Abstract
The human serine protease inhibitor (serpin) gene cluster at 14q32.1 contains a number of genes that are specifically expressed in hepatic cells. Cell-specific enhancers have been identified in several of these genes, but elements involved in locus-wide gene and chromatin control have yet to be defined. To identify regulatory elements in this region, we prepared a series of mutant chromosomal alleles by homologous recombination and transferred the specifically modified human chromosomes to hepatic cells for functional tests. We report that deletion of an 8-kb DNA segment upstream of the human α1-antitrypsin gene yields a mutant serpin allele that fails to be activated in hepatic cells. Within this region, a 2.3-kb DNA segment between kb −8.1 and −5.8 contains a previously unrecognized control region that is required not only for serpin gene activation but also for chromatin remodeling of the entire locus.
Chromatin is the template for transcription in vivo. Alterations in the chromatin organization of specific loci generally accompany changes in gene activity, and these chromatin effects can be monitored in several ways. For example, the accessibility of specific chromosomal loci to exogenous nucleases generally reflects their transcriptional activity (48). Similarly, the formation of expression-associated DNase I-hypersensitive sites (DHSs) (39), covalent modifications of histones and other chromosomal proteins (25), and nuclear compartmentalization of specific loci (43) all change during eukaryotic gene activation. This suggests that regulatory mechanisms that govern gene activity are linked to those that control chromatin structure. It is a fundamental challenge for mammalian genetics to identify and characterize these regulatory interactions in their native genomic environments.
The human serine protease inhibitor (serpin) gene cluster at 14q32.1 is a useful model system for studying the cell-specific regulation of gene expression and chromatin structure. The serpin locus is located at approximately 92.3 megabases (Mb) along chromosome 14q's 104-Mb length (http://www.ncbi.nlm.nih.gov/). There are 11 functionally diverse serpin genes within this ∼400-kb region, and the genes are organized into three discrete subclusters of four, three, and four genes each. The proximal subcluster, about 107 kb in length, is the best characterized; it includes the well-studied α1-antitrypsin gene (α1AT, SERPINA1), an antitrypsin-related pseudogene (ATR, SERPINA2) ∼13 kb downstream, the corticosteroid-binding globulin gene (CBG, SERPINA6) ∼68 kb downstream of α1AT (37), and the recently identified protein Z inhibitor gene (ZPI, SERPINA10) ∼100 kb downstream of α1AT (20). These serpin genes are expressed in hepatic cells, but they are repressed in most other cell types.
Regulation of human α1AT gene expression has been studied in detail (8, 13, 21, 29, 32, 33, 45). Human α1AT mRNA is one of the most abundant transcripts in hepatic cells, but the gene is not expressed in most other cell types. These large differences in the transcriptional activities of α1AT are correlated with cell-specific chromatin structures of the locus in expressing and nonexpressing cells. For example, there are 29 DHSs in the ∼130-kb region from ∼25 kb upstream of α1AT to ∼105 kb downstream in expressing cells, but only seven of these DHSs are present in nonexpressing cells (39). As DHSs often mark sites of important regulatory interactions, these observations suggest that the α1AT locus is rich in regulatory information. However, specific roles for any of the 22 expression-associated DHSs in controlling gene activity and/or chromatin structure remain to be defined.
Transfection experiments have identified DNA elements that are required in cis for cell-specific activation of α1AT gene expression in cultured cells and transgenic animals. An α1AT promoter fragment of only 137 bp was sufficient to drive liver-specific expression of reporter genes in transient-transfection assays (13, 21, 32). This 137-bp sequence contains the minimal α1AT promoter plus a cell-specific enhancer located between bp −75 and −115. The enhancer contains binding sites for the liver-enriched transactivators HNF-1α (at bp −75) and HNF-4 (at bp −115), and site-specific mutagenesis of the binding sites indicated that these factors activated α1AT transcription 100- and 10-fold, respectively (13). When the same HNF-1α and/or HNF-4 binding site mutations were introduced into constructs containing an additional 5 kb of upstream sequence, disruption of the HNF-1α binding site reduced liver-specific expression of α1AT ∼100-fold in transgenic mice, but disruption of the HNF-4 binding site did not significantly affect human α1AT expression in the transgenic animals (47).
Other experiments have revealed complexities in the tissue-specific patterns of human α1AT expression in transgenic mice. For example, transgenes containing ∼9 or ∼7 kb of upstream sequence expressed human α1AT mRNA not only in liver but also in kidney, small intestine, lung, and spleen (26, 27, 42). A more restricted pattern of gene expression was observed when shorter transgenes were used: expression of transgenes with only 2 or 1.2 kb of 5′ flanking sequence was restricted to liver and kidney (45, 46), while expression from a 348-bp promoter/enhancer construct was restricted to liver (49). High-level expression of all these transgenes in liver is likely due to the strong hepatic promoter/enhancer, and expression at other sites may involve the macrophage-specific promoter, which is located about 2 kb upstream (29, 42).
All of the transgenes analyzed in the experiments described above were integrated at ectopic chromosomal sites, where chromosomal position effects could affect expression levels in different transgenic lines (15, 46, 49). Furthermore, transgene copy number was uncontrolled in these lines, and this could affect gene activity. Finally, most transgenic lines contained multicopy transgene arrays (46), which can be subject to transcriptional interference (17). All of these factors have conspired to make the various transgene expression phenotypes difficult to interpret.
To circumvent the inherent limitations of transgene experiments, we sought to identify regulatory elements in human α1AT by making specific modifications of the locus within its normal context on human chromosome 14. To do this, human chromosome 14 was transferred to recombination-proficient chicken DT40 cells (5) by microcell transfer, and the chicken-human hybrids were used to make precise modifications in human α1AT by homologous recombination (14). The expression and chromatin organization of the various mutant alleles were then assessed after transfer of the mutant chromosomes to hepatic cells. These studies identified regulatory elements upstream of the α1AT gene that are necessary for cell-specific gene activation and chromatin remodeling of the proximal serpin subcluster. The activities of these regulatory elements had not been apparent previously in other experimental tests.
MATERIALS AND METHODS
Cell lines and culture conditions.
DT40 is an avian leukosis virus-induced chicken B-cell line (5); DT40-2 is a subclone of DT40 that is sensitive to 1.25 mg of Geneticin (Gibco BRL)/ml. Both lines were cultured as described previously (14).
HDm-5 is a murine 3T6 cell line that contains a neo-tagged human chromosome 14 derived from human diploid fibroblasts (30). MEL is a murine erythroleukemia cell line. M(h14n)8 is a microcell hybrid that was prepared by transferring human chromosome 14 from HDm-5 to MEL cells by microcell fusion. These cells were cultured in Dulbecco's modified Eagle's medium with 10% bovine calf serum (HyClone) and 500 μg of Geneticin/ml. D(h14n)F is a DT40-2 derivative that contains a single, intact, neo-tagged human chromosome 14 derived from M(h14n)8.
Fao-1 is a rat hepatoma cell line derived from H4IIEC3 (12). F(14n)14 is a microcell hybrid prepared by transferring human chromosome 14 from HDm-5 to Fao-1 cells, as described previously (44).
Murine A9 cells and Rat2 fibroblasts were obtained from the American Type Culture Collection. Fao-1, Rat2, A9, and HDm-5 were propagated in 1:1 Ham's F12-Dulbecco's modified Eagle's medium with 5% fetal bovine serum (Gibco BRL). Five hundred micrograms of Geneticin/ml was used to select for the neo-tagged human chromosome 14 in the various hybrid cell lines.
Transfer of human chromosome 14.
The M(h14n)8 microcell hybrid was generated by micronucleating and enucleating HDm-5 donor cells (neoR HPRT−) as described previously (44). The resulting microcells were fused with MEL recipients (neoS HPRT+) in suspension, and hybrid clones were selected in 500 μg of Geneticin/ml and 20 μg of 2,6-diaminopurine (Sigma)/ml, as described previously (14).
The D(h14n)F microcell hybrid was generated as described previously (14) by using donor M(h14n)8 cells (neoR APRT−) and recipient DT40-2 cells (neoS APRT+). Hybrid clones were selected in medium containing adenine-aminopterin-thymidine (AAT) and 1.25 mg of Geneticin/ml.
R(Δ8.0) series hybrids were generated by whole-cell fusions between donor D(Δ8.0) cell lines (neoR ouabainS), which contained various modified human chromosomes 14, and recipient Rat2 cells (neoS ouabainR) by using a suspension-monolayer fusion protocol (30). Hybrid clones were selected in 500 μg of Geneticin/ml and 20 μM ouabain. The A9(Δ4.3), A9(Δ2.0), F(Δ2.3), and F(Δ3.5) series hybrids were generated by whole-cell fusions between donor DT40 chicken-human hybrids and recipient A9 (neoS ouabainR) or Fao-1 (neoS ouabainR) cells by using similar methods.
F(Δ8.0) series hybrids were generated by microcell fusion between donor R(Δ8.0) cells (neoR ouabainS) and recipient Fao-1 cells (neoS ouabainR). Hybrid clones were selected in 500 μg of Geneticin/ml and 3 mM ouabain. F(Δ4.3) and F(Δ2.0) series hybrids were generated by microcell fusion between the donor A9(Δ4.3) (neoR APRT−) or A9(Δ2.0) (neoR APRT−) cells, respectively, and recipient Fao-1 cells (neoS APRT+). Hybrid clones were selected in 500 μg of Geneticin/ml and AAT.
Homologous-recombination substrates.
The pLAHL-PGKDipA targeting vector (Fig. 1) contained unique XhoI and SalI sites for the insertion of 5′ and 3′ arms of homology adjacent to each loxP site. HisD gene expression was driven by the chicken actin promoter (14). A counterselection cassette, containing the diphtheria toxin A-chain gene (DipA) driven by the murine phosphoglycerate kinase promoter, was located adjacent to the SalI insertion site.
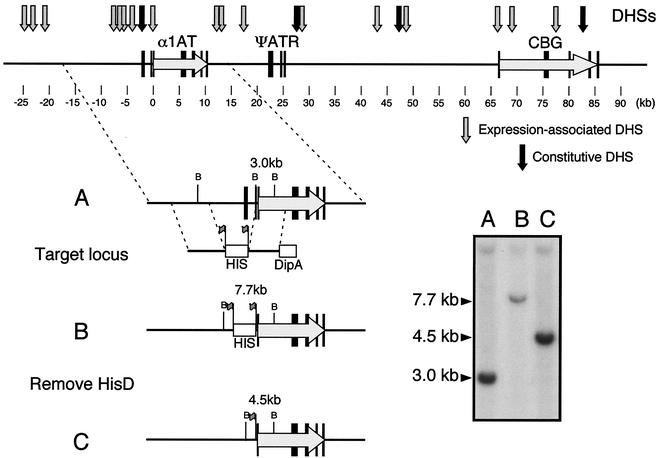
FIG. 1.
Structure and modification of the proximal serpin subcluster. At top is the proximal serpin subcluster, showing the locations of the human α1AT, ATR, and CBG genes, with exons indicated by black bars. Arrows indicate the positions of 21 DHSs that are present on wild-type human chromosome 14 in Fao-1 rat hepatoma hybrids; 18 of these sites are expression associated (gray arrows) and 4 are constitutive (black arrows). The bottom portion shows excision of an 8.0-kb DNA segment upstream of the 315-bp α1AT hepatic promoter in DT40 hybrids. (A) The locus is targeted with the recombination substrate containing two ∼4-kb arms of homology, a hisD selection cassette flanked by loxP sites (flags), and a DipA counterselection cassette. (B) Homologous recombinants are identified by Southern hybridization using a diagnostic BglII band shift from the wild-type 3.0-kb DNA fragment to the mutant fragment of 7.7 kb. (C) The hisD selection cassette is removed by transient transfection with Cre recombinase. Cre recombinants were identified by a diagnostic 4.5-kb BglII fragment after Southern hybridization.
Homology segments for the Δ8.0 targeting vector were as follows: 5′ arm, a 7.0-kb XbaI-NcoI fragment from p7.7XbaIcos10 (37); 3′ arm, a 4.9-kb PstI-BamHI fragment subcloned from αATc1 (26), where the PstI site was within exon IB of α1AT, 315 bp upstream of the hepatic transcription initiation site.
Arms of homology for the Δ3.4, Δ4.3, Δ2.0, and Δ2.3 constructs were amplified from cosmid Ycos72, which contains the entire human α1AT gene and 23 kb of 5′ flanking sequence (37). The following primer pairs contained either SalI or XhoI tails (underlined) for cloning directly into the targeting vector: Δ3.4 5′-arm primers, TAAGTCGACAGACCCCCAAAAGTCTTATG and AATCTCGAGACTGCAGCTAGGTGGACTTG; Δ3.4 3′-arm primers, TAAGTCGACCAGTGAGAGCAGAGGGCCAG and TTAGTCGACCCAAATGGAACAGACCACAC; Δ4.3 5′-arm primers, TATGTCGACTTCTCATAGCTTCCCATCTC and ATTCTCGAGCAGCATAGAGAAGACACTTC; Δ4.3 3′-arm primers, TTAGTCGACCAAGTCCACCTAGCTGCAGTC and TATGTCGACAGTCATTGTACCTGGCTCAG; Δ2.0 5′-arm primers, TTACTCGAGGGATCAGCTGACACCACCCAG and ATTCTCGAGAACCGGAGGGCAGGGACTGTG; Δ2.0 3′-arm primers, these primers were the same as those used for the 3′ arm of Δ4.3; Δ2.3 5′-arm primers, these primers were the same as those used for the 5′ arm of Δ4.3; and Δ2.3 3′-arm primers, ATTCTCGAGAACACAGTCCCTGCCCTCCGG and TTTCTCGAGTGCTCTCCTCAAGCTCTCCTC. Recombination substrates were linearized at a unique PvuI site in the vector backbone prior to electroporation into D(h14n)F cells.
DNA transfections.
Stable and transient transfections were performed as described previously (14). Stable transfectants were selected in DT40-conditioned medium containing 1.5 mg of l-histidinol (Sigma)/ml.
Nucleic acid isolation and blot hybridization.
Isolation of genomic DNA and Southern blot analysis were performed as described previously (37). The probes and restriction endonucleases used for the identification of homologous recombinants were as follows.
(i) D(Δ8.0) chromosome.
XbaI genomic digests were probed with a 1.27-kb BamHI-XhoI fragment from the subclone p4.7BamHI derived from αATc1 (26) that contains most of α1AT intron I and a hisD probe, which detected both wild-type (13.7 kb) and specifically modified (9.0 kb) mutant alleles.
(ii) D(Δ3.4), D(Δ4.3), and D(Δ2.0) chromosomes.
For 5′-end analysis, SpeI and XhoI genomic digests were probed with a 0.6-kb XbaI-ApaI fragment from the subclone p7.7XbaIcos10 (37), which detected both wild-type (19.9 kb) and specifically modified (15.4, 11.1, and 13.4 kb, respectively) mutant alleles. For 3′-end analysis, EcoRV and SalI genomic digests were probed with a 1.27-kb BamHI-XhoI fragment from p4.7BamHI, which detected both wild-type (19.5 kb) and specifically modified (8.0, 11.4, and 11.4 kb, respectively) mutant alleles.
(iii) D(Δ2.3) chromosome.
For 5′-end analysis, SpeI and MluI genomic digests were probed with a 0.6-kb XbaI-ApaI fragment from p7.7XbaIcos10, which detected both wild-type (19.9 kb) and specifically modified (12.8 kb) mutant alleles. For 3′-end analysis, EcoRV and XhoI genomic digests were probed with a 1.27-kb BamHI-XhoI fragment from p4.7BamHI, which detected both wild-type (13.0 kb) and specifically modified (10.0 kb) mutant alleles.
The blots were washed sequentially in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS in two 5-min washes at room temperature, in 0.2× SSC-0.1% SDS for 30 min at room temperature, and in 0.1× SSC-0.1% SDS for 30 min at 65°C. Autoradiography was carried out for 3 to 10 days at −70°C by using Kodak XAR film. Autoradiographs were scanned for reproduction by using a Umax PowerLook II scanner and Adobe Photoshop 5.5 (Adobe Systems, Inc.).
Cytoplasmic RNA was isolated from cell monolayers by using Trizol (Gibco BRL) as directed by the manufacturer. Northern blot hybridization was performed as described previously (39). The probes used for RNA blots were a human α1AT exon II probe, a 1.46-kb EcoRI fragment containing full-length human CBG cDNA (39), a rat α1AT probe amplified from rat α1AT exon II sequences, and rat cyclophilin cDNA (11).
DHS mapping.
DHS mapping assays were performed as described previously (39). Each DHS is specified by its position in kilobases on the physical map of the proximal serpin cluster relative to the α1AT hepatic transcription start site (33); this position is ∼1.9 kb 3′ of the EcoRI site previously defined as zero (39). In cases where sequences had been deleted from the various mutant chromosomes, the following alternative strategies were used for DHS mapping. The probes used have been described previously (37, 39) and are as follows: (i) for the Δ4.3 chromosome, DHS at kb −2.1, HindIII genomic digestion, 0.91 SpeI-HindIII probe, 6.0- to 3.8-kb band shift; DHS at kb −0.06, HindIII genomic digestion, 0.91 SpeI-HindIII probe, 6.0- to 1.8-kb band shift; (ii) for the Δ2.3 chromosome, DHS at kb −5.4, BglII genomic digestion, 0.77 EcoRI probe, 7.3- to 4.9-kb band shift, or HindIII genomic digestion, 2.2 HindIII probe, 1.56- to 0.87- and 0.68-kb band shift, or EcoRI genomic digestion, 2.2 HindIII probe, 2.1- to 0.9-kb band shift; DHS at kb −4.2, BglII genomic digestion, 0.77 EcoRI probe, 7.3- to 3.7-kb band shift; (iii) for the Δ3.4 chromosome, DHS at kb −4.2, BglII genomic digestion, 0.42 HindIII-ClaI probe, 6.3- to 2.9-kb band shift; DHS at kb −0.06, HindIII genomic digestion, 0.91 SpeI-HindIII probe, 2.9- to 1.8-kb band shift; (iv) for the Δ2.0 chromosome, DHS at kb −7.5, SacI genomic digestion, 0.42 HindIII-ClaI probe, 4.4- to 1.1-kb band shift; DHS at kb −6.1, BglII genomic digestion, 0.42 HindIII-ClaI probe, 4.7- to 1.1-kb band shift, or EcoRI genomic digestion, 2.2 HindIII probe, 4.7- to 2.3-kb band shift, or HindIII genomic digestion, 0.42 HindIII-ClaI probe, 6.68- to 0.89-kb band shift; DHS at kb −2.1, HindIII genomic digestion, 0.91 SpeI-HindIII probe, 6.7- to 3.8-kb band shift; DHS at kb −0.06, HindIII genomic digestion, 0.91 SpeI-HindIII probe, 6.7- to 1.8-kb band shift; (v) for the Δ8.0 chromosome, DHS at kb −0.06, HindIII genomic digestion, 0.91 SpeI-HindIII probe, 2.2- to 1.8-kb band shift.
RESULTS
Transfer of human chromosome 14 to DT40.
We used the DT40 chromosome shuttle system (14) to make precise changes in human α1AT chromosomal sequences and to analyze the effects of those modifications on gene activity and chromatin structure. To do this, we first transferred human chromosome 14 to MEL cells, which micronucleate efficiently and can be processed in large numbers, to create the mouse-human hybrid M(h14n)8. Subsequent transfer of human chromosome 14 from M(h14n)8 to DT40 cells yielded nine independent avian-human hybrids. Three of these hybrids contained human chromosome 14 rearrangements as assessed by sequence-tagged site (STS) marker analysis and fluorescent in situ hybridization (FISH) karyotyping, but six clones retained human chromosome 14 in an intact, apparently unrearranged form. One of these recombination-proficient clones, D(h14n)F, was used in all further experiments.
Deletion of sequences upstream of the α1AT hepatic promoter.
The HNF-1α/HNF-4-dependent enhancer just upstream (bp −115 to −75) of the human α1AT gene activates transcription ∼1,000-fold in hepatic cells (6, 8, 13, 21, 29, 32). According to the results of transfection tests, DNA sequences further upstream seem to play little, if any, role in cell-specific gene activation (41). However, minimal promoter/enhancer transgenes do not recapitulate fully the regulation of the corresponding chromosomal alleles (19, 22). Furthermore, the ∼8-kb region upstream of α1AT contains a cluster of four evolutionarily conserved, expression-associated DHSs (39). To test whether these sites were important for cell-specific regulation of α1AT, we deleted chromosomal sequences between kb −8.4 and −0.32.
The structure of the p(Δ8.0) targeting vector used in this experiment is shown in Fig. 1. D(h14n)F cells were transfected with linearized p(Δ8.0) by electroporation, and Geneticin-resistant transfectants were screened by Southern hybridization to identify homologous recombinants. One of the 22 Geneticin-resistant clones analyzed contained a homologous insertion (Fig. 1). This transfectant was transiently transfected with a plasmid encoding Cre recombinase, histidinol-sensitive revertants were obtained, and clones with precise hisD deletions were identified by Southern hybridization (Fig. 1). Transfectant D(Δ8.0)41, which contained the desired 8.0-kb deletion, served as a donor for transfer of the mutant chromosome to rat hepatoma cells.
Five independent hybrids were obtained after fusion of D(Δ8.0)41 with Fao-1 rat hepatoma recipient cells. Three of these hybrids contained intact human chromosomes 14 as assessed by STS marker analysis, FISH, and long-range restriction mapping within the serpin locus (Fig. 2).
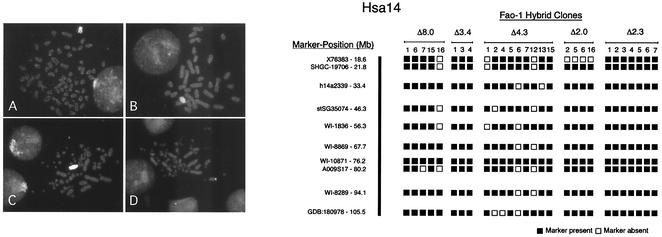
FIG. 2.
Karyotypic and marker analysis of hepatoma hybrid clones. Panels at left show FISH of hybrid clones containing human chromosome 14. Shown are karyotypes of the mouse fibroblast hybrid HDm-5 (A), mouse erythroleukemia hybrid M(h14n)8 (B), and avian hybrid D(h14n)F (C) probed with labeled total human genomic DNA and D(h14n)F (D) probed with a human α1AT locus-specific cosmid probe, αATc1. Shown at right are STS markers present in Fao-1 hybrid clones containing the modified human chromosomes 14. Five independent Δ8.0 clones, three independent Δ3.4 clones, nine independent Δ4.3 clones, four independent Δ1.9 clones, and seven Δ2.3 clones were tested. The number above each column identifies the clone within a series. Filled boxes indicate that the marker was present, and open boxes indicate that it was absent.
Expression phenotype of the Δ8.0-kb mutant allele.
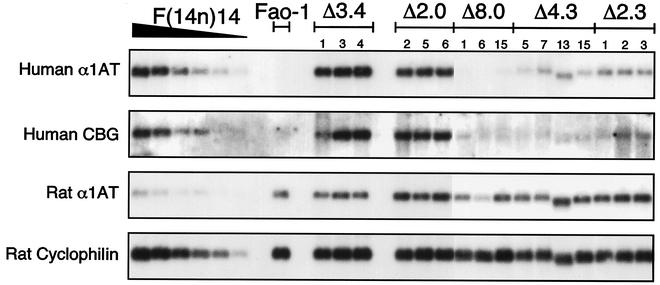
Human α1AT and CBG transcription is activated when a wild-type copy of human chromosome 14 is transferred from nonexpressing cells to rat hepatoma recipients, and the chromatin structure of the entire ∼125-kb region is reorganized to an expressing cell-typical state (39). For example, F(14n)14 cells, an Fao-1 derivative that contains a single wild-type human chromosome 14, express high levels of both human α1AT and CBG mRNAs (Fig. 3, lane 1). In marked contrast, α1AT gene activation failed to occur when the Δ8.0-kb mutant chromosome was transferred to Fao-1 cells (Fig. 3, lanes 3 to 5). These hybrids continued to express rat α1AT mRNA at parental levels, demonstrating that all of the trans-acting factors necessary for α1AT gene expression were present and active in the hybrid cells. Thus, the hybrids' inability to express human α1AT was an allele-specific phenotype. Furthermore, expression of the human CBG gene, which is ∼65 kb downstream of α1AT, was also consistently and strikingly reduced in the Δ8.0-kb chromosome-containing hybrids (Fig. 3). We estimate that human α1AT and CBG mRNA levels in these hybrids were reduced ∼20- to 50-fold relative to those in the wild type. These data indicate that chromosomal sequences in the region between kb −8.37 and −0.32 upstream of the human α1AT gene are required not only for activation of human α1AT transcription but also for CBG gene activation. This dramatic down-regulation phenotype was not observed previously in stable cosmid transfectants that did and did not contain the relevant 8.0-kb region (41). Thus, the mutant expression phenotype of the Δ8.0-kb hybrids provides evidence for the existence of DNA regulatory elements whose functions are apparent only in their normal chromosomal contexts.
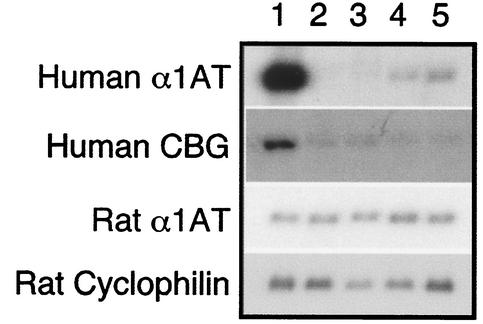
FIG. 3.
Serpin gene expression in Δ8.0-kb hybrids. RNA blot hybridization of Fao-1 hybrids containing the Δ8.0 kb chromosome. Lane designations: 1, F(14n)14; 2, Fao-1; 3, F(Δ8.0)1; 4, F(Δ8.0)6; 5, F(Δ8.0)7. The probes for human α1AT, human CBG, rat α1AT, and rat cyclophilin detected RNA transcripts of ∼1,600, 1,500, 1,400, and 1,000 nucleotides, respectively.
Chromatin structure of the Δ8.0-kb mutant allele.
The Δ8.0-kb deletion described above affects gene expression in a region of at least 100 kb, and this phenotype is sensitive to chromosomal context. These observations suggested that the chromatin structure of the mutant allele might differ from wild type. To test this possibility, DHS mapping experiments were performed.
There are 22 expression-associated and 7 constitutive DHSs in the ∼125-kb region around the human α1AT and CBG genes (39). Transfer of human chromosome 14 from nonexpressing cells to rat hepatoma cells results not only in human α1AT and CBG activation but also in the de novo formation of most of the 22 expression-associated DHSs (39). For example, Fig. 4 shows that an activated serpin allele on wild-type human chromosome 14 displays 12 DHSs in the region from kb ∼−25 to +68. Ten of these DHSs are expression associated, and two (at kb −2.1 and +48.1) are found in all cell types (38-40). In contrast, none of the expression-associated DHSs were formed when the Δ8.0-kb mutant chromosome was transferred to rat hepatoma cells (Fig. 4). The DNA sequences of five of these DHSs had been deleted from the Δ8.0 kb mutant chromosome, but there still remained the potential to form six other expression-associated DHSs at kb −24.2, −22.9, −20.9, −0.06, +11.4, and +66.9. These expression-associated DHSs failed to form on the Δ8.0-kb mutant chromosome.
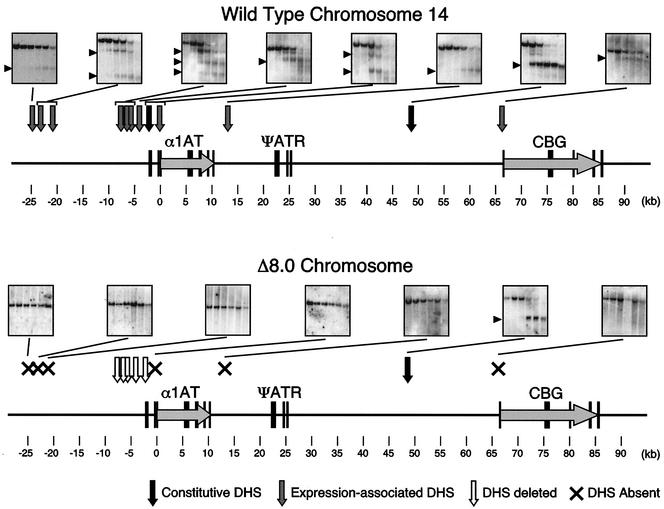
FIG. 4.
Chromatin organization of wild-type and Δ8.0-kb mutant alleles. The upper portion shows DHSs on wild-type human chromosome 14 in Fao-1 cells. The Southern blots used to assay specific DHSs are shown linked to the relevant site(s) on the map. Gray arrows denote expression-associated DHSs, and black arrows denote constitutive DHSs. The lower portion shows DHSs on the Δ8.0-kb mutant chromosome in Fao-1 cells. Only a single constitutive DHS was present (black arrow). DHSs that were removed by the deletion are shown as light arrows. All other expression-associated DHSs in the region failed to form on the Δ8.0-kb mutant chromosome (X's).
These results indicate that sequences within the Δ8.0-kb deletion are required not only for cell-specific activation of human α1AT and CBG transcription but also for the establishment of a cell-specific chromatin state, at the level of DHS formation, over a region of at least 100 kb.
Localization of regulatory elements within the 8-kb region.
The dramatic gene expression and chromatin reorganization phenotype of the Δ8.0-kb deletion raised the possibility that multiple regulatory elements might be involved. To explore this possibility and to localize the relevant regulatory regions, a series of subdeletions within the 8.0-kb interval was prepared.
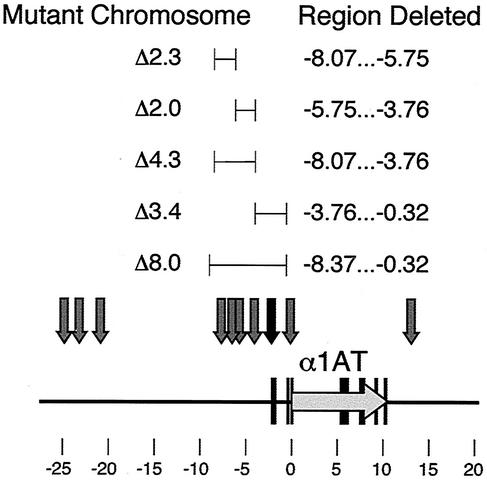
Two mutant chromosomes that subdivided the 8.0-kb interval into nearly equal halves were generated (Fig. 5). The Δ3.4-kb deletion removed the proximal half of the 8.0-kb region; it had the same 3′ boundary as the original Δ8.0-kb deletion but removed only 3.44 kb of genomic DNA upstream, including the constitutive DHS at kb −2.1. The Δ4.3-kb deletion removed the distal half of the 8.0-kb region; its 3′ boundary was at the 5′ edge of the Δ3.4-kb deletion and it extended 4.3 kb upstream. The Δ4.3 mutant allele removed all four expression-associated DHSs in the region, at kb −7.5, −6.1, −5.4, and −4.2. Two additional mutant chromosomes subdivided the 4.3-kb region further: the Δ2.0-kb deletion, which removed the two most proximal DHSs, and the Δ2.3-kb deletion, which removed the two most distal expression-associated DHSs (Fig. 5).
FIG. 5.
Schematic diagram of the human α1AT locus showing the extents of subdeletions within the Δ8.0-kb region. Gray arrows denote expression-associated DHSs; the black arrow denotes the position of a constitutive DHS.
The homology segments and targeting efficiencies of the recombination templates used to prepare the various subdeletions are summarized in Table 1. All of the recombination templates had 5′ and 3′ homology segments of ∼4 kb, and targeting efficiencies ranged from 2 to 20%.
TABLE 1.
Targeting efficiencies of recombination templates
| Targeting vector | Deletion size (kb) | 5′ arm (kb) | 3′ arm (kb) | Targeting efficiency (%) |
|---|---|---|---|---|
| p(Δ4.29) | 4.29 | 4.5 | 3.6 | 1/48 (2.0) |
| p(Δ2.30) | 2.30 | 4.5 | 3.9 | 7/44 (15.9) |
| p(Δ3.44) | 3.44 | 4.1 | 4.0 | 7/132 (5.3) |
| p(Δ1.99) | 1.99 | 3.6 | 3.9 | 9/45 (20.0) |
The hisD selection cassette in each of the four modified chromosomes was removed by Cre-mediated recombination, and the modified chromosomes were then transferred to Fao-1 cells. The integrity of the modified human chromosomes in the various hybrids was assessed by STS marker analysis, FISH, and restriction mapping within each mutant serpin allele (Fig. 2). At least three independent Fao-1 hybrids containing each mutant human chromosome 14 were analyzed further
Expression phenotypes of the Δ3.4-, Δ4.3-, Δ2.0-, and Δ2.3-kb mutant alleles.
The Δ3.4-kb deletion removed sequences immediately upstream of the hepatic α1AT promoter/enhancer (kb −3.76 to −0.32), a region that contains a constitutive DHS at kb −2.1 (Fig. 5). Each of three Fao-1 hybrids containing the Δ3.4-kb mutant chromosome expressed human α1AT and CBG mRNAs at levels comparable to wild type (Fig. 6). In contrast, the Δ4.3 deletion removed the kb −8.07 to −3.76 region (Fig. 5). Each of the four Fao-1 hybrids containing the Δ4.3-kb mutant chromosome that were analyzed expressed human α1AT and CBG mRNAs at levels that were significantly reduced from wild type. We estimate that α1AT and CBG mRNAs were reduced approximately 10- to 20-fold in these hybrid clones (Fig. 6). These observations demonstrate that regulatory elements necessary for high-level expression of human α1AT and CBG reside between kb −8.07 and −3.76, a region that contains a cluster of four expression-associated DHSs.
FIG. 6.
Serpin gene expression in Fao-1 hybrids containing various mutant serpin alleles. A serial twofold dilution series of cytoplasmic RNA from F(14n)14, a rat hepatoma hybrid containing a wild-type human chromosome 14, is shown in the lanes on the left. RNAs from parental Fao-1 cells and from three to four independent hybrid clones containing the Δ3.4, Δ2.0, Δ8.0, Δ4.3, and Δ2.3 mutant chromosomes are also shown. The number above each column identifies the clone within a series. Human α1AT and CBG expression was down-regulated in the Δ8.0, Δ4.3, and Δ2.3 series hybrids, but rat α1AT was expressed in all the hybrid clones. Rat cyclophilin served as an RNA loading control.
Two subdeletions of the 4.3-kb region were prepared: a Δ2.0-kb deletion that removed sequences between kb −5.75 and −3.76, and a Δ2.3-kb deletion that removed sequences between kb −8.07 and −5.75. The Δ2.0-kb deletion removed two expression-associated DHSs, at kb −4.2 and −5.4 (Fig. 5). However, all three Fao-1 hybrids that contained the Δ2.0-kb mutant chromosome expressed human α1AT and CBG mRNAs at wild-type levels (Fig. 6). The Δ2.3 deletion (kb −8.07 to −5.75) also removed two expression-associated DHSs, at kb −6.1 and −7.5. In three hybrids containing the Δ2.3 mutant chromosome, α1AT and CBG mRNA levels were reduced ∼10-fold relative to wild type (Fig. 6). We conclude that DNA sequences in the region between kb −8.07 and −5.75 contain regulatory elements that are required for cell-specific activation of the human α1AT and CBG genes in their normal chromosomal contexts.
Chromatin structures of the Δ3.4-, Δ4.3-, Δ2.0-, and Δ2.3-kb mutant alleles.
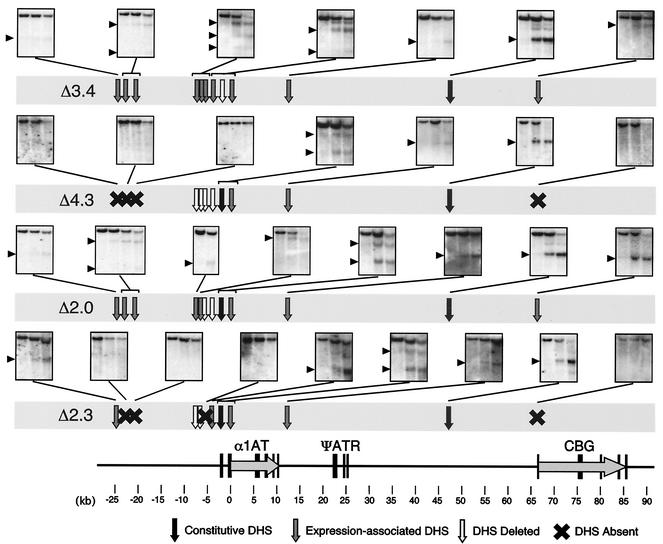
The Δ8.0-kb deletion prevented the formation of six expression-associated DHSs in the region from approximately kb −25 to +68 (Fig. 4). To map the elements required for expression-associated DHS formation, the chromatin configurations of the various subdeletions were compared.
The Δ3.4-kb deletion (kb −3.76 to −0.32) and the Δ4.3-kb deletion (kb −8.07 to −3.76) separated the 8-kb region into proximal and distal halves. Deletion of the proximal half of the 8-kb region (Δ3.4 kb) had no effect on α1AT and CBG gene expression (Fig. 6), and this mutant allele displayed a full complement of expression-associated DHSs (Fig. 7). In contrast, deletion of the distal half of the 8-kb region (Δ4.3 kb) resulted in a marked decrease in α1AT and CBG expression, although this decrease was somewhat less than that of the Δ8.0-kb allele (Fig. 6). Significantly, the Δ4.3-kb allele displayed a pattern of DHS formation that was intermediate between that of the Δ8.0-kb allele and wild type (Fig. 7). Specifically, the Δ8.0-kb allele failed to form six expression-associated DHSs, at kb −24.2, −22.9, −20.9, −0.06, +11.4, and +66.9. The Δ4.3-kb chromosome failed to display expression-associated sites far upstream (kb −24.2, −22.9, and −20.9) and downstream (kb +66.9) of α1AT, but it did form two expression-associated DHSs immediately upstream (kb −0.06, at the hepatic promoter/enhancer) and downstream (kb +11.4) of the α1AT gene. These data suggest that the Δ8.0-kb region contains multiple regulatory elements: elements within the Δ4.3-kb region are necessary for high-level α1AT and CBG transcription and for the formation of far upstream and downstream DHSs, but other elements within the Δ3.4-kb region are also required to encode the full gene expression and chromatin structure phenotype of the Δ8.0-kb allele.
FIG. 7.
Chromatin organization of the Δ3.4, Δ4.3, Δ2.0, and Δ2.3 mutant alleles. Expression-associated DHSs that were present are indicated by gray arrows, constitutive DHSs are indicated by black arrows, DHSs that were deleted are indicated by open arrows, and DHSs that failed to form are indicated by X's. The Southern blots used to assay specific DHSs are shown linked to the relevant site(s) on the map. The Δ3.4 and Δ2.0 mutant alleles had wild-type patterns of DHSs throughout the region, but the Δ4.3 and Δ2.3 mutant alleles each affected formation of a distinctive set of DHSs.
The Δ4.3-kb region was further subdivided into the Δ2.0-kb (kb −5.75 to −3.76) and Δ2.3-kb (kb −8.07 to −5.75) deletions, each of which removed two expression-associated DHSs. The Δ2.0-kb deletion transcribed the human α1AT and CBG genes at wild-type levels (Fig. 6), and all 10 expression-associated DHSs were formed on the Δ2.0-kb mutant allele (Fig. 7). In contrast, the Δ2.3-kb deletion down-regulated α1AT and CBG transcription to levels similar to those of the Δ4.3-kb allele (Fig. 6), and the Δ2.3-kb chromosome failed to form expression-associated DHSs at kb −22.9, −20.9, −5.4, and +66.9.
We conclude that the 8-kb region upstream of the human α1AT gene contains multiple regulatory elements that are necessary for transcription and chromatin remodeling of the entire proximal serpin subcluster. These elements are necessary for high-level transcription of both α1AT and CBG, as well as for the formation of expression-associated DHSs over a region of at least 100 kb. The most dominant regulatory functions map to the 2.3-kb interval between kb −8.07 and −5.75.
DISCUSSION
Human genetic loci comprise both coding sequences for specific gene products and cis elements that encode regulatory information necessary for establishing and maintaining temporal and tissue-specific gene control. These control elements are the genomic targets to which specific regulatory proteins bind, further recruiting other factors that remodel chromatin and enable gene transcription. Identifying and characterizing regulatory elements involved in both processes are important goals for postgenomics research.
The human α1AT gene has been used to study the molecular biology of tissue-specific transcription for many years, and its cell-specific enhancer and associated transcriptional activators remain important paradigms in the field (13, 21, 32). More recently, regulation of α1AT gene expression has been studied in a larger genomic context, as α1AT is now known to reside in a cluster of related serpin genes that spans a region of nearly 400 kb on human chromosome 14q32.1 (37). Many of these serpin genes are coordinately activated during hepatocyte differentiation, suggesting that they might share regulatory elements that function in transcriptional activation and/or chromatin remodeling. To explore these issues, we used a chromosome shuttle approach (14) to prepare and characterize a series of mutant serpin alleles from which specific chromosomal sequences had been deleted by homologous recombination.
The ∼8.0-kb region upstream of the minimal hepatic promoter/enhancer of α1AT contains a cluster of four evolutionarily conserved, expression-associated DHSs (39). This observation suggested that the region might contain important regulatory information. To test this possibility, we prepared a mutant chromosome 14 in which sequences between kb −8.4 and −0.32 had been deleted. When this mutant chromosome was transferred to hepatic cells, α1AT gene activation failed to occur. Furthermore, the Δ8.0-kb mutant allele failed to express human CBG, which is ∼65 kb downstream. The chromatin structure of the Δ8.0-kb mutant allele also differed from wild type, as expression-associated DHSs in the ∼100-kb region around α1AT and CBG did not form on the mutant chromosome in hepatic cells. These allele-specific phenotypes were in marked contrast to the behavior of wild-type human chromosome 14 (39).
These observations indicate that regulatory elements upstream of α1AT are required for activation of α1AT and CBG transcription and for the formation of an expressing cell-specific chromatin state in the entire ∼100-kb region. This is the first evidence that different genes in the serpin cluster share regulatory information. The serpin gene family has arisen largely by gene duplication (31), and shared regulatory elements in other gene families, including apoE, α-globin, and β-globin, have been described (1, 16, 28). As the upstream control elements in the serpin cluster affect a large genomic region, these elements may function in locus-wide regulation of gene activity and chromatin structure.
Subdeletions within the 8.0-kb segment were used to localize the implicated regulatory elements more precisely. These mutant alleles provided evidence that the major determinants of both gene activation and DHS formation mapped to a 2.3-kb region between kb −8.07 and −5.75. However, both expression data and DHS mapping experiments suggested that elements in other regions were able to interact with control elements in the 2.3-kb region, resulting in the full mutant phenotype displayed by the original Δ8.0-kb allele. For example, all of the mutations that affected α1AT expression, CBG expression, and DHS formation had the Δ2.3-kb region deleted, and deletions of other regions had no effects on gene expression or DHS formation. However, the Δ2.3-kb mutation, as well as the Δ4.3-kb mutation that contained it, did not down-regulate α1AT and CGB expression levels as dramatically as did the original Δ8.0-kb allele. Furthermore, each of these mutant alleles displayed a distinctive pattern of DHSs. Thus, it appears that there exist regulatory sequences in other regions whose effects are only apparent in combination with the Δ2.3-kb mutation. This suggests that some of the elements in the region are functionally redundant. The concept of functional redundancy and the notion that the effects of individual elements can be additive have been suggested previously in studies of both the murine (4, 18) and human (34) β-globin loci, where deletions of individual DHSs cause little change in phenotype. Further deletions of mutant serpin alleles should allow these interacting regulatory elements to be defined.
The different phenotypes encoded by the Δ8.0-, Δ4.3-, and Δ2.3-kb alleles are particularly noteworthy with respect to DHS formation, because each of these mutant alleles had a distinctive pattern of DHSs around α1AT and CBG. For example, deletion of the Δ2.3-kb region, which itself contained two DHSs, prevented the formation of DHSs at kb −22.9, −20.9, −5.4, and +66.9, but two expression-associated DHSs immediately upstream (kb −0.06) and downstream (kb +11.4) of the α1AT transcription unit were present on this mutant allele. However, all six of these expression-associated DHSs failed to form on Δ8.0-kb chromatin. To our knowledge, this is the first instance in which the formation of expression-associated DHSs in an activated locus appears to occur in a hierarchical manner.
Since the hepatic promoter/enhancer is sufficient to activate α1AT transcription in transgene experiments and removing the 8-kb upstream region causes inhibitory effects on gene expression, one function of these elements may be to establish and maintain a permissive chromosomal environment for α1AT and CBG gene expression. At other loci, notably human and mouse β-globin, the regulatory elements necessary for DHS formation and transcriptional activation seem to be distinct (35). These kinds of regulatory functions are not generally apparent in transgene experiments. For example, the dramatic gene expression and DHS formation phenotypes encoded by chromosomal sequences in the Δ8.0-kb region were not observed previously in cosmid transfectants that contained various portions of the α1AT locus (41). In these studies, the α1AT transgenes underwent positive selection via a linked expression cassette, so the transgenes were necessarily integrated into permissive chromosomal environments. This may have masked the function of the upstream control region that is required for expression and DHS formation of the chromosomal locus. These results provide another indication of the importance of studying regulatory elements in their native chromosomal contexts.
Locus-wide regulation of gene expression and chromatin structure has been well-studied at the β-globin locus. The β-globin locus control region (LCR) affects the expression of genes throughout the locus and was initially defined by its ability to confer high level, position-independent, erythroid-specific expression to linked transgenes (reviewed in reference 28). More recently, targeted mutagenesis of the murine β-globin LCR has been used to study its regulatory functions. The murine β-globin LCR consists of six DHSs distributed over a region of ∼24 kb. Deletions of individual DHSs in this region resulted in modest decreases in globin gene expression (3, 4, 18, 24), but none of the deletions affected the formation of other LCR DHSs (2). This situation is different from the human α1AT control region, which affects the formation of multiple expression-associated DHSs.
The human growth hormone (hGH) locus also contains an LCR, which is composed of five DHSs upstream of the growth hormone gene cluster. Recently, Ho et al. (23) demonstrated that deletion of the most proximal DHS of an 87-kb hGH transgene resulted in down-regulation of hGH-N gene expression in the pituitary glands of transgenic mice. This mutation also decreased histone acetylation of an ∼32-kb domain that included the hGH LCR and hGH-N promoter. Thus, like the upstream element in the human serpin locus, a dominant activity localized to a specific regulatory site. In contrast, however, the hGH DHS deletion did not affect the formation of other hGH DHSs.
The 2.3-kb region that contains the upstream regulatory element of the human serpin locus contains two expression-associated DHSs. Sequence analysis of this region (GenBank accession number NT_026437) shows that it contains consensus binding sites for C/EBP, HNF4, and HNF3. One putative HNF3 binding site maps near the DHS at kb −4.1, and it is contained within a larger 25-bp DNA element that is also found upstream of the liver-specific fetuin gene. HNF3 (FOXA) has a role in chromatin remodeling, and it is an essential factor for the transcription of many hepatic genes (10). In the serum albumin enhancer, HNF3 can affect chromatin structure directly by displacing linker histones (9), thus facilitating the binding of other transcription factors via chromatin decondensation. Other work suggests that FoxA may bind to the nucleosome directly to form an enhancersome (7). HNF3 also induces DNase I hypersensitivity in regulatory elements of the vitellogenin B1 gene when transcription is activated (36). It will be interesting to determine whether any of these mechanisms are involved in α1AT and CBG activation through the upstream control region.
Alterations in transcription and DHS formation are but two ways in which mutant serpin alleles may have changed. Other changes in chromatin structure are likely to have occurred, such as methylation, acetylation, and phosphorylation of histones and other non-histone chromosomal proteins. Furthermore, the association of specific trans-acting factors with their target sites in vivo may be affected in various mutant alleles, as may be the sequestration of altered chromosomal sequences into specific nuclear compartments. The generation and analysis of specifically modified human chromosomal genes provides an experimental inroad into all of these regulatory processes.
Acknowledgments
We thank Mark Groudine and Barb Trask for their comments on the manuscript.
This study was supported by grant GM26449 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Allan, C. M., S. Taylor, and J. M. Taylor. 1997. Two hepatic enhancers, HCR.1 and HCR.2, coordinate the liver expression of the entire human apolipoprotein E/C-I/C-IV/C-II gene cluster. J. Biol. Chem. 272:29113-29119. [DOI] [PubMed] [Google Scholar]
- 2.Bender, M. A., M. G. Mehaffey, A. Telling, B. Hug, T. J. Ley, M. Groudine, and S. Fiering. 2000. Independent formation of DNase I hypersensitive sites in the murine beta-globin locus control region. Blood 95:3600-3604. [PubMed] [Google Scholar]
- 3.Bender, M. A., A. Reik, J. Close, A. Telling, E. Epner, S. Fiering, R. Hardison, and M. Groudine. 1998. Description and targeted deletion of 5′ hypersensitive site 5 and 6 of the mouse beta-globin locus control region. Blood 92:4394-4403. [PubMed] [Google Scholar]
- 4.Bender, M. A., J. N. Roach, J. Halow, J. Close, R. Alami, E. E. Bouhassira, M. Groudine, and S. N. Fiering. 2001. Targeted deletion of 5′HS1 and 5′HS4 of the beta-globin locus control region reveals additive activity of the DNase I hypersensitive sites. Blood 98:2022-2027. [DOI] [PubMed] [Google Scholar]
- 5.Buerstedde, J. M., and S. Takeda. 1991. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell 67:179-188. [DOI] [PubMed] [Google Scholar]
- 6.Bulla, G. A. 1999. Extinction of α1-antitrypsin expression in cell hybrids is independent of HNF1α and HNF4 and involves both promoter and internal DNA sequences. Nucleic Acids Res. 27:1190-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaya, D., T. Hayamizu, M. Bustin, and K. S. Zaret. 2001. Transcription factor FoxA (HNF3) on a nucleosome at an enhancer complex in liver chromatin. J. Biol. Chem. 276:44385-44389. [DOI] [PubMed] [Google Scholar]
- 8.Ciliberto, G., L. Dente, and R. Cortese. 1985. Cell-specific expression of a transfected human alpha 1-antitrypsin gene. Cell 41:531-540. [DOI] [PubMed] [Google Scholar]
- 9.Cirillo, L. A., and K. S. Zaret. 1999. An early developmental transcription factor complex that is more stable on nucleosome core particles than on free DNA. Mol. Cell 4:961-969. [DOI] [PubMed] [Google Scholar]
- 10.Costa, R. H., D. R. Grayson, and J. E. J. Darnell. 1989. Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and alpha 1-antitrypsin genes. Mol. Cell. Biol. 9:1415-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danielson, P. E., S. Forss-Petter, M. A. Brow, L. Calavetta, J. Douglass, R. J. Milner, and J. G. Sutcliffe. 1988. p1B15: a cDNA clone of the rat mRNA encoding cyclophilin. DNA 7:261-267. [DOI] [PubMed] [Google Scholar]
- 12.Deschatrette, J., and M. C. Weiss. 1975. Extinction of liver-specific functions in hybrids between differentiated and dedifferentiated rat hepatoma cells. Somatic Cell Genet. 1:279-292. [DOI] [PubMed] [Google Scholar]
- 13.De Simone, V., G. Ciliberto, E. Hardon, G. Paonessa, F. Palla, L. Lundberg, and R. Cortese. 1987. cis- and trans-acting elements responsible for the cell-specific expression of the human alpha 1-antitrypsin gene. EMBO J. 6:2759-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dieken, E. S., E. M. Epner, S. Fiering, R. E. K. Fournier, and M. Groudine. 1996. Efficient modification of human chromosomal alleles using recombination-proficient chicken/human microcell hybrids. Nat. Genet. 12:174-182. [DOI] [PubMed] [Google Scholar]
- 15.Dycaico, M. J., S. G. Grant, K. Felts, W. S. Nichols, S. A. Geller, J. H. Hager, A. J. Pollard, S. W. Kohler, H. P. Short, F. R. Jirik, D. Hanahan, and J. A. Sorge. 1988. Neonatal hepatitis induced by alpha 1-antitrypsin: a transgenic mouse model. Science 242:1409-1412. [DOI] [PubMed] [Google Scholar]
- 16.Engel, J. D., and K. Tanimoto. 2000. Looping, linking, and chromatin activity: new insights into beta-globin locus regulation. Cell 100:499-502. [DOI] [PubMed] [Google Scholar]
- 17.Eszterhas, S. K., E. E. Bouhassira, D. I. Martin, and S. Fiering. 2002. Transcriptional interference by independently regulated genes occurs in any relative arrangement of the genes and is influenced by chromosomal integration position. Mol. Cell. Biol. 22:469-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiering, S., E. Epner, K. Robinson, Y. Zhuang, A. Telling, M. Hu, D. I. Martin, T. Enver, T. J. Ley, and M. Groudine. 1995. Targeted deletion of 5′HS2 of the murine beta-globin LCR reveals that it is not essential for proper regulation of the beta-globin locus. Genes Dev. 9:2203-2213. [DOI] [PubMed] [Google Scholar]
- 19.Grosveld, F. 1999. Activation by locus control regions? Curr. Opin. Genet. Dev. 9:152-157. [DOI] [PubMed] [Google Scholar]
- 20.Han, X., Z. F. Huang, R. Fiehler, and G. J. Broze, Jr. 1999. The protein Z-dependent protease inhibitor is a serpin. Biochemistry 38:11073-11078. [DOI] [PubMed] [Google Scholar]
- 21.Hardon, E. M., M. Frain, G. Paonessa, and R. Cortese. 1988. Two distinct factors interact with the promoter regions of several liver-specific genes. EMBO J. 7:1711-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgs, D. R., J. A. Sharpe, and W. G. Wood. 1998. Understanding alpha globin gene expression: a step towards effective gene therapy. Semin. Hematol. 35:93-104. [PubMed] [Google Scholar]
- 23.Ho, Y., F. Elefant, N. Cooke, and S. Liebhaber. 2002. A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Mol. Cell 9:291-302. [DOI] [PubMed] [Google Scholar]
- 24.Hug, B. A., R. L. Wesselschmidt, S. Fiering, M. A. Bender, E. Epner, M. Groudine, and T. J. Ley. 1996. Analysis of mice containing a targeted deletion of beta-globin locus control region 5′ hypersensitive site 3. Mol. Cell. Biol. 16:2906-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imhof, A., and A. P. Wolffe. 1998. Transcription: gene control by targeted histone acetylation. Curr. Biol. 8:R422-R424. [DOI] [PubMed] [Google Scholar]
- 26.Kelsey, G. D., S. Povey, A. E. Bygrave, and R. H. Lovell-Badge. 1987. Species- and tissue-specific expression of human alpha 1-antitrypsin in transgenic mice. Genes Dev. 1:161-171. [DOI] [PubMed] [Google Scholar]
- 27.Koopman, P., S. Povey, and R. H. Lovell-Badge. 1989. Widespread expression of human alpha 1-antitrypsin in transgenic mice revealed by in situ hybridization. Genes Dev. 3:16-25. [DOI] [PubMed] [Google Scholar]
- 28.Li, Q., S. Harju, and K. R. Peterson. 1999. Locus control regions: coming of age at a decade plus. Trends Genet. 15:403-408. [DOI] [PubMed] [Google Scholar]
- 29.Li, Y., L. Zhou, S. S. Twinings, J. Sugar, and B. Y. J. T. Yue. 1998. Involvement of Sp1 elements in the promoter activity of the α1-proteinase inhibitor gene. J. Biol. Chem. 273:9959-9965. [DOI] [PubMed] [Google Scholar]
- 30.Lugo, T. G., B. Handelin, A. M. Killary, D. E. Housman, and R. E. K. Fournier. 1987. Isolation of microcell hybrid clones containing retroviral vector insertions into specific human chromosomes. Mol. Cell. Biol. 7:2814-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marshall, C. J. 1993. Evolutionary relationships among the serpins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 342:101-119. [DOI] [PubMed] [Google Scholar]
- 32.Monaci, P., A. Nicosia, and R. Cortese. 1988. Two different liver-specific factors stimulate in vitro transcription from the human alpha 1-antitrypsin promoter. EMBO J. 7:2075-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perlino, E., R. Cortese, and G. Ciliberto. 1987. The human alpha 1-antitrypsin gene is transcribed from two different promoters in macrophages and hepatocytes. EMBO J. 6:2767-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterson, K. R., C. H. Clegg, P. A. Navas, E. J. Norton, T. G. Kimbrough, and G. Stamatoyannopoulos. 1996. Effect of deletion of 5′HS3 or 5′HS2 of the human beta-globin LCR on the developmental regulation of globin expression in beta-YAC transgenic mice. Proc. Natl. Acad. Sci. USA 93:6605-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reik, A., A. Telling, G. Zitnik, D. Cimbora, E. Epner, and M. Groudine. 1998. The locus control region is necessary for gene expression in the human beta-globin locus but not in the maintenance of an open chromatin structure in erythroid cells. Mol. Cell. Biol. 18:5992-6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robyr, D., A. Gegonne, A. P. Wolffe, and W. Wahli. 2000. Determinants of vitellogenin B1 promoter architecture. J. Biol. Chem. 275:28291-28300. [DOI] [PubMed] [Google Scholar]
- 37.Rollini, P., and R. E. K. Fournier. 1997. A 370-kb cosmid contig of the serpin gene cluster on human chromosome 14q32.1: molecular linkage of the genes encoding alpha 1-antichymotrypsin, protein C inhibitor, kallistatin, alpha 1-antitrypsin, and corticosteroid-binding globulin. Genomics 46:409-415. [DOI] [PubMed] [Google Scholar]
- 38.Rollini, P., and R. E. K. Fournier. 2000. Differential regulation of gene activity and chromatin structure within the human serpin gene cluster at 14q32.1 in macrophage microcell hybrids. Nucleic Acids Res. 28:1767-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rollini, P., and R. E. K. Fournier. 1999. Long-range chromatin reorganization of the human serpin gene cluster at 14q32.1 accompanies gene activation and extinction in microcell hybrids. Genomics 56:22-30. [DOI] [PubMed] [Google Scholar]
- 40.Rollini, P., L. Xu, and R. E. K. Fournier. 1999. Partial activation of gene activity and chromatin remodeling of the human 14q32.1 serpin gene cluster by HNF-1α and HNF-4 in fibroblast microcell hybrids. Somat. Cell Mol. Genet. 25:207-221. [DOI] [PubMed] [Google Scholar]
- 41.Rollini, P., L. Xu, and R. E. K. Fournier. 2000. Stable expression and cell-specific chromatin structure of human α1-antitrypsin cosmid transgenes in rat hepatoma cells. Nucleic Acids Res. 28:3605-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruther, U., M. Tripodi, R. Cortese, and E. F. Wagner. 1987. The human α1-antitrypsin gene is efficiently expressed from two tissue-specific promoters in transgenic mice. Nucleic Acids Res. 15:7519-7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schubeler, D., C. Francastel, D. M. Cimbora, A. Reik, D. I. K. Martin, and M. Groudine. 2000. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 14:940-950. [PMC free article] [PubMed] [Google Scholar]
- 44.Shapero, M. H., A. A. Langston, and R. E. K. Fournier. 1994. Tissue-specific extinguisher loci in the human genome: a screening study based on random marking and transfer of human chromosomes. Somat. Cell Mol. Genet. 20:215-231. [DOI] [PubMed] [Google Scholar]
- 45.Shen, R. F., S. M. Clift, J. L. DeMayo, M. J. Finegold, and S. L. Woo. 1989. Tissue-specific regulation of human alpha 1-antitrypsin gene expression in transgenic mice. DNA 8:101-108. [DOI] [PubMed] [Google Scholar]
- 46.Sifers, R. N., J. A. Carlson, S. M. Clift, F. J. DeMayo, D. W. Bullock, and S. L. Woo. 1987. Tissue specific expression of the human α1-antitrypsin gene in transgenic mice. Nucleic Acids Res. 15:1459-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tripodi, M., C. Abbott, N. Vivian, R. Cortese, and R. Lovell-Badge. 1991. Disruption of the LF-A1 and LF-B1 binding sites in the human α1-antitrypsin gene has a differential effect during development in transgenic mice. EMBO J. 10:3177-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weintraub, H., and M. Groudine. 1976. Chromosomal subunits in active genes have an altered conformation. Science 193:848-856. [DOI] [PubMed] [Google Scholar]
- 49.Yull, F. E., R. M. Wallace, and A. J. Clark. 1995. Restricted tissue-specific but correct developmental expression mediated by a short human alpha 1AT promoter fragment in transgenic mice. Transgenic Res. 4:70-74. [DOI] [PubMed] [Google Scholar]