W. S. Dynan, J. P. Hoeffler, and J. K. Nyborg. 1993. Transactivation by the human T-cell leukemia virus Tax protein is mediated through enhanced binding of activating transcription factor-2 (ATF-2), ATF-2 response and cAMP element-binding protein (CREB). J. Biol. Chem. 268:21225-21231. [PubMed] [Google Scholar]
- 17.Garcia-Ramirez, M., C. Rocchini, and J. Ausio. 1995. Modulation of chromatin folding by histone acetylation. J. Biol. Chem. 270:17923-17928. [DOI] [PubMed] [Google Scholar]
- 18.Georges, S. A., W. L. Kraus, K. Luger, J. K. Nyborg, and P. J. Laybourn. 2002. p300-mediated Tax transactivation from recombinant chromatin: histone tail deletion mimics coactivator function. Mol. Cell. Biol. 22:127-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giebler, H. A., J. E. Loring, K. van Orden, M. A. Colgin, J. E. Garrus, K. W. Escudero, A. Brauweiler, and J. K. Nyborg. 1997. Anchoring of CREB binding protein to the human T-cell leukemia virus type 1 promoter: a molecular mechanism of Tax transactivation. Mol. Cell. Biol. 17:5156-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godde, J. S., Y. Nakatani, and A. P. Wolffe. 1995. The amino-terminal tails of the core histones and the translational position of the TATA box determine TBP/TFIIA association with nucleosomal DNA. Nucleic Acids Res. 23:4557-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 22.Goren, I., O. J. Semmes, K.-T. Jeang, and K. Moelling. 1995. The amino terminus of Tax is required for interaction with the cyclic AMP response element binding protein. J. Virol. 69:5806-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen, J. C. 2002. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 31:361-392. [DOI] [PubMed] [Google Scholar]
- 24.Hansen, J. C., C. Tse, and A. P. Wolffe. 1998. Structure and function of the core histone N-termini: more than meets the eye. Biochemistry 37:17637-17641. [DOI] [PubMed] [Google Scholar]
- 25.Harrod, R., Y. L. Kuo, Y. Tang, Y. Yao, A. Vassilev, Y. Nakatani, and C. Z. Giam. 2000. p300 and p300/cAMP-responsive element-binding protein associated factor interact with human T-cell lymphotropic virus type-1 Tax in a multi-histone acetyltransferase/activator-enhancer complex. J. Biol. Chem. 275:11852-11857. [DOI] [PubMed] [Google Scholar]
- 26.Harrod, R., Y. Tang, C. Nicot, H. S. Lu, A. Vassilev, Y. Nakatani, and C.-Z. Giam. 1998. An exposed KID-like domain in human T-cell lymphotropic virus type 1 Tax is responsible for the recruitment of coactivators CBP/p300. Mol. Cell. Biol. 18:5052-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernandez, F., L. Lopez-Alarcon, C. Puerta, and E. Palacian. 1998. Transcriptional inhibitory role of the tail domains of histone (H3 × H4)2 tetramers. Arch. Biochem. Biophys. 358:98-103. [DOI] [PubMed] [Google Scholar]
- 28.Ito, T., M. Bulger, R. Kobayashi, and J. T. Kadonaga. 1996. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol. Cell. Biol. 16:3112-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito, T., M. E. Levenstein, D. V. Fyodorov, A. K. Kutach, R. Kobayashi, and J. T. Kadonaga. 1999. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 13:1529-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 31.Jiang, H., H. Lu, R. L. Schiltz, C. A. Pise-Masison, V. V. Ogryzko, Y. Nakatani, and J. N. Brady. 1999. PCAF interacts with Tax and stimulates Tax transactivation in a histone acetyltransferase-independent manner. Mol. Cell. Biol. 19:8136-8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kashanchi, F., J. F. Duvall, R. P. Kwok, J. R. Lundblad, R. H. Goodman, and J. N. Brady. 1998. The coactivator CBP stimulates human T-cell lymphotrophic virus type I Tax transactivation in vitro. J. Biol. Chem. 273:34646-34652. [DOI] [PubMed] [Google Scholar]
- 33.Kelly, T. J., S. Qin, D. E. Gottschling, and M. R. Parthun. 2000. Type B histone acetyltransferase Hat1p participates in telomeric silencing. Mol. Cell. Biol. 20:7051-7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimzey, A. L., and W. S. Dynan. 1998. Specific regions of contact between human T-cell leukemia virus type I Tax protein and DNA identified by photocross-linking. J. Biol. Chem. 273:13768-13775. [DOI] [PubMed] [Google Scholar]
- 35.Krajewski, W. A., and J. Ausio. 1996. Modulation of the higher-order folding of chromatin by deletion of histone H3 and H4 terminal domains. Biochem. J. 316:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraus, W. L., and J. T. Kadonaga. 1998. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 12:331-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kraus, W. L., E. T. Manning, and J. T. Kadonaga. 1999. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol. 19:8123-8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kundu, T. K., V. B. Palhan, Z. Wang, W. An, P. A. Cole, and R. G. Roeder. 2000. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell 6:551-561. [DOI] [PubMed] [Google Scholar]
- 39.Kwok, R. P., M. E. Laurance, J. R. Lundblad, P. S. Goldman, H. Shih, L. M. Connor, S. J. Marriott, and R. H. Goodman. 1996. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature 380:642-646. [DOI] [PubMed] [Google Scholar]
- 40.Kwok, R. P., J. R. Lundblad, J. C. Chrivia, J. P. Richards, H. P. Bachinger, R. G. Brennan, S. G. Roberts, M. R. Green, and R. H. Goodman. 1994. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 370:223-226. [DOI] [PubMed] [Google Scholar]
- 41.Lau, O. D., T. K. Kundu, R. E. Soccio, S. Ait-Si-Ali, E. M. Khalil, A. Vassilev, A. P. Wolffe, Y. Nakatani, R. G. Roeder, and P. A. Cole. 2000. HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol. Cell 5:589-595. [DOI] [PubMed] [Google Scholar]
- 42.Lemasson, I., and J. K. Nyborg. 2001. Human T-cell leukemia virus type I Tax repression of p73beta is mediated through competition for the C/H1 domain of CBP. J. Biol. Chem. 276:15720-15727. [DOI] [PubMed] [Google Scholar]
- 43.Lenzmeier, B. A., E. E. Baird, P. B. Dervan, and J. K. Nyborg. 1999. The tax protein-DNA interaction is essential for HTLV-I transactivation in vitro. J. Mol. Biol. 291:731-744. [DOI] [PubMed] [Google Scholar]
- 44.Lenzmeier, B. A., H. A. Giebler, and J. K. Nyborg. 1998. Human T-cell leukemia virus type 1 Tax requires direct access to DNA for recruitment of CREB binding protein to the viral promoter. Mol. Cell. Biol. 18:721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu, H., C. A. Pise-Masison, T. M. Fletcher, R. L. Schiltz, A. K. Nagaich, M. Radonovich, G. Hager, P. A. Cole, and J. N. Brady. 2002. Acetylation of nucleosomal histones by p300 facilitates transcription from Tax-responsive human T-cell leukemia virus type 1 chromatin template. Mol. Cell. Biol. 22:4450-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luger, K., T. J. Rechsteiner, A. J. Flaus, M. M. Waye, and T. J. Richmond. 1997. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 272:301-311. [DOI] [PubMed] [Google Scholar]
- 47.Luger, K., T. J. Rechsteiner, and T. J. Richmond. 1999. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304:3-19. [DOI] [PubMed] [Google Scholar]
- 48.Lundblad, J. R., R. P. Kwok, M. E. Laurance, M. S. Huang, J. P. Richards, R. G. Brennan, and R. H. Goodman. 1998. The human T-cell leukemia virus-1 transcriptional activator Tax enhances cAMP-responsive element-binding protein (CREB) binding activity through interactions with the DNA minor groove. J. Biol. Chem. 273:19251-19259. [DOI] [PubMed] [Google Scholar]
- 49.Manning, E. T., T. Ikehara, T. Ito, J. T. Kadonaga, and W. L. Kraus. 2001. p300 forms a stable, template-committed complex with chromatin: role for the bromodomain. Mol. Cell. Biol. 21:3876-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naar, A. M., P. A. Beaurang, K. M. Robinson, J. D. Oliner, D. Avizonis, S. Scheek, J. Zwicker, J. T. Kadonaga, and R. Tjian. 1998. Chromatin, TAFs, and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes Dev. 12:3020-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 52.Okada, M., and K.-T. Jeang. 2002. Differential requirements for activation of integrated and transiently transfected human T-cell leukemia virus type 1 long terminal repeat. J. Virol. 76:12564-12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Polach, K. J., P. T. Lowary, and J. Widom. 2000. Effects of core histone tail domains on the equilibrium constants for dynamic DNA site accessibility in nucleosomes. J. Mol. Biol. 298:211-223. [DOI] [PubMed] [Google Scholar]
- 54.Polesskaya, A., I. Naguibneva, L. Fritsch, A. Duquet, S. Ait-Si-Ali, P. Robin, A. Vervisch, L. L. Pritchard, P. Cole, and A. Harel-Bellan. 2001. CBP/p300 and muscle differentiation: no HAT, no muscle. EMBO J. 20:6816-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Protacio, R. U., G. Li, P. T. Lowary, and J. Widom. 2000. Effects of histone tail domains on the rate of transcriptional elongation through a nucleosome. Mol. Cell. Biol. 20:8866-8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robzyk, K., J. Recht, and M. A. Osley. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287:501-504. [DOI] [PubMed] [Google Scholar]
- 57.Schiltz, R. L., C. A. Mizzen, A. Vassilev, R. G. Cook, C. D. Allis, and Y. Nakatani. 1999. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 274:1189-1192. [DOI] [PubMed] [Google Scholar]
- 58.Scoggin, K. E. S., A. Ulloa, and J. K. Nyborg. 2001. The oncoprotein Tax binds the SRC-1-interacting domain of CBP/p300 to mediate transcriptional activation. Mol. Cell. Biol. 21:5520-5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 61.Sun, Z. W., and C. D. Allis. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104-108. [DOI] [PubMed] [Google Scholar]
- 62.Tse, C., and J. C. Hansen. 1997. Hybrid trypsinized nucleosomal arrays: identification of multiple functional roles of the H2A/H2B and H3/H4 N-termini in chromatin fiber compaction. Biochemistry 36:11381-11388. [DOI] [PubMed] [Google Scholar]
- 63.van Leeuwen, F., P. R. Gafken, and D. E. Gottschling. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109:745-756. [DOI] [PubMed] [Google Scholar]
- 64.Vitolo, J. M., C. Thiriet, and J. J. Hayes. 2000. The H3-H4 N-terminal tail domains are the primary mediators of transcription factor IIIA access to 5S DNA within a nucleosome. Mol. Cell. Biol. 20:2167-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan, J. P., J. E. Garrus, H. A. Giebler, L. A. Stargell, and J. K. Nyborg. 1998. Molecular interactions between the coactivator CBP and the human T-cell leukemia virus Tax protein. J. Mol. Biol. 281:395-400. [DOI] [PubMed] [Google Scholar]
- 66.Yoshida, M. 2001. Multiple viral strategies of HTLV-1 for dysregulation of cell growth control. Annu. Rev. Immunol. 19:475-496. [DOI] [PubMed] [Google Scholar]
- 67.Zhao, L. J., and C. Z. Giam. 1992. Human T-cell lymphotropic virus type I (HTLV-I) transcriptional activator, Tax, enhances CREB binding to HTLV-I 21-base-pair repeats by protein-protein interaction. Proc. Natl. Acad. Sci. USA 89:7070-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao, L. J., and C. Z. Giam. 1991. Interaction of the human T-cell lymphotrophic virus type I (HTLV-I) transcriptional activator Tax with cellular factors that bind specifically to the 21-base-pair repeats in the HTLV-I enhancer. Proc. Natl. Acad. Sci. USA 88:11445-11449. [DOI] [PMC free article] [PubMed] [Google Scholar]