Abstract
Activating signal cointegrator 2 (ASC-2), a cancer-amplified transcriptional coactivator of nuclear receptors and many other transcription factors, contains two LXXLL-type nuclear receptor interaction domains. Interestingly, the second LXXLL motif is highly specific to the liver X receptors (LXRs). In cotransfection, DN2, an ASC-2 fragment encompassing this motif, exerts a potent dominant-negative effect on transactivation by LXRs, which is rescued by ectopic coexpression of the full-length ASC-2 but not by other LXXLL-type coactivators, such as SRC-1 and TRAP220. In contrast, DN2/m, in which the LXXLL motif is mutated to LXXAA to abolish the interactions with LXRs, is without any effect. Accordingly, expression of DN2, but not DN2/m, in transgenic mice results in phenotypes that are highly homologous to those previously observed with LXRα−/− mice, including a rapid accumulation of large amounts of cholesterol and down-regulation of the known lipid-metabolizing target genes of LXRα in the liver upon being fed a high-cholesterol diet. These results identify ASC-2 as a physiologically important transcriptional coactivator of LXRs and demonstrate its pivotal role in the liver lipid metabolism.
The nuclear receptor superfamily is a group of proteins that regulate, in a ligand-dependent manner, transcriptional initiation of target genes by binding to specific DNA sequences called hormone response elements (31). Genetic studies have indicated that transcription coactivators without specific DNA-binding activity are essential for transcriptional activation, which led to the identification of many proteins interacting with the C-terminal ligand-dependent transactivation domain of nuclear receptors (23, 33, 40). These coactivators, including the p160 family, CBP/p300, p/CAF, TRAP/DRIP, and many others, bridge transcription factors and the basal transcription apparatus and/or remodel the chromatin structures.
Activating signal cointegrator 2 (ASC-2), also called AIB3, TRBP, RAP250, NRC, and PRIP, is a recently isolated transcriptional coactivator molecule which is gene amplified and overexpressed in human cancers and stimulates transactivation by nuclear receptors, AP-1, NF-κB, SRF, and numerous other transcription factors (5, 19, 24-26, 30, 46, 58). In particular, single-cell microinjection results with ASC-2 antibody have demonstrated that endogenous ASC-2 is required for transactivation by nuclear receptors and AP-1 (24, 25). More recently, ASC-2 was found to exist in a 2-MDa steady-state complex which also contains histone H3-lysine 4-specific methyltransferase enzymes (12). Interestingly, ASC-2 contains two nuclear receptor interaction domains (26), both of which are dependent on the integrity of their core LXXLL sequences (14, 48). The C-terminal LXXLL motif specifically interacts with liver X receptor α (LXRα) and LXRβ, whereas the N-terminal motif binds a broad range of nuclear receptors (26).
LXRα (NR1H3) and LXRβ (NR1H2) are known to control cholesterol and fatty acid metabolism. LXRα is expressed mainly in the liver, whereas LXRβ is ubiquitously expressed (47, 53). The physiological ligands for LXRs are oxysterols, and the most potent ones are 22(R)-hydroxycholesterol, 24(S)-hydroxycholesterol, and 24(S),25-epoxycholesterol (27). Activation of LXR in macrophages results in increased expression of genes encoding the ATP-binding cassette (ABC) cholesterol transporters ABCA1 (9, 37, 42, 50) and ABCG1 (21, 51) and apolipoprotein E (21) that are involved with cholesterol efflux from macrophages toward high-density lipoproteins. In the liver, LXR is involved in transcriptional control of Cyp7A1, encoding a critical enzyme in the conversion of cholesterol into bile acids (27, 35), as well as ABCG5/ABCG8 (3, 38), encoding ABC transporters implicated in biliary cholesterol excretion. Induction of intestinal ABCA1, ABCG5, and ABCG8 expression upon LXR activation accelerates fecal cholesterol disposal by reducing the efficiency of cholesterol absorption (37). LXR has also been reported to control genes that encode proteins involved in de novo lipogenesis. In particular, induced transcription has been reported for the gene encoding SREBP-1c (39, 41, 56), a transcription factor that regulates the expression of various lipogenic genes, including those encoding acetyl-coenzyme A (CoA) carboxylase and fatty acid synthase (15). In addition, LXR is known to directly influence the transcription of genes encoding fatty acid synthase (17), lipoprotein lipase (57), cholesterol ester transfer protein (29), and stearoyl-CoA desaturase 1 (28).
Gene-targeting approaches to elucidate the roles of many coactivators in mice have often been hampered by early embryonic lethality or functional redundancy. In particular, deletion of the ASC-2 gene also resulted in early embryonic lethality (20, 59). As an alternative approach, we have recently expressed a dominant-negative fragment of ASC-2 encompassing the N-terminal LXXLL motif (i.e., DN1) in mice, which inhibited recruitment of the endogenous ASC-2 to nuclear receptors. These DN1-transgenic (TG) mice exhibited a plethora of developmental and phenotypic abnormalities, including problems with the eye and heart, motor activities, and fat metabolism in the liver (18). Importantly, these mice were significantly compromised in the ability to respond to exogenous ligands, including retinoids (reference 18 and our unpublished results).
In this study, we took a similar approach and established transgenic mice expressing DN2, a dominant-negative fragment of ASC-2 that encompasses the LXR-specific second LXXLL motif and potently represses transactivation by LXRs in cotransfections. Accordingly, these DN2-TG mice exhibited phenotypes that are highly homologous to those previously observed with LXRα−/− mice (35). Together with the DN1-TG mouse results (18), these results strongly suggest that ASC-2 is a physiologically pivotal transcriptional coactivator protein of LXRs and other nuclear receptors in vivo.
MATERIALS AND METHODS
Generation of transgenic mice.
For construction of transgenic mice, hemagglutinin (HA)-tagged DN2 and DN2/m (in which the LXXLL motif is mutated to LXXAA to abolish the interactions with LXRs) were cloned into a mammalian expression vector, pCAGGS (34), containing the chicken β-actin promoter linked to a human cytomegalovirus (CMV) immediate-early enhancer. These plasmids were microinjected into fertilized mouse eggs of the C57BL/6 strain. The genotype was determined by PCR and Southern analysis of genomic DNA from tail biopsies. The primers used for genotyping were 5′-CCGCTCGAGATGGCCTCCTACCCTTAT-3′ and 5′-GAAGATCTTCATGTAAGCCCAGGGGG-3′. Three and two lines of independent transgenic founders expressing DN2 and DN2/m, respectively, in various tissues were obtained, as demonstrated for transgenic expression by Western blot analysis or immunohistochemistry with anti-HA (αHA) antibody.
Transfections.
HeLa and CV-1 cells were grown in 24-well plates with medium supplemented with 10% fetal calf serum for 24 h and transfected with 100 ng of the lacZ expression vector pRSV-β-gal and 100 ng of LXRE-LUC reporter gene, along with appropriate amounts of mammalian expression vectors for LXRα, ASC-2, DN2, DN2/m, SRC-1, and TRAP220. The total amounts of expression vectors were kept constant by adding pcDNA3. Transfections and luciferase assays were done as described previously (24), and the results were normalized to lacZ expression. Similar results were obtained in more than two similar experiments.
GST pull-down and yeast two-hybrid assays.
Glutathione S-transferase (GST) fusions or GST alone was expressed in Escherichia coli, bound to glutathione-Sepharose 4B beads (Pharmacia, Piscataway, N.J.) in binding buffer (25 mM HEPES [pH 7.8], 0.2 mM EDTA, 20% glycerol, 100 mM KCl, and 0.1% NP-40), and incubated with labeled proteins expressed by in vitro translation by using the TNT-coupled transcription-translation system (Promega, Madison, Wis.), with conditions as described by the manufacturer. Specifically bound proteins were eluted from beads with 40 mM reduced glutathione in 50 mM Tris (pH 8.0) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography as described previously (24). Yeast two-hybrid assays were done as described previously (24). A yeast expression vector encoding LexA fused to the full-length LXRα was obtained from Young-Chul Lee at Chonnam National University. Yeast expression vectors encoding LexA-LXRβ and B42-DN2 were as previously described (26).
Chromatin immunoprecipitation.
293T cells transfected with DN2 or DN2/m expression vector were treated with 5 μM T0901317 for 40 min. Soluble chromatin from these cells was prepared and immunoprecipitated with appropriate antibodies, as recently described (44). The final DNA extractions were amplified using pairs of primers that encompass the LXR-responsive element of the SREBP-1c promoter region and generate a 320-bp PCR product. The primers used were 5′-AAGGGCCAGGAGTGGGTAAAC-3′ and 5′-CGCGCCGCGCCCCATTAGG-3′.
Histological examination and immunohistochemistry.
Organs were excised, frozen, sectioned in 10-μm-thick slices, and stained with hematoxylin and eosin and oil red O, as described previously (43). The organs examined were the eyes, liver, kidney, heart, lung, spleen, stomach, brain, pituitary gland, gall bladder, and adrenal gland from mouse embryos and postnatal animals.
Cholesterol and triglyceride measurement.
Plasma from euthanized mice was prepared using standard centrifugation techniques and enzymatically analyzed for cholesterol and triglyceride as previously described (16). Hepatic lipids were extracted from 0.2 g of liver and analyzed for triglyceride and cholesterol as described previously (4, 55).
DNA microarrays.
The mouse cDNA microarrays consisted of 8,000 cDNAs, including mouse clones from Incyte Genomics, Inc. (Palo Alto, Calif.), as well as housekeeping genes; tissue-specific genes; positive, negative, ratio, and sensitivity controls; and other controls. The PCR-amplified cDNAs were printed on CMT-GAPS II silane slide glass (Corning) and processed according to the manufacturer's protocol. Total RNA extracted from the treated and untreated livers of wild-type and DN2-TG mice was reverse transcribed to fluorescence-labeled cDNA probes using SuperScript II reverse transcriptase (Gibco BRL, Life Technologies, Grand Island, N.Y.) and Cy3- or Cy5-labeled dCTP (NEN Life Science Products Inc). The Cy3- and Cy5-labeled cDNAs were placed on the slide and hybridized in a hybridization chamber. The hybridized slides were scanned with the Axon Instruments (Union City, Calif.) GenePix 4000B. Finally, the scanned images were analyzed with GenePix Pro 3.0 (Axon) and GeneSpring (Silicon Genetics, Redwood City, Calif.). The signals of GAPDH and β-actin housekeeping genes were used for normalization.
RESULTS
DN2 as a specific dominant-negative mutant of LXR.
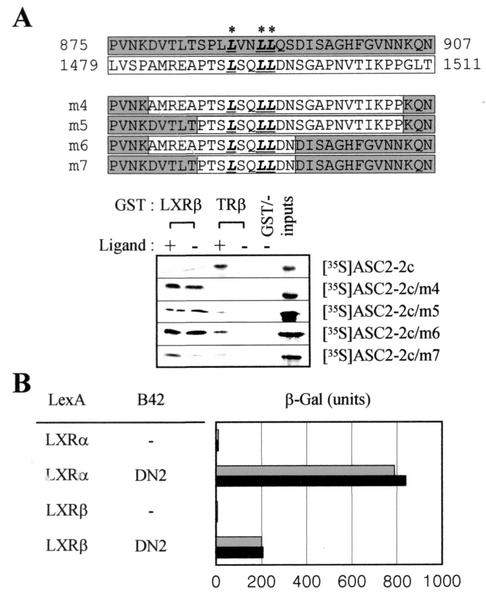
The second LXXLL motif of ASC-2 has been characterized as a specific interaction interface with LXRs (26). In an effort to identify important residues for this specificity, we constructed a series of chimeric proteins that incorporate various residues of the second LXXLL motif into ASC2-2c. Radiolabeled ASC2-2c itself, which consists of the ASC-2 residues 849 to 1057, containing the first LXXLL motif, bound to GST protein fused to TRβ but not LXRβ in a ligand-dependent manner, was previously described (26) (Fig. 1A). However, introduction of as little as 10 residues surrounding the second LXXLL motif was sufficient to change the binding specificity of the resulting chimeric protein (i.e., ASC2-2c/m7) to bind GST-LXRβ (Fig. 1A). Consistent with these results, specific residues within this 10-amino-acid region in ASC2-2c/m7 were identified as the residues most critical for LXR binding, as demonstrated by our recent random mutagenic analysis of an ∼100-amino-acid region surrounding the second LXXLL motif of ASC-2 (our unpublished results). It is interesting that the sequences of this second LXXLL motif were unique, and we could not find any similar motif in different protein databases. Similarly, DN2, previously called ASC2-4LR (26), which encodes the ASC-2 residues 1431 to 1511 and contains the second LXXLL motif, interacted with LXRα (Fig. 1B).
FIG. 1.
The second LXXLL motif specifically binds to LXRα and LXRβ. (A) A series of chimeric proteins containing amino acids of the second LXXLL motif region within the context of ASC2-2c (i.e., the ASC-2 residues 849 to 1057) were constructed using two-step PCR procedures and synthesized as labeled proteins expressed by in vitro translation using the TNT-coupled transcription-translation system, with conditions as described by the manufacturer. These proteins were incubated with GST fusion to TRβ and LXRβ in the absence and presence of either 0.1 μM T3 or 10 μM T0901317, as previously reported (20). Twenty percent of the total reaction mixture was loaded as input. The ASC-2 residues 875 to 907 (shaded) and 1479 to 1511 (open), containing the first and second LXXLL motif, respectively, are shown. (B) The indicated B42 and LexA plasmids were transformed into a yeast strain containing an appropriate lacZ reporter gene, as described previously (24). The shaded and solid bars indicate the absence and presence of 1 μM of 22(R)-hydroxycholesterol, respectively. Better ligand responsiveness was observed with higher doses of 22(R)-hydroxycholesterol (data not shown). −, B42 alone.
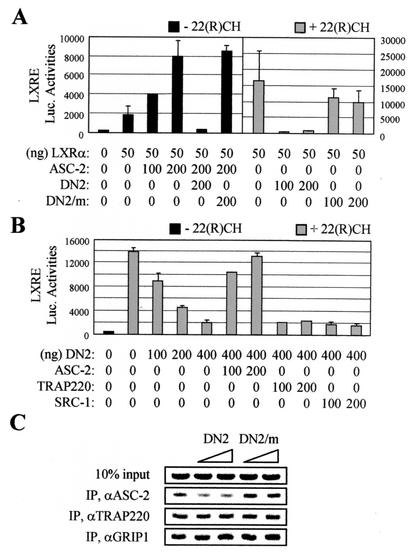
It was previously reported that DN2 represses transactivation by LXRβ (26). Similarly, DN2 was a potent dominant-negative mutant of LXRα in cotransfections in both the absence and presence of its ligand, 22(R)-hydroxycholesterol (Fig. 2A). In contrast, DN2/m, in which the LXXLL motif was mutated to LXXAA, had no effect. It should be noted that DN2 is inert with transactivation by other nuclear receptors that do not bind this second LXXLL motif, including thyroid hormone receptors and retinoic acid receptor α (data not shown). Importantly, the inhibition was rescued by coexpressed ASC-2 but not by two well-characterized LXXLL-type coactivators, SRC-1 and TRAP220 (Fig. 2B). These results were further confirmed in chromatin immunoprecipitation assays with 293T cells cotransfected with vectors encoding DN2 and DN2/m. As shown in Fig. 2C, the LXRE region of SREBP-1c was occupied by ASC-2, TRAP220, and GRIP1. DN2 specifically inhibited the recruitment of ASC-2 without interfering with the recruitment of TRAP220 and GRIP1. In contrast, DN2/m had no significant effect on the recruitment of any of these factors. Consistent with the reported relatively higher basal activities of LXRs under these conditions (47, 53), similar results were also obtained in the absence of exogenous ligand (data not shown). The exact mechanism responsible for this specificity is not clear. Nonetheless, these results demonstrate that DN2 can serve as an excellent tool to study the LXR-specific function of ASC-2, i.e., DN2 specifically inhibits LXR transactivation by blocking the recruitment of endogenous ASC-2 to LXRs bound to the target promoters.
FIG. 2.
DN2 as a specific dominant-negative repressor of LXR transactivation. (A and B) An oxysterol-responsive LXRE-LUC reporter construct was cotransfected into HeLa cells, along with lacZ expression vector (100 ng) and expression vectors for ASC-2, DN2 (i.e., the ASC-2 residues 1431 to 1511), DN2/m, TRAP220, and SRC-1, as indicated. The solid and shaded bars indicate the absence and presence of 10 μM 22(R)-hydroxycholesterol, respectively. Normalized luciferase expressions from triplicate samples were calculated relative to the lacZ expression. Similar results were obtained with CV-1 cells (data not shown). The error bars indicate standard deviations. (C) ASC-2 recruitment to the LXRE region of SREBP-1c. 293T cells were cotransfected with expression vectors for DN2 (50 and 100 ng) and DN2/m (50 and 100 ng) in the presence of 5 μM T0901317, as indicated. Chromatin from these cells was isolated and immunoprecipitated (IP) by the indicated antibodies. The endogenous SREBP-1c-LXRE region present in the immunoprecipitated samples was amplified by PCR, and the input PCR (10%) is shown for loading controls. Similar results were also obtained in the absence of ligand (data not shown).
Construction of DN2-TG mice and their phenotypic analysis.
Based on the results presented above, we hypothesized that any phenotype in DN2-expressing transgenic mice not observed with DN2/m-expressing mice should reflect the in vivo function of ASC-2 specific to LXRs. Thus, we exploited the ubiquitously active β-actin promoter to obtain three independent transgenic founder mouse lines (designated DN2-TG) that express HA-tagged DN2 in various tissues examined, as well as two HA-DN2/m-expressing founder lines (designated DN2/m-TG). Interestingly, DN2-TG mice did not show any apparent phenotype, in contrast to a variety of pathological anomalies recently reported with DN1-TG mice (18) (Table 1). However, it should be noted that this lack of phenotype was consistent with the specificity of DN2 to LXRs and the absence of any apparent developmental function ascribed to LXRs (47, 53).
TABLE 1.
Phenotypes exhibited by DN1 and DN2-TG
| Abnormality | Presence in transgenic line no.a
|
|||||||
|---|---|---|---|---|---|---|---|---|
| DN1
|
DN2
|
|||||||
| 54 | 71 | 84 | 87 | 104 | 4 | 18 | 32 | |
| Eye | ||||||||
| Embryonic eye | ||||||||
| Microphthalmia | + | − | + | NA | − | − | − | − |
| Retinal dysplasia | + | − | + | NA | − | − | − | − |
| Sclera locally thinner or missing | + | − | + | NA | − | − | − | − |
| Retrolenticular membrane | + | − | + | NA | − | − | − | − |
| PHPVb | + | − | + | NA | − | − | − | − |
| Cataracts | + | − | + | NA | − | − | − | − |
| Adult eye | ||||||||
| Posterior lenticonus | + | − | + | NA | − | − | − | − |
| Cataracts | + | + | + | + | + | − | − | − |
| Microphthalmia | + | + | + | + | + | − | − | − |
| Retinal dysplasia | + | − | + | NA | − | − | − | − |
| Sclera locally thinner or missing | + | − | + | NA | − | − | − | − |
| Retrolenticular membrane | + | − | + | NA | − | − | − | − |
| PHPV | + | − | + | NA | − | − | − | − |
| Heart | ||||||||
| Embryonic heart | ||||||||
| Ventricular septal defects | ND | ND | + | ND | ND | ND | ND | ND |
| Adult heart | ||||||||
| Atrial thrombosis | − | + | − | + | + | − | − | − |
| Hypertrophy | − | + | − | + | + | − | − | − |
| Conduction defects | − | + | − | ND | + | − | − | − |
| Respiratory tract | ND | ND | ND | |||||
| Lung hypoplasia | − | + | − | + | + | |||
| Lung congestion | − | + | − | + | + | |||
| Lung inflammation | − | + | − | + | + | |||
| Liver | ||||||||
| Fatty liver on normal diet | − | + | − | + | + | − | − | − |
| Fatty liver with high-cholesterol diet | ND | ND | ND | ND | ND | + | + | + |
| Thymic atrophy (agenesis) | − | + | − | + | + | − | − | − |
| Spleen atrophy | − | + | − | + | + | − | − | − |
| Gall bladder hydrops | − | + | − | + | + | − | − | − |
| Kidney hypoplasia | − | + | − | + | + | − | − | − |
| Pituitary gland redness | − | + | − | + | + | ND | ND | ND |
| Adrenal gland redness | − | + | − | + | + | − | − | − |
| Motor defects | + | ND | + | ND | ND | ND | ND | ND |
+, present; −, absent; NA, not applicable; ND, not determined.
PHPV, persistent hyperproliferation of primary vitreous.
Impaired cholesterol metabolism with DN2-TG mice on a high-cholesterol diet.
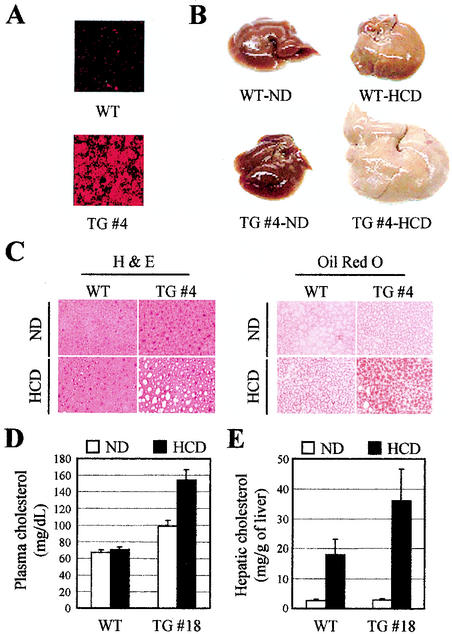
As the expected lack of apparent phenotypes was confirmed, we next explored the status of the known signaling pathways of LXRs in DN2-TG mice. Among the three major physiological targets of LXRs with regard to cholesterol and fatty acid metabolism (i.e., the liver, macrophages, and the intestine), we decided to focus on the detailed phenotypic analysis of the liver in this report. The results with macrophages and intestines will be described elsewhere. First, the expressions of DN2 and DN2/m in the livers of DN2-TG and DN2/m-TG mice, respectively, were confirmed by indirect immunofluorescence assays (Fig. 3A and data not shown). It should be noted that mice lacking LXRα were previously shown to lose the ability to respond normally to dietary cholesterol and are unable to tolerate any amount of cholesterol in excess of that which they synthesize de novo (29). Thus, we tested the effect of a diet rich in cholesterol (chow supplemented with 2% cholesterol). Remarkably, there were dramatic morphological, histological, and chemical changes in the livers of DN2-TG mice fed the same diet. Within 7 days of beginning the 2% cholesterol diet, and chronically worsening over a 90-day period, there were prominent color and size changes in the DN2-TG livers (Fig. 3B). Consistent with the known ability of wild-type animals to adapt quickly to cholesterol-rich diets, the livers of wild-type animals maintained relatively normal appearance and function. As expected, at least two independent DN2/m-TG lines were phenotypically indistinguishable from wild-type mice, allowing us to use both DN2/m-TG and wild-type mice as controls. Histological examination of the DN2-TG mouse livers revealed the presence of a time-dependent increase in the number and size of intracellular vacuoles, characteristic of lipid deposits (Fig. 3C, left). Oil red O staining of these liver sections verified the deposition of lipid (Fig. 3C, right). Plasma cholesterol levels rose significantly (>200%) in DN2-TG mice over the course of the 90-day high-cholesterol diet, while wild-type mice characteristically showed no change (Fig. 3D). Chemical analysis showed that the accumulated lipid within the livers of DN2-TG mice is in the form of cholesterol (Fig. 3E). In wild-type animals, there were relatively fewer significant changes in liver histology, size, and hepatic cholesterol levels (Fig. 3B, C, and E and data not shown). Overall, these results support the importance of ASC-2 as a specific coactivator of LXRs in vivo.
FIG. 3.
Morphology and histology of livers from DN2-TG (TG) and wild-type (WT) mice on high-cholesterol diets. (A) Expression of DN2 was assessed with indirect immunofluorescence with αHA antibody. TG #4 indicates the DN2-TG line no. 4 (Table 1). (B) Gross morphology of livers from male DN2-TG and wild-type mice fed chow supplemented with 2% cholesterol (HCD, high-cholesterol diet) for 90 days. ND, normal chow diet. The development of fatty livers in the DN2-TG mice is evident after 7 days on the high-cholesterol diet. (C) Sections from livers shown in panel B were prepared for histology and stained with oil red O, as indicated. The unstained vacuoles visible in the hematoxylin and eosin (H & E)-stained sections of the livers from DN2-TG mice on the high-cholesterol diet stain positive (red) for lipids with oil red O. (D and E) Measurements of plasma and hepatic cholesterol quantitated enzymatically from extracts of the livers shown in panel B. All values are expressed as means plus standard errors of the mean; n = 5.
Impaired de novo lipid synthesis in the livers of DN2-TG mice.
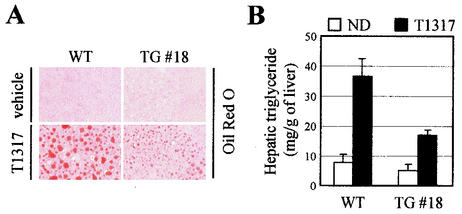
LXR has been reported to control genes that encode proteins involved in de novo lipogenesis. These include SREBP-1c (15, 39, 41, 56), fatty acid synthase (17), lipoprotein lipase (57), cholesterol ester transfer protein (29), and stearoyl-CoA desaturase-1 (28). Furthermore, it was recently shown that induction of these lipogenic genes with the synthetic LXR ligand T0901317 in mice is associated with an increased secretion of very low density lipoprotein-triglyceride and massive hepatic steatosis along the entire liver lobule (13). Consistent with these results, DN2-TG mice fed T0901317 showed reduced amounts of lipid accumulation in the liver relative to wild-type mice (Fig. 4A). Similarly, examination of hepatic triglyceride levels revealed a significant T0901317-dependent increase with wild-type mice but not with DN2-TG mice (Fig. 4B). These results further strengthen the importance of ASC-2 as a specific coactivator of LXRs in vivo.
FIG. 4.
Impaired lipogenesis in DN2-TG mice. (A) Liver sections from wild-type (WT) and DN2-TG (TG) mice treated orally with vehicle or 50 mg of T0901317/kg for 6 days were prepared for histology and stained with oil red O, as indicated. #18, DN2-TG line no. 18. (B) Measurements of hepatic triglyceride quantitated enzymatically from extracts of the livers shown in panel A. All values are expressed as means plus standard errors of the mean; n = 5.
A subset of LXR target genes altered in expression with DN2-TG.
It is important to note that ASC-2 also has a multitude of other target nuclear receptors and transcription factors (5, 19, 24-26, 30, 46, 58). Thus, our results, which demonstrated the validity of DN2-TG mice as a novel animal model system to study the LXR-specific function of ASC-2 in vivo, attest to the beauty of our transgenic approach over a general deletion of the ASC-2 gene (20, 59). To further confirm the LXR specificity of DN2-TG mice, as well as to identify additional hepatic target genes of the LXR ligand T0901317 in mice, we analyzed the T0901317-dependent gene expression profile of the DN2-TG liver by employing DNA microarray experiments. The microarrays we analyzed consisted of ∼8,000 mouse cDNAs, as well as various control genes. For these experiments, three wild-type mice were treated with vehicle alone, while six wild-type and three DN2-TG mice were orally fed 50 mg of T0901317/kg of body weight for 6 days, after which their livers were excised to isolate total RNA. Samples from these animals were randomly paired to generate three sets of vehicle-treated wild-type mice versus T0901317-treated wild-type mice, as well as three sets of wild-type mice versus DN2-TG mice, both treated with T0901317. The first three sets were designed to isolate all the T0901317-responsive genes, while the second three sets were designed to identify genes responsive to both DN2 and T0901317. First, the data were remarkably consistent with each other within each set. In particular, the results with a group of genes most responsive to T0901317 and DN2 were highly reproducible both in rank order and fold induction or repression (Tables 2, 3, and 4). Second, it was interesting that only a subset of genes either up- or down-regulated by T0901317 in wild-type mice were affected by the presence of DN2 (Tables 2 and 3). For instance, among the 38 genes most up-regulated by T0901317, only 12 genes (i.e., ∼32%) were significantly down-regulated in DN2-TG mice relative to wild-type mice (Table 2). Similarly, among the 38 genes most down-regulated by T0901317, only 7 genes (i.e., ∼18%) were up-regulated in DN2-TG mice (Table 3). These results strongly suggest that the presence of LXR-responsive elements, either direct or indirect, is not sufficient to confer ASC-2 responsiveness. This notion is consistent with the general requirement of highly regulated mammalian genes for multiple cis elements and the resultant assembly of a higher-order transcription enhancer complex (i.e., an enhanceosome) consisting of multiple coregulatory proteins and transcription factors occupying these cis elements (reviewed in reference 32). Table 4 lists the most up- and down-regulated genes in T0901317-treated DN2-TG mice relative to T0901317-treated wild-type mice. Importantly, 55 and 60% of these genes were down- and up-regulated, respectively, by T0901317 in wild-type mice (Table 4). These results reinforce the notion that the major group of genes affected in T0901317-treated DN2-TG mice is indeed genuine LXR-target genes. It is also interesting that there is not a strict correlation between the rank order and the potency (i.e., fold induction or repression). For instance, AA473153 and AA213017 are repressed to 0.78- and 0.48-fold, respectively, by T0901317 in wild-type mice, i.e., AA213017 is more responsive to T0901317 (Table 4). However, AA473153 is up-regulated 5.00-fold in ligand-treated DN2-TG mice over ligand-treated wild-type mice, whereas AA213017 is up-regulated only 2.20-fold. Thus, a gene less responsive to ligand (i.e., AA473153) proves to be more responsive to DN2. Overall, these results validate the utility of our DN2-TG mice as a model system to study the LXR-specific function of ASC-2 in vivo.
TABLE 2.
Genes up-regulated by T0901317
| Gene | Name | Fold changea
|
|
|---|---|---|---|
| +WT/ −WT | +DN2-TG/ +WT | ||
| AA109684 | P450, 4A10 | 7.87 | NA |
| AA197454 | EST | 6.05 | 0.32 |
| AI893661 | P450, 4A14 | 6.03 | NA |
| AA200989 | EST | 5.46 | 0.38 |
| AA288170 | EST | 4.20 | 3.68 |
| AA270506 | EST | 4.18 | 0.98 |
| AI464827 | EST | 3.98 | 0.36 |
| AA444946 | insig-2 | 3.89 | 0.36 |
| AA009268 | Myelocytomatosis oncogene | 3.83 | 0.95 |
| W98251 | EST | 3.79 | 0.36 |
| AA239254 | EST | 3.47 | 1.81 |
| AA175346 | EST | 3.46 | 0.83 |
| AA458178 | CD36 | 3.42 | 0.99 |
| AA466026 | EST | 3.40 | 0.63 |
| AA067967 | EST | 3.21 | 1.06 |
| AA268120 | P450, 3A11 | 2.96 | 0.94 |
| W64388 | Myeloid response gene 118 | 2.94 | 1.25 |
| AA238875 | Aledehyde dehydrogenase 4 | 2.77 | 1.15 |
| AA122542 | EST | 2.74 | 1.34 |
| AA122814 | Aledehyde dehydrogenase 1 | 2.66 | 1.04 |
| AA261287 | Peroxisomal acyl-CoA oxidase | 2.59 | 0.84 |
| AA267605 | PPARγ | 2.57 | 0.49 |
| AA467249 | EST | 2.53 | 1.28 |
| W89594 | EST | 2.48 | 0.81 |
| AA271522 | EST | 2.46 | 1.40 |
| AA108340 | C3f gene | 2.45 | 1.06 |
| AA276752 | EST | 2.42 | 0.96 |
| A1587794 | EST | 2.40 | 1.01 |
| AA087206 | EST | 2.36 | 1.22 |
| AA106263 | EST | 2.28 | 1.24 |
| AA271043 | EST | 2.28 | 0.94 |
| AA097860 | CDC-like kinase 2 | 2.21 | 1.02 |
| AA259400 | EST | 2.20 | 0.71 |
| AA275203 | EST | 2.17 | 0.96 |
| AA097194 | EST | 2.15 | 0.97 |
| AA245078 | Fatty acid binding protein | 2.09 | 0.87 |
| AA458273 | EST | 2.08 | 1.12 |
| W87950 | EST | 2.06 | 1.01 |
+, treated with T0901317; −, treated with vehicle; italics, significantly down-regulated in DN2-TG mice.
TABLE 3.
Genes down-regulated by T0901317
| Gene | Name | Fold changea
|
|
|---|---|---|---|
| +WT/ −WT | +DN2-TG/ +WT | ||
| AA108438 | EST | 0.37 | 0.86 |
| AA239479 | EST | 0.43 | 0.69 |
| W66757 | EST | 0.46 | 1.16 |
| A1604749 | EST | 0.47 | NA |
| AA014384 | EST | 0.47 | 2.01 |
| AA213017 | Monooxygenase 3 | 0.48 | 2.20 |
| AA271451 | C-reactive protein, pretexin related | 0.49 | 1.46 |
| AA285921 | Major urinary protein 2 | 0.50 | 1.00 |
| AA245847 | EST | 0.50 | 1.00 |
| W83009 | EST | 0.50 | 1.08 |
| AA238208 | EST | 0.51 | 1.00 |
| AA154452 | EST | 0.51 | 1.10 |
| AA542013 | Fibroblast growth factor receptor | 0.52 | 0.96 |
| AI385792 | Sulfotransferase-related protein | 0.52 | 0.69 |
| AA218405 | 1KKɛ | 0.52 | 1.00 |
| W11846 | EST | 0.53 | 1.27 |
| AA239480 | EST | 0.54 | 1.13 |
| AA259979 | Angiotensinogen | 0.54 | 1.26 |
| AA213062 | EST | 0.54 | 1.12 |
| W18484 | MEL91 | 0.55 | NA |
| AA260520 | Ets variant gene 6 | 0.56 | 1.09 |
| AA476030 | EST | 0.57 | 0.80 |
| AA212899 | Deiodinase, iodothyronine, type | 0.57 | 0.96 |
| AA277314 | Angiogenin-3 | 0.57 | 1.55 |
| W11759 | EST | 0.57 | NA |
| AA241936 | EST | 0.57 | 0.98 |
| AI893937 | Protein tyrosine phosphatase | 0.58 | 1.00 |
| AA414106 | EST | 0.58 | NA |
| AA153024 | EST | 0.58 | 1.13 |
| AI605748 | EST | 0.59 | 0.93 |
| AA266146 | EST | 0.59 | 1.01 |
| W47974 | EST | 0.59 | NA |
| W34018 | EST | 0.60 | NA |
| AA062129 | Inter-αtrypsin inhibitor, hc 3 | 0.60 | 0.80 |
| AA122791 | Histocompatibility 2, Q locus 7 | 0.60 | 1.15 |
| W10072 | IGF 1 | 0.60 | 0.89 |
| AI390830 | EST | 0.60 | 0.90 |
| AA174320 | EST | 0.60 | 1.77 |
+, treated with T0901317; −, treated with vehicle; italics, significantly up-regulated in DN2-TG mice.
TABLE 4.
Genes most up- and down-regulated by T0901317 in transgenic versus wild-type mice
| Gene | Name | Fold changea
|
|
|---|---|---|---|
| +WT/, −WT | +DN2-TG/ +WT | ||
| AA473153 | EST | 0.78 | 5.00 |
| AA288170 | EST | 4.20 | 4.57 |
| AA060979 | EST | 0.62 | 4.17 |
| AA020307 | VLDL receptor | 0.96 | 3.21 |
| AA244820 | EST | 0.64 | 3.06 |
| AA238618 | EST | 1.04 | 2.94 |
| AI323180 | Cyclin D1 | 0.90 | 2.82 |
| AA269533 | Cyp2B9 | 0.87 | 2.71 |
| AA048915 | G nucleotide BP, beta-2 related | 0.86 | 2.68 |
| AI510113 | EST | 0.84 | 2.68 |
| AA125367 | Protein tyrosine phosphtase 16 | 0.92 | 2.65 |
| W36002 | EST | 1.01 | 2.35 |
| AA144169 | EST | 0.94 | 2.24 |
| AA268608 | Squalene epoxidase | 0.72 | 2.23 |
| AA276003 | Prolactin receptor-related 1 | 0.73 | 2.16 |
| AA388607 | EST | 1.13 | 2.01 |
| AA014384 | EST | 0.57 | 2.01 |
| AA542160 | EST | 1.25 | 1.98 |
| AA213017 | Monooxygenase 3 | 0.48 | 2.20 |
| AA268587 | Serum amyloid P component | 0.78 | 1.80 |
| AA290107 | EST | 1.13 | 2.02 |
| AA153205 | EST | 0.81 | 1.77 |
| AA472154 | EST | 1.03 | 0.29 |
| W88005 | EST | 2.00 | 0.31 |
| AA197454 | EST | 6.05 | 0.32 |
| AA097421 | Apolipoprotein A-IV | 1.98 | 0.32 |
| AA255171 | EST | 0.87 | 0.32 |
| W98251 | EST | 3.79 | 0.36 |
| AA444946 | insig-2 | 3.89 | 0.36 |
| AA108370 | GST, pi 2 | 1.43 | 0.37 |
| AA008579 | ABCG2 | 1.71 | 0.38 |
| AA177549 | EST | 1.12 | 0.38 |
| AA200989 | EST | 5.46 | 0.38 |
| AA466026 | EST | 4.00 | 0.39 |
| AA221226 | EST | 2.69 | 0.41 |
| AA254921 | Esterase 31 | 0.46 | 0.42 |
| AA177717 | Interleukin 1 receptor, type 1 | 1.06 | 0.43 |
| AI464827 | EST | 3.98 | 0.44 |
| AI386390 | Translation initiation factor 2-3 | 1.95 | 0.44 |
| AA064236 | EST | 4.43 | 0.45 |
| AA145237 | EST | 0.69 | 0.45 |
| AI893893 | Cytokine-inducible SH2 protein | 0.94 | 0.51 |
| AA245505 | Cyokine-inducible SH2 protein 2 | 1.16 | 0.53 |
| AI322465 | Caseinolytic protease X (Escherichia coli) | 1.56 | 0.54 |
| AA049246 | EST, NEDD4 | 1.26 | 0.56 |
| AA175618 | EST | 1.10 | 0.57 |
| AA230822 | EST | 1.05 | 0.57 |
+, treated with T0901317; −, treated with vehicle; italics, significantly up- or down-regulated in wild-type mice.
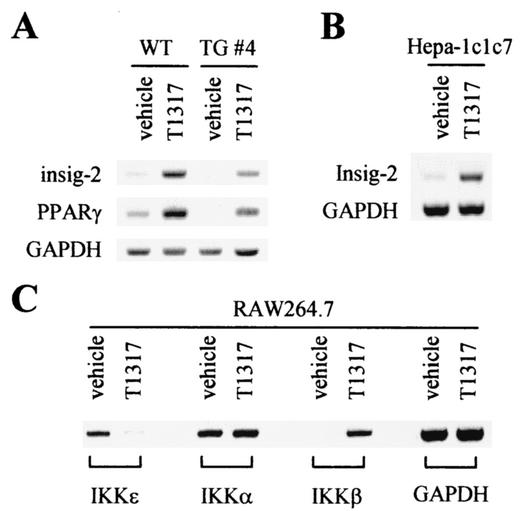
These cDNA microarray results allowed us to directly identify a series of novel hepatic genes targeted by ASC-2 and LXRs. For instance, genes encoding peroxisome proliferator-activated receptor γ (PPARγ) and insig-2 (54) were found to be positively regulated by T0901317 in an ASC-2-dependent manner (Tables 2 and 3). These results were further confirmed by reverse transcription (RT)-PCR (Fig. 5A). Expression of insig-2 was also induced by T0901317 in the murine hepatoma cell line hepa-1c1c7 (Fig. 5B). Like the previously described insig-1, insig-2 binds to SREBP cleavage-activating protein in a sterol-regulated fashion and prevents the proteolytic processing of SREBPs by Golgi enzymes, thereby blocking cholesterol-lipid synthesis (54). Both PPARγ and insig-2 may play important roles in the liver lipid metabolism mediated by LXRs. A novel IκB kinase complex containing PMA-inducible IKKɛ, distinct from the well-characterized high-molecular-weight IκB kinase complex containing IKKα, IKKβ, and IKKγ, was recently reported (36). Interestingly, IKKɛ was identified as a T0901317-repressible gene from the cDNA microarray experiments, although it was not a direct target gene of ASC-2 (Tables 2 and 3). In the macrophage cell line RAW264.7, IKKɛ was indeed repressed by T0901317 (Fig. 5C). However, T0901317 did not affect expression of IKKα, while it significantly induced IKKβ. These results suggest the presence of a complex cross talk between two distinct signaling pathways (i.e., LXRs and NF-κB). Overall, these results demonstrate the credibility of our cDNA microarray results. More importantly, studies of these and other identified genes in the liver lipid metabolism will provide an important insight into the complex hepatic signaling pathways mediated by ASC-2 and LXRs in vivo.
FIG. 5.
Confirmation of cDNA microarray results. Total RNA was prepared using Trizol reagent (Life Technologies), according to the instructions given by the manufacturer, from wild-type and DN2-TG mice treated with vehicle alone or 50 mg of T0901317/kg for 6 days (A), as well as Hepa-1c1c7 (B) and RAW264.7 (C) cells treated with either vehicle alone or 10 μM T0901317 for 16 h, as indicated. Semiquantitative RT-PCR was used to determine the relative levels of mRNA for insig-2, PPARγ, IKKɛ, IKKα, IKKβ, and GAPDH. #4, DN2-TG mouse line no. 4.
Expression of lipid-metabolizing LXR target genes in DN2-TG mice.
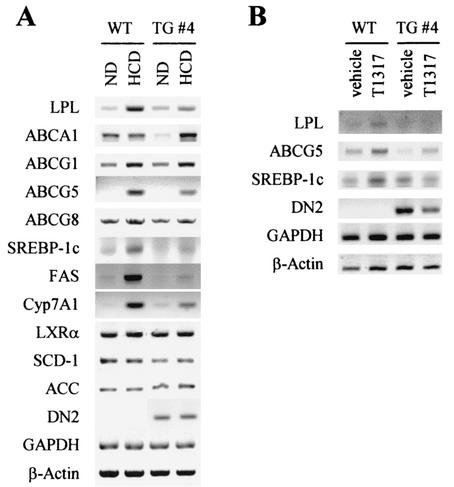
Unfortunately, our cDNA microarray chips did not include most of the known target genes of LXRs, particularly ones involved with reverse cholesterol transport and lipogenesis in the liver. Thus, we employed RT-PCR to directly assess the expression patterns for some of these genes in the livers of DN2-TG mice. Interestingly, these experiments revealed that only a subset of the genes examined are impaired in DN2-TG mice (Fig. 6A), consistent with our cDNA microarray results (Tables 2 through 4). For instance, the LXR target genes ABCA1, ABCG1, and ABCG8 were less sensitive to the expression of DN2 in DN2-TG mice fed high-cholesterol diets. However, high-cholesterol-diet-induced expression of LPL, ABCG5, FAS, SREBP-1c, and Cyp7A1 genes was significantly impaired in DN2-TG mice. Similarly, T0901317-induced expression of LPL, SREBP-1c, and ABCG5 was also impaired in DN2-TG mice (Fig. 6B). These results, along with our cDNA microarray results, suggest that ASC-2 likely regulates only a selective set of the LXR target genes in vivo, further supporting the importance of the context of the individual target gene and cell type in ASC-2 responsiveness (32). Likewise, ASC-2 is predicted to regulate only a subset of target genes of other nuclear receptors.
FIG. 6.
RT-PCR analyses of LXR target genes in DN2-TG mice. Total RNA was prepared from wild-type (WT) and DN2-TG (TG) mice untreated or treated with either a high-cholesterol diet for 60 days (A) or 50 mg of T0901317/kg for 6 days (B). Semiquantitative RT-PCR was used to determine the relative levels of mRNA for lipoprotein lipase (LPL), ABCA1, ABCG1, ABCG5, ABCG8, SREBP-1c, fatty acid synthase (FAS), Cyp7A1, LXRα, SCD-1, acetyl-CoA carboxylase (ACC), DN2, GAPDH, and β-actin.
DISCUSSION
Our results clearly demonstrate that expression of at least a subset of LXR-target genes is impaired in the livers of DN2-TG mice, implicating ASC-2 as an essential coactivator of LXRs in vivo. The rate-limiting step in the classical bile acid synthesis pathway utilizes the liver-specific enzyme Cyp7A1, which converts cholesterol into 7α-hydroxycholesterol (39, 41, 56). The down-regulation of this gene (Fig. 6) should lead to more accumulation of cholesterol concomitant with less conversion of hepatic cholesterol to bile acids in DN2-TG mice. The impaired expression of SREBP-1c (Fig. 6), a transcription factor that regulates the expression of various lipogenic genes, including those encoding acetyl-CoA carboxylase and fatty acid synthase (15), is likely responsible for the decreased de novo lipogenesis in the liver (Fig. 4). The down-regulation of ABCG5 (3, 38), encoding an ABC transporter implicated in biliary cholesterol excretion, may also lead to the observed cholesterol accumulation in the livers of DN2-TG mice. These results are correlated with the observed phenotypes of DN2-TG mice with regard to dietary-cholesterol-mediated regulation of hepatic and plasma cholesterol levels (Fig. 3D and E), as well as T0901317-mediated regulation of hepatic trigylceride levels (Fig. 4B). However, we observed at least two lines of data inconsistent with the previous reports about the regulatory function of LXRs in lipid metabolism. Hepatic triglyceride levels were significantly increased in DN2-TG mice fed high-cholesterol diets, unlike LXRα−/− mice (35). In addition, plasma triglyceride levels were significantly increased with T0901317-treated DN2-TG mice, while LXRα/β double-knockout mice had only a slight increase (41). A few possibilities exist. First, DN2 may bind still other uncharacterized nuclear receptors, and the impairment of the signaling pathway for these receptors may lead to the observed triglyceride phenotypes. Consistent with this possibility, many genes that are not responsive to T0901317 in wild-type mice were altered in expression in DN2-TG mice (AA238618, W36002, AA472154, and AA177717 [Table 4], for instance). We are currently investigating whether DN2 binds to other receptors, particularly those known to be involved with lipid metabolism, such as PPARδ (1), PXR (10), RORα (52), and Rev-erbAα (6). One likely group of physiological target genes of these putative non-LXR-type receptors that lead to the altered triglyceride levels may involve genes regulating triglyceride clearance. In this regard, it was interesting that the gene for very low density lipoprotein receptor (45), which was not significantly regulated by T0901317 in wild-type mice, was identified as a gene targeted by DN2 in DN2-TG mice. Secondly, it should be noted that DN2-TG mice are clearly different from LXRα/β double-knockout mice. In the former mice, the functional endogenous ASC-2 still exists, so LXR signaling can function at least to some extent, while LXR signaling should be completely off in the latter mice. In particular, in the presence of higher doses of T0901317, a subset of LXR target genes were significantly up-regulated in DN2-TG mice, likely through the ability of this potent synthetic LXR ligand to overcome the repressive effect of DN2 (data not shown).
Our cDNA microarray experiments suggested a number of novel roles of LXRs and ASC-2 in the murine liver (Tables 2 through 4). It was noted that many genes previously described as constitutive androstane receptor (CAR) and/or PPARα target genes, including those encoding Cyp4A10, Cyp4A14, and Cyp3A11 (2, 49), were modulated by T0901317 in wild-type mice, suggesting a novel cross talk between LXRs and CAR/PPARα signaling pathways. The gene for angiogenin-3, a potent activator of angiogenesis (11), emerged as a down-regulated target gene of ASC-2 and T0901317. Consistent with these results, our preliminary results indicated that T0901317 exhibits antiangiogenic effects with cultured human umbilical vein endothelial cells. As shown in Fig. 5C, T0901317 also showed a complex regulation of different IκB kinases. Since various NF-κB-activating signals may differentially utilize different IκB kinases (36), these results suggest a novel regulatory role of LXRs in NF-κB transactivation. It was also surprising to observe the up-regulation of PPARγ by T0901317 (Fig. 5A). Consistent with the previous work implicating PPARγ in the regulation of CD36 expression and macrophage uptake of oxidized LDL (7), CD36 also turned up as a T090317-induced gene in our microarray assays (Tables 2 and 3). It was recently shown that PPARγ induces ABCA1 expression and cholesterol removal from macrophages through a transcriptional cascade mediated by the nuclear receptor LXRα (8). Thus, these results suggest a possible autoregulatory loop between PPARγ and LXRs present in the murine liver. Finally, it was intriguing that insig-2 was up-regulated by not only T0901317 (Tables 2 and 3 and Fig. 5) but also agonists of other metabolically important receptors, such as PPARα, PPARγ, and vitamin D receptor (our unpublished results). These results implicate insig-2 as a central sensor protein for various cellular conditions that call for a shutdown of cholesterol-lipid synthesis in vivo. Notably, our cDNA microarray experiments identified many additional genes that may unravel yet other exciting and unexpected functions of LXRs and ASC-2.
Recently, LXRs were shown to be endogenous inhibitors of atherogenesis (22). We found that DN2 was also expressed in macrophages of DN2-TG mice. Thus, bone marrow transplantations were exploited to assess the effect of selectively eliminating the LXR function of macrophages (i.e., bone marrow transplantation of macrophages from DN2-TG mice into murine models of atherosclerosis). These mice with transplants mimicked many aspects of Tangier disease, a human high-density lipoprotein deficiency, including aberrant regulation of cholesterol transporter expression (i.e., ABCA1 and ABCG1), lipid accumulation in macrophages, and increased atherosclerosis (our unpublished results). These results, along with the results presented in this report, strongly suggest that ASC-2 is a bona fide coactivator of LXRs in vivo.
In conclusion, we established novel transgenic mouse lines expressing a dominant-negative fragment of ASC-2 that specifically blocks the recruitment of endogenous ASC-2 to LXRs bound to the target promoters. We demonstrated that these mice were indeed impaired in the ability to respond to dietary cholesterol and the synthetic LXR ligand T0901317. These results, along with the recent results with DN1-TG mice (18), led us to conclude that ASC-2 is a physiologically important transcriptional coactivator of LXRs and other nuclear receptors in vivo. From the livers of these mice, we further isolated a series of novel genes that are targeted by ASC-2 and T0901317. Studies of these genes will provide further insights into the molecular mechanisms for the roles of ASC-2 with regard to LXRs in dietary-cholesterol metabolism and lipogenesis in the liver.
Acknowledgments
We thank Young-Chul Lee for the yeast LXRα plasmids.
This work was supported by grants from Vascular System Research of KOSEF (Y.Y.K) and Critical Technology 21 on Life Phenomena and Function Research of the Ministry of Science and Technology (H.K.), as well as POSTECH Biotech Center and GenoCheck, Inc. (J.W.L.), and 21c Frontier Functional Proteomics Project from the Ministry of Science and Technology (J.W.L.). Jungyeob Ham was supported by the BK21 program.
Seung-Whan Kim and Keunhee Park contributed equally to this study.
REFERENCES
- 1.Barak, Y., D. Liao, W. He, E. S. Ong, M. C. Nelson, J. M. Olefsky, R. Boland, and R. M. Evans. 2002. Effects of peroxisome proliferator-activated receptor delta on placentation, adiposity, and colorectal cancer. Proc. Natl. Acad. Sci. USA 99:303-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barclay, T. B., J. M. Peters, M. B. Sewer, L. Ferrari, F. J. Gonzalez, and E. T. Morgan. 1999. Modulation of cytochrome P-450 gene expression in endotoxemic mice is tissue specific and peroxisome proliferator-activated receptor-alpha dependent. J. Pharmacol. Exp. Ther. 290:1250-1257. [PubMed] [Google Scholar]
- 3.Berge, K. E., H. Tian, G. A. Graf, L. Yu, N. V. Grishin, J. Schultz, P. Kwiterovich, B. Shan, R. Barnes, and H. H. Hobbs. 2000. Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent ABC transporters. Science 290:1771-1775. [DOI] [PubMed] [Google Scholar]
- 4.Bucolo, G., and H. David. 1973. Quantitative determination of serum triglycerides by the use of enzymes. Clin. Chem. 19:476-482. [PubMed] [Google Scholar]
- 5.Caira, F., P. Antonson, M. Pelto-Huikko, E. Treuter, and J. A. Gustafsson. 2000. Cloning and characterization of RAP250, a novel nuclear receptor coactivator. J. Biol. Chem. 275:5308-5317. [DOI] [PubMed] [Google Scholar]
- 6.Chawla, A., and M. A. Lazar. 1993. Induction of Rev-ErbA alpha, an orphan receptor encoded on the opposite strand of the alpha-thyroid hormone receptor gene, during adipocyte differentiation. J. Biol. Chem. 268:16265-16269. [PubMed] [Google Scholar]
- 7.Chawla, A., Y. Barak, L. Nagy, D. Liao, P. Tontonoz, and R. M. Evans. 2001. PPAR-gamma dependent and independent effects on macrophage-gene expression in lipid metabolism and inflammation. Nat. Med. 7:48-52. [DOI] [PubMed] [Google Scholar]
- 8.Chawla, A., W. A. Boisvert, C. H. Lee, B. A. Laffitte, Y. Barak, S. B. Joseph, D. Liao, L. Nagy, P. A. Edwards, L. K. Curtiss, R. M. Evans, and P. Tontonoz. 2001. A PPAR gamma-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol. Cell 7:161-171. [DOI] [PubMed] [Google Scholar]
- 9.Costet, P., Y. Luo, N. Wang, and A. R. Tall. 2000. Sterol-dependent transactivation of the ABC1 promoter by the liver X receptor/retinoid X receptor. J. Biol. Chem. 275:28240-28245. [DOI] [PubMed] [Google Scholar]
- 10.Edwards, P. A., H. R. Kast, and A. M. Anisfeld. 2002. BAREing it all: the adoption of LXR and FXR and their roles in lipid homeostasis. J. Lipid Res. 43:2-12. [PubMed] [Google Scholar]
- 11.Fu, X., W. G. Roberts, V. Nobile, R. Shapiro, and M. P. Kamps. 1999. mAngiogenin-3, a target gene of oncoprotein E2a-Pbx1, encodes a new angiogenic member of the angiogenin family. Growth Factors 17:125-137. [DOI] [PubMed] [Google Scholar]
- 12.Goo, Y. H., Y. C. Sohn, D. H. Kim, S. W. Kim, M. J. Kang, D. J. Jung, E. Kwak, N. A. Barlev, S. L. Berger, V. Y. Chow, R. G. Roeder, D. O. Azorsa, P. S. Meltzer, P. G. Suh, E. J. Song, K. J. Lee, Y. C. Lee, and J. W. Lee. 2003. Activating signal cointegrator-2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol. Cell. Biol. 23:140-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grefhorst, A., B. M. Elzinga, P. J. Voshol, T. Plosch, T. Kok, V. W. Bloks, F. H. van der Sluijs, L. M. Havekes, J. A. Romijn, H. J. Verkade, and F. Kuipers. 2002. Stimulation of lipogenesis by pharmacological activation of the liver X receptor leads to production of large, triglyceride-rich very low density lipoprotein particles. J. Biol. Chem. 277:34182-34190. [DOI] [PubMed] [Google Scholar]
- 14.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 15.Horton, J. D., I. Shimomura, M. S. Brown, R. E. Hammer, J. L. Goldstein, and H. Shimano. 1998. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J. Clin. Investig. 101:2331-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishibashi, S., M. S. Brown, J. L. Goldstein, R. D. Gerrard, R. E. Hammer, and J. Herz. 1993. Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery. J. Clin. Investig. 92:883-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joseph, S. B., B. A. Laffitte, P. H. Patel, M. A. Watson, K. E. Matsukuma, R. Walczak, J. L. Collins, T. F. Osborne, and P. Tontonoz. 2002. Direct and indirect mechanisms for regulation of fatty acid synthase gene expression by liver X receptors. J. Biol. Chem. 277:11019-11025. [DOI] [PubMed] [Google Scholar]
- 18.Kim, S. W., C. Cheong, Y. C. Sohn, Y. H. Goo, W. J. Oh, J. H. Park, S. Y. Joe, H. S. Kang, D. K. Kim, C. Kee, J. W. Lee, and H. W. Lee. 2002. Multiple developmental defects derived from impaired recruitment of ASC-2 to nuclear receptors in mice: implication for posterior lenticonus with cataract. Mol. Cell. Biol. 22:8409-8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ko, L., G. R. Cardona, and W. W. Chin. 2000. Thyroid hormone receptor-binding protein, an LXXLL motif-containing protein, functions as a general coactivator. Proc. Natl. Acad. Sci. USA 97:6212-6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuang, S. Q., L. Liao, H. Zhang, F. A. Pereira, Y. Yuan, F. J. DeMayo, L. Ko, and J. Xu. 2002. Deletion of the cancer-amplified coactivator AIB3 results in defective placentation and embryonic lethality. J. Biol. Chem. 277:45356-45360. [DOI] [PubMed] [Google Scholar]
- 21.Laffitte, B. A., J. J. Repa, S. B. Joseph, D. C. Wilpitz, H. R. Kast, D. J. Mangelsdorf, and P. Tontonoz. 2001. LXRs control lipid-inducible expression of the apolipoprotein E gene in macrophages and adipocytes. Proc. Natl. Acad. Sci. USA 98:507-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laffitte, B. A., and P. Tontonoz. 2002. Orphan nuclear receptors find a home in the arterial wall. Curr. Atheroscler. Rep. 4:213-221. [DOI] [PubMed] [Google Scholar]
- 23.Lee, J. W., Y. C. Lee, S. Y. Na, D. J. Jung, and S. K. Lee. 2001. Transcriptional coregulators of the nuclear receptor superfamily: coactivators and corepressors. Cell. Mol. Life Sci. 58:289-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, S. K., S. L. Anzick, J. E. Choi, L. Bubendorf, X. Y. Guan, Y. K. Jung, O. P. Kallioniemi, J. Kononen, J. M. Trent, D. Azorsa, B. H. Jhun, J. H. Cheong, Y. C. Lee, P. S. Meltzer, and J. W. Lee. 1999. A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J. Biol. Chem. 274:34283-34293. [DOI] [PubMed] [Google Scholar]
- 25.Lee, S. K., S. Y. Na, S. Y. Jung, J. E. Choi, B. H. Jhun, J. Cheong, P. S. Meltzer, Y. C. Lee, and J. W. Lee. 2000. Activating protein-1, nuclear factor-κB, and serum response factor as novel target molecules of the cancer-amplified transcription coactivator ASC-2. Mol. Endocrinol. 14:915-925. [DOI] [PubMed] [Google Scholar]
- 26.Lee, S. K., S. Y. Jung, Y. S. Kim, S. Y. Na, Y. C. Lee, and J. W. Lee. 2001. Two distinct nuclear receptor-interaction domains and CREB-binding protein-dependent transactivation function of activating signal cointegrator-2. Mol. Endocrinol. 15:241-254. [DOI] [PubMed] [Google Scholar]
- 27.Lehmann, J. M., S. A. Kliewer, L. B. Moore, T. A. Smith-Oliver, B. B. Oliver, J. L. Su, S. S. Sundseth, D. A. Winegar, D. E. Blanchard, T. A. Spencer, and T. M. Willson. 1997. Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J. Biol. Chem. 272:3137-3140. [DOI] [PubMed] [Google Scholar]
- 28.Liang, G., J. Yang, J. D. Horton, R. E. Hammer, J. L. Goldstein, and M. S. Brown. 2002. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J. Biol. Chem. 277:9520-9528. [DOI] [PubMed] [Google Scholar]
- 29.Luo, Y., and A. R. Tall. 2000. Sterol upregulation of human CETP expression in vitro and in transgenic mice by an LXR element. J. Clin. Investig. 105:513-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahajan, M. A., and H. H. Samuels. 2000. A new family of nuclear receptor coregulators that integrate nuclear receptor signaling through CREB-binding protein. Mol. Cell. Biol. 20:5048-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, and R. M. Evans. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maniatis, T., J. V. Falvo, T. H. Kim, T. K. Kim, C. H. Lin, B. S. Parekh, and M. G. Wathelet. 1998. Structure and function of the interferon-beta enhanceosome. Cold Spring Harbor Symp. Quant. Biol. 63:609-620. [DOI] [PubMed] [Google Scholar]
- 33.McKenna, N. J., and B. W. O'Malley. 2000. From ligand to response: generating diversity in nuclear receptor coregulator function. J. Steroid Biochem. Mol. Biol. 74:351-356. [DOI] [PubMed] [Google Scholar]
- 34.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 35.Peet, D. J., S. D. Turley, W. Ma, B. A. Janowski, J.-M. A. Lobaccaro, R. E. Hammer, and D. J. Mangelsdorf. 1998. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell 93:693-704. [DOI] [PubMed] [Google Scholar]
- 36.Peters, R. T., S. M. Liao, and T. Maniatis. 2000. IKKɛ is part of a novel PMA-inducible IκB kinase complex. Mol. Cell 5:513-522. [DOI] [PubMed] [Google Scholar]
- 37.Repa, J. J., S. D. Turley, J.-M. A. Lobaccaro, J. Medina, L. Li, K. Lustig, B. Shan, R. A. Heyman, J. M. Dietschy, and D. J. Mangelsdorf. 2000. Regulation of absorption and ABC1-mediated efflux of cholesterol by RXR heterodimers. Science 289:1524-1529. [DOI] [PubMed] [Google Scholar]
- 38.Repa, J. J., K. E. Berge, C. Pomajzl, J. A. Richardson, H. Hobbs, and D. J. Mangelsdorf. 2002. Regulation of ATP-binding cassette sterol transporters ABCG5 and ABCG8 by the liver X receptors alpha and beta. J. Biol. Chem. 277:18793-18800. [DOI] [PubMed] [Google Scholar]
- 39.Repa, J. J., G. Liang, J. Ou, Y. Bashmakov, J.-M. A. Lobaccaro, I. Shimomura, B. Shan, M. S. Brown, J. L. Goldstein, and D. J. Mangelsdorf. 2000. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev. 14:2819-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenfeld, M. G., and C. K. Glass. 2001. Coregulator codes of transcriptional regulation by nuclear receptors. J. Biol. Chem. 276:36865-36868. [DOI] [PubMed] [Google Scholar]
- 41.Schultz, J. R., H. Tu, A. Luk, J. J. Repa, J. C. Medina, L. Li, S. Schwender, S. Wang, M. Thoolen, D. J. Mangelsdorf, K. D. Lustig, and B. Shan. 2000. Role of LXRs in control of lipogenesis. Genes Dev. 14:2831-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwartz, K., R. M. Lawn, and D. P. Wade. 2000. ABC1 gene expression and ApoA-I-mediated cholesterol efflux are regulated by LXR. Biochem. Biophys. Res. Commun. 274:794-802. [DOI] [PubMed] [Google Scholar]
- 43.Serrano, M., H. Lee, L. Chin, C. Cordon-Cardo, D. Beach, and R. A. DePinho. 1996. Role of the INK4a locus in tumor suppression and cell mortality. Cell 85:27-37. [DOI] [PubMed] [Google Scholar]
- 44.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 45.Tacken, P. J., M. H. Hofker, L. M. Havekes, and K. W. van Dijk. 2001. Living up to a name: the role of the VLDL receptor in lipid metabolism. Curr. Opin. Lipidol. 12:275-279. [DOI] [PubMed] [Google Scholar]
- 46.Tanner, M. M., M. Tirkkonen, A. Kallioniemi, J. Isola, T. Kuukasjarvi, C. Collins, D. Kowbel, X. Y. Guan, J. Trent, J. W. Gray, P. Meltzer, and O. P. Kallioniemi. 1996. Independent amplification and frequent co-amplification of three nonsyntenic regions on the long arm of chromosome 20 in human breast cancer. Cancer Res. 56:3441-3445. [PubMed] [Google Scholar]
- 47.Teboul, M., E. Enmark, Q. Li, J.-A. Gustafsson, M. Pelto-Huikko, and J.-Å. Gustafsson. 1995. OR-1, a member of the nuclear receptor superfamily that interacts with the 9-cis-retinoic acid receptor. Proc. Natl. Acad. Sci. USA 92:2096-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torchia, J., D. W. Rose, J. Inostroza, Y. Kamei, S. Westin, C. K. Glass, and M. G. Rosenfeld. 1997. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387:677-684. [DOI] [PubMed] [Google Scholar]
- 49.Ueda, A., H. K. Hamadeh, H. K. Webb, Y. Yamamoto, T. Sueyoshi, C. A. Afshari, J. M. Lehmann, and M. Negishi. 2002. Diverse roles of the nuclear orphan receptor CAR in regulating hepatic genes in response to phenobarbital. Mol. Pharmacol. 61:1-6. [DOI] [PubMed] [Google Scholar]
- 50.Venkateswaran, A., B. A. Laffitte, S. B. Joseph, P. A. Mak, D. C. Wilpitz, P. A. Edwards, and P. Tontonoz. 2000. Control of cellular cholesterol efflux by the nuclear oxysterol receptor LXR alpha. Proc. Natl. Acad. Sci. USA 97:12097-12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venkateswaran, A., J. J. Repa, J.-M. A. Lobaccaro, A. Bronson, D. J. Mangelsdorf, and P. A. Edwards. 2000. Human white/murine ABC8 mRNA levels are highly induced in lipid-loaded macrophages. A transcriptional role for specific oxysterols. J. Biol. Chem. 275:14700-14707. [DOI] [PubMed] [Google Scholar]
- 52.Vu-Dac, N., P. Gervois, T. Grotzinger, P. De Vos, K. Schoonjans, J. C. Fruchart, J. Auwerx, J. Mariani, A. Tedgui, and B. Staels. 1997. Transcriptional regulation of apolipoprotein A-I gene expression by the nuclear receptor RORα. J. Biol. Chem. 272:22401-22404. [DOI] [PubMed] [Google Scholar]
- 53.Willy, P. J., K. Umesono, E. S. Ong, R. M. Evans, R. A. Heyman, and D. J. Mangelsdorf. 1995. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev. 9:1033-1045. [DOI] [PubMed] [Google Scholar]
- 54.Yabe, D., M. S. Brown, and J. L. Goldstein. 2002. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc. Natl. Acad. Sci. USA 99:12753-12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yokode, M., R. E. Hammer, S. Ishibashi, M. S. Brown, and J. L. Goldstein. 1990. Diet-induced hypercholesterolemia in mice: prevention by overexpression of LDL receptors. Science 250:1273-1275. [DOI] [PubMed] [Google Scholar]
- 56.Yoshikawa, T., H. Shimano, M. Amemiya-Kudo, N. Yahagi, A. H. Hasty, T. Matsuzaka, H. Okazaki, Y. Tamura, Y. Iizuka, K. Ohashi, J. I. Osuga, K. Harada, T. Gotoda, S. Kimura, S. Ishibashi, and N. Yamada. 2001. Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol. Cell. Biol. 21:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, Y., J. J. Repa, K. Gauthier, and D. J. Mangelsdorf. 2001. Regulation of lipoprotein lipase by the oxysterol receptors, LXRα and LXRβ. J. Biol. Chem. 276:43018-43024. [DOI] [PubMed] [Google Scholar]
- 58.Zhu, Y., L. Kan, C. Qi, Y. S. Kanwar, A. V. Yeldandi, M. S. Rao, and J. K. Reddy. 2000. Isolation and characterization of peroxisome proliferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR. J. Biol. Chem. 275:13510-13516. [DOI] [PubMed] [Google Scholar]
- 59.Zhu, Y. J., S. E. Crawford, V. Stellmach, R. S. Dwivedi, M. S. Rao, F. J. Gonzalez, C. Qi, and J. K. Reddy. 2003. Coactivator PRIP, the peroxisome proliferator-activated receptor-interacting protein, is a modulator of placental, cardiac, hepatic and embryonic development. J. Biol. Chem. 278:1986-1990. [DOI] [PubMed] [Google Scholar]