Abstract
Tumor necrosis factor alpha (TNF-α) expression is regulated by transcriptional as well as posttranscriptional mechanisms, the latter including the control of mRNA decay through an AU-rich element (ARE) in the 3′ untranslated region (UTR). Using two mutant cell lines deficient for ARE-mediated mRNA decay, we provide evidence for a second element, the constitutive decay element (CDE), which is also located in the 3′ UTR of TNF-α. In stably transfected RAW 264.7 macrophages stimulated with lipopolysaccharide (LPS), the CDE continues to target a reporter transcript for rapid decay, whereas ARE-mediated decay is blocked. Similarly, the activation of p38 kinase and phosphatidylinositol 3-kinase in NIH 3T3 cells inhibits ARE-mediated but not CDE-mediated mRNA decay. The CDE was mapped to an 80-nucleotide (nt) segment downstream of the ARE, and point mutation analysis identified within the CDE a conserved sequence of 15 nt that is required for decay activity. We propose that the CDE represses TNF-α expression by maintaining the mRNA short-lived, thereby preventing excessive induction of TNF-α after LPS stimulation. Thus, CDE-mediated mRNA decay is likely to be an important mechanism limiting LPS-induced pathologic processes.
Tumor necrosis factor alpha (TNF-α) is a major mediator of inflammation and plays a pivotal role during the early phase of a host's reaction to infection or injury. Localized and transient secretion of TNF-α is physiological, whereas persistent production occurs in certain autoimmune diseases and can lead to severe tissue damage. Systemically released TNF-α is a potent mediator of septic shock induced by gram-negative bacteria. Macrophages stimulated by bacterial lipopolysaccharide (LPS) are regarded as the major source of TNF-α during this process (16, 40). Regulatory mechanisms that interfere at virtually every step of its synthesis ensure that the production of this potentially harmful cytokine is tightly controlled.
Transcriptional activation of TNF-α occurs in response to LPS through the binding of NF-κB and other transcription factors to the TNF-α promoter (9, 25, 35, 41, 49). Lymphocytes store a pool of unspliced TNF-α pre-mRNAs which are spliced only upon cell activation in a protein kinase R-dependent manner (15, 27, 48). The TNF-α transcript contains in its 3′ untranslated region (UTR) an AU-rich element (ARE) which interacts with a variety of ARE-binding proteins that regulate the transport of mRNA, its decay rate in the cytoplasm, and translation efficiency (1, 34, 43). In nonstimulated macrophages, spliced TNF-α mRNA appears to be retained in the nucleus by an ARE-dependent mechanism. LPS specifically induces export of the mRNA into the cytoplasm through a Tpl2- and extracellular signal-regulated kinase (ERK)-dependent pathway, as demonstrated with Tpl2-deficient mice, which exhibit a defective LPS response with the nuclear retention of TNF-α mRNA (11).
Once the mRNA is in the cytoplasm, its rate of decay is stringently controlled. In lymphocytes stimulated by the CD3 receptor complex, TNF-α mRNA is very labile, while costimulation by CD3 and CD28 leads to marked stabilization (20). In macrophages, the issue is controversial, as some investigators have reported stabilization of the mRNA upon LPS treatment (2, 14), whereas others have not seen differences in the mRNA half-life (13, 29). When inserted into reporter constructs, the ARE of TNF-α induces processive deadenylation of the mRNA and efficiently targets the body of the transcript for rapid degradation (6, 39). Decay is mediated through binding of the zinc finger protein tristetraprolin (TTP) to the ARE, as shown with TTP-deficient mice, which suffer from elevated levels of TNF-α due to an increase in the mRNA half-life (5).
In addition to regulating mRNA decay, the ARE can cause silencing at the level of translation (13, 29). TIA-1, an RNA-binding protein with a high affinity for the TNF-α ARE, is required for translational silencing. This requirement was shown in macrophages from TIA-1-deficient mice; compared to control macrophages, these macrophages expressed similar mRNA levels but significantly higher TNF-α protein levels (28). Notably, TNF-α mRNA was shifted from the monosomal ribosome fractions to the polysomal fractions in TIA-1−/− macrophages, while the rate of mRNA decay was not altered. Translational silencing is overcome in LPS-stimulated macrophages through a mitogen-activated protein kinase-activated protein (MAPKAP) kinase 2-dependent signaling pathway (18). Finally, an inhibitory mechanism has been found in macrophages subjected to prolonged treatment with LPS; a second exposure to LPS fails to induce TNF-α secretion. As intracellular mRNA and protein levels are still upregulated, repression appears to occur at the level of secretion during the refractory phase (50).
With the aim of characterizing mechanisms that regulate the stability of cytokine mRNAs, three mutant cell lines (slowA, slowB, and slowC) defective for ARE-mediated mRNA decay were previously isolated from mutagenized human HT1080 fibrosarcoma cells (38). Mutant slowC was recently identified to harbor a frameshift mutation in both alleles of the gene encoding butyrate response factor 1 (BRF1). BRF1 is a zinc finger protein homologous to TTP, and it also promotes ARE-dependent mRNA decay (36). Mutant slowA was classified by cell fusion into a complementation group different from that of slowB or slowC, but its lesion is currently unknown. Both slowA and slowC fail to degrade reporter transcripts containing AREs of various cytokines (including TNF-α, interleukin 2 [IL-2], IL-3, IL-6, and granulocyte-macrophage colony-stimulating factor [GM-CSF]), implying that the mutants define a common ARE-dependent decay pathway (39). An unexpected observation was that the entire 3′ UTR of TNF-α was still capable of stimulating decay in the mutant background (38). This finding indicated that the TNF-α 3′ UTR is the target of a second mRNA degradation pathway which is independent of ARE-mediated decay. In this report, we identified and characterized the corresponding mRNA-destabilizing element. It is located immediately downstream of the ARE and is unresponsive to signals which stabilize ARE-containing mRNAs. For this reason, we designated the novel element a constitutive decay element (CDE).
MATERIALS AND METHODS
Plasmid construction.
Plasmids puroMXβglobin, puroMXβ-IL-3-ARE, and puroMXβ-TNF-α-ARE (termed puroMXβ-TNF-α-ARE53) were described elsewhere (39). Construct neoMXβ-TNF-α-UTR has been published as neoMXβglobin-TNF-α (38). To generate neoMXβ-IL-3-UTR, the hph resistance gene was excised as an XmnI-ClaI fragment from MXh-β-globin-IL-3 (22) and replaced by the neomycin resistance gene-containing XmnI-AccI fragment of pGEMneo (38). To generate MXh-β-TNF-α-UTR, the murine TNF-α 3′ UTR was amplified with primers M1750 and M1751 (38), subcloned into pSP73 (Promega), and subsequently ligated into the BglII sites of MXh-β-globin-IL-3, thereby replacing the IL-3 3′ UTR.
To generate the β-globin reporter constructs, different portions of the murine TNF-α 3′ UTR were amplified from plasmid neoMXβ-TNF-α-UTR by PCR with upstream BamHI and downstream BglII linker primers. The fragments were ligated into the BglII site of puroMXβglobin, a process which placed the elements in between the stop codon and the 70-nucleotide (nt) β-globin 3′ UTR. The following primers were used: M2161 and M2162 for plasmid puroMXβ-TNF-α-I, M2163 and M2164 for puroMXβ-TNF-α-II, M2182 and M2164 for puroMXβ-TNF-α-A, M2183 and M2164 for puroMXβ-TNF-α-B, M2184 and M2164 for puroMXβ-TNF-α-C, M2163 and M2185 for puroMXβ-TNF-α-D, M2163 and M2187 for puroMXβ-TNF-α-F, M2183 and M2185 for puroMXβ-TNF-α-I, M2163 and M2201 for puroMXβ-TNF-α-J, M2163 and M2297 for puroMXβ-TNF-α-K, M2198 and M2297 for puroMXβ-TNF-α-KΔAU, M2206 and M2297 for puroMXβ-TNF-α-Km2, M2163 and TV41 for puroMXβ-TNF-α-Km5, M2163 and TV38 for puroMXβ-TNF-α-Km6, and M2163 and TV39 for puroMXβ-TNF-α-Km7. For puroMXβ-TNF-α-L, oligonucleotides M2354 and M2355 were annealed and ligated directly into the BglII site of puroMXβglobin. For puroMXβ-TNF-α-JΔ15, the sequences upstream and downstream of the deletion were amplified by PCR with puroMXβ-TNF-α-J as a template and primer pairs M2016-M2253 and M2256-M1505. The fragments were digested with EcoRI/XbaI and XbaI/BglII, respectively, and cloned into the EcoRI/BglII sites of puroMXβglobin in a single step. For puroMXβ-TNF-α-IIΔ15, primers M2256 and M2164 were used to amplify the downstream portion. The fragment was digested with XbaI/BglII and ligated together with the EcoRI-XbaI fragment of puroMXβ-TNF-α-JΔ15 into the EcoRI/BglII sites of puroMXβglobin.
For the green fluorescent protein (GFP) reporter constructs, a SacII-SmaI fragment from pLXSN (Clontech) containing the Moloney murine leukemia virus (MMLV) promoter was first ligated into the SacII/SmaI sites of pEGFP-N1 (Clontech). From this plasmid, MMLV-GFP was ligated as a HindIII-XbaI fragment into the HindIII/XbaI sites of MXhIL3-wt (37) to generate plasmid MoGFPwt containing the hph selection marker. The EcoRI-SalI fragment of puroMXβglobin was then blunt-end ligated into the XbaI/ClaI sites of MoGFPwt to generate MXhGFP-control. In the same way, the EcoRI-SalI fragments of puroMXβ-TNF-α-II and puroMXβ-TNF-α-ARE were used to construct MXhGFP-TNF-α-II and MXhGFP-TNF-α-ARE, respectively. For pBABEpuro-GFP-control, pBABEpuro-GFP-TNF-α-II, and pBABEpuro-GFP-TNF-α-ARE, the BamHI-EcoRI fragments were excised from MXhGFP-control, MXhGFP-TNF-α-II, and MXhGFP-TNF-α-ARE, respectively, and ligated into the BamHI/EcoRI sites of pBABEpuro (24).
Primers.
The primers used were as follows: M1505 (5′-AAGGGGCTTCATGATGTCC-3′), M1687 (5′-TGACCTCAGCGCTGAGTTGG-3′), M1689 (5′-ACAAGGCTGCCCCGACTACG-3′), M2016 (5′-GTGCTGGTTATTGTGCTG-3′), M2161 (5′-ATGGATCCAGGGAATGGGTG-3′), M2162 (5′-ATGGATCCGGGGGGCTG-3′), M2163 (5′-ATGGATCCCTTATGAATGTATT-3′), M2164 (5′-ATAGATCTCAATTGACTGTAG-3′), M2182 (5′-ATGGATCCAGACATGTTTTCTG-3′), M2183 (5′-ATGGATCCCCTCCTCTTTTGC-3′), M2184 (5′-ATGGATCCCGCTACATCACTG-3′), M2185 (5′-ATAGATCTACAGCCTGGTCAC-3′), M2187 (5′-ATAGATCTTGTCTGAAGACAG-3′), M2198 (5′-ATGGATCCGGAAGGCCGG-3′), M2201 (5′-ATTGGGTTAGATAGATCTTTTG-3′), M2206 (5′-ATGGATCCCTTATGAATGTAGGTAGGTGGAAG-3′), M2253 (5′-ATTCTAGAATGTCTGTCTGAAGAC-3′), M2354 (5′-GATCCAGACATGTTTTCTGTGAAAACGGAGCTGA-3′), M2355 (5′-GATCTCAGCTCCGTTTTCACAGAAAACATGTCTG-3′), M2256 (5′-ATTCTAGAGGAGCTGAGCTGTC-3′), M2297 (5′-TGGGGAGATCTCAGCTCCG-3′), TV38 (5′-AAAGATCTCAGCTCCGTTTTCACAGTTATCATGTC-3′), TV39 (5′-AAAGATCTCAGCTCCGATAACACAGAAAACATGTC-3′), and TV41 (5′-AAAGATCTCAGCTCCGTTTTCTGAGAAAACATGTC-3′).
Cell lines and transfection.
The cell line termed wt is a human HT1080 fibrosarcoma cell line which expresses a hybrid GFP- IL-3 reporter gene (38). Mutants slowA and slowC are derivatives of cell line wt and were described previously (38). Transfection of HT1080 and RAW 264.7 cells was performed with Lipofectamine 2000 and Optimem medium (Life Technologies) in accordance with the manufacturer's protocol. Puromycin (2 μg/ml) or G418 (1 mg/ml) was added 48 h later to select for stably transfected cells. After selection, RAW 264.7 cells were subcloned in microtiter plates, and clones with detectable GFP expression were identified by fluorescence-activated cell sorting (FACS) analysis (38). All cells were maintained in Iscove's modified Dulbecco medium supplemented with 10% fetal calf serum, 50 μM 2-mercaptoethanol, 2 mM glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml.
NIH 3T3 B2A2 cells (47) were transiently transfected by calcium phosphate precipitation (33). At 12 to 16 h after transfection, the medium was changed to a low serum concentration (0.5%) for another 24 h prior to RNA extraction. For the activation of signal transduction pathways, β-globin reporter constructs were transiently cotransfected with vector alone, plasmid SRα3-MEK6DD (22), rCD2-p110 (30), or v-H-ras (26). Alternatively, cells were treated with12-O-tetradecanoylphorbol-13-acetate (TPA) (10 ng/ml) or ionomycin (5 μM) 30 min prior to the addition of actinomycin D.
Actinomycin D chase and Northern blot analysis.
RAW 264.7 macrophages were stimulated with LPS (5 μg/ml) purified from Escherichia coli O26:B6 (Sigma L3755). Upon the addition of actinomycin D (5 μg/ml), total cytoplasmic RNA was extracted from the cells at different times and subjected to Northern blot analysis as described previously (38). A 32P-labeled SP6 probe from the 86-bp BglII-EcoRI fragment of rabbit β-globin was used for the detection of β-globin reporter mRNA. Blots were rehybridized with an SP6 probe from a 260-bp fragment of murine β-actin (39), a randomly primed probe from a 640-bp fragment of the puromycin N-acetyltransferase gene (39), or an SP6 probe from a 96-bp EcoRI-PstI fragment of the hph gene (37). An SP6 probe for murine TNF-α was transcribed from a 250-nt fragment of exon 4 that was amplified with primers M1689 and M1687. The template for the GM-CSF SP6 probe was obtained from A. P. K. Nair (unpublished data). All blots were exposed on a phosphorimager (Bio-Rad), and signal intensities were quantified with the help of Quantity One software. β-Globin mRNA half-lives were calculated by linear regression of values normalized to the level of expression of the actin, puromycin N-acetyltransferase, or hph gene.
RESULTS
mRNA decay mediated by the TNF-α 3′ UTR involves an ARE-independent pathway.
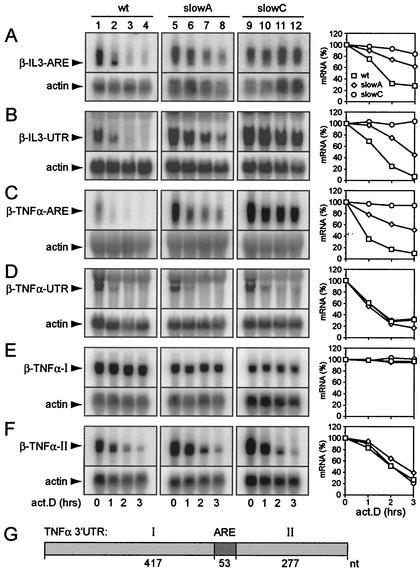
In order to systematically analyze mRNA decay mediated by regulatory sequences of murine IL-3 and TNF-α, we compared the stabilities of β-globin reporter transcripts carrying either the entire 3′ UTR of the two cytokines (β-TNF-α-UTR and β-IL-3-UTR) or their AREs only (β-TNF-α-ARE and β-IL-3-ARE). The reporter constructs were stably transfected into wt cells as well as into mutants slowA and slowC, both of which are deficient for ARE mRNA decay. Degradation of the reporter transcripts was examined by actinomycin D chase experiments and Northern blot analysis. Both the β-IL-3-ARE and the β-IL-3-UTR transcripts were partially stabilized in slowA and strongly stabilized in slowC (Fig. 1A and B). The β-TNF-α-ARE transcript showed the same degree of stabilization in the mutants (Fig. 1C). The β-TNF-α-UTR transcript, however, remained labile in both slowA and slowC (Fig. 1D). These results indicated that the TNF-α 3′ UTR contains an additional destabilizing activity.
FIG. 1.
Comparison of reporter mRNA degradation mediated by regulatory sequences in the 3′ UTRs of IL-3 and TNF-α. (A to F) HT1080 wt cells, as well as cells of slowA and slowC, two derivative mutants deficient for ARE-dependent mRNA decay, were stably transfected with the following β-globin reporter constructs: puroMXβ-IL-3-ARE (A), neoMXβ-IL-3-UTR (B), puroMXβ-TNF-α-ARE (C), neoMXβ-TNF-α-UTR (D), puroMXβ-TNF-α-I (E), and puroMXβ-TNF-α-II (F). The mRNA decay rates were determined by actinomycin D (act.D) chase experiments and Northern blot analysis. Twenty-five micrograms of total RNA per time point was resolved by formaldehyde- 1.1% agarose gel electrophoresis. The reporter transcripts were detected by using a radiolabeled SP6 probe specific for β-globin exon 3. The membranes were rehybridized with a β-actin probe to control for loading. β-Globin mRNA signal intensities were quantified by phosphorimaging, normalized to the actin signal intensity, and plotted as a percentage of the initial value against time (graphs at right). (G) Schematic representation of the 747-nt 3′ UTR of murine TNF-α divided into region I, the ARE, and region II.
To locate the predicted cis-acting element, the 3′ UTR of TNF-α was divided into three regions: the centrally located ARE, 5′-terminal 417-nt region I, and 3′-terminal 277-nt region II (Fig. 1G). Regions I and II were inserted separately into the 3′ UTR of the β-globin reporter gene to examine their effects on mRNA decay. No degradation was observed with the reporter transcript containing region I (Fig. 1E). On the other hand, region II did promote decay of the reporter transcript (Fig. 1F). Importantly, the β-TNF-α-II transcript was also rapidly degraded in the mutant cell lines, demonstrating that decay induced by region II is ARE independent. Taken together, these results indicated that the turnover of TNF-α mRNA is controlled by two distinct decay pathways.
GFP reporter gene expression in RAW 264.7 macrophages.
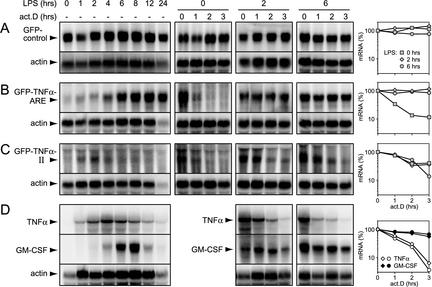
HT1080 fibrosarcoma cells and their derivative mutants slowA and slowC may not be an appropriate model for posttranscriptional mechanisms that regulate the physiological expression of TNF-α. Therefore, we sought evidence for an ARE-independent mRNA decay pathway in the murine macrophage line RAW 264.7, which has been widely used to study the induction of TNF-α in response to LPS. A GFP reporter gene was linked to the β-globin 3′ UTR into which either no decay element (control), region II, or the ARE had been inserted. Upon stable transfection into RAW 264.7 cells, subclones in which GFP expression was readily detectable were selected. The expression of GFP control mRNA was not altered appreciably over a 24-h period of LPS treatment (Fig. 2A, left panel). This result implied that the MMLV long terminal repeat promoter was neither activated nor repressed by LPS; these characteristics made the construct suitable for monitoring posttranscriptional changes in expression. Introduction of the ARE greatly reduced the steady-state level of reporter mRNA and allowed for a strong induction of expression by LPS treatment (Fig. 2B, left panel). Induced levels reached a plateau after 6 to 8 h and remained high for up to 24 h. Upon blocking of transcription with actinomycin D, the mRNA decayed rapidly in untreated cells (Fig. 2B, middle panel [0 h LPS]). Note that the blots had to be overexposed to visualize decay of the mRNA in untreated cells. Stimulation with LPS for 2 or 6 h caused strong stabilization of the ARE mRNA (Fig. 2B, middle and right panels). With region II, a drastic reduction of steady-state mRNA expression again was noted. In response to LPS, however, a much weaker, earlier (maximum after 2 h), and transient induction was observed (Fig. 2C, left panel). Importantly, the mRNA was labile not only in nonstimulated cells but also after 2 or 6 h of LPS treatment (Fig. 2C, middle and right panels). Again, the blots had to be overexposed to monitor mRNA decay, resulting in a higher background and making quantification more difficult. Nevertheless, it was apparent that region II targets the mRNA for degradation throughout the entire period of induction. The same results were obtained with another ARE clone and two other region II clones (data not shown), excluding the possibility that the differences seen in Fig. 2 were due to clonal variations. We concluded that region II contains a destabilizing sequence that is also active in macrophages. Because of its constitutive decay activity during LPS stimulation, we termed it the CDE.
FIG. 2.
Analysis of ARE- and CDE-dependent mRNA expression in LPS-stimulated RAW 264.7 macrophages. (A to C) Cells were stably transfected with the following reporter constructs: pBABEpuro-GFP-control, in which GFP is linked to the β-globin 3′ UTR alone (A); pBABEpuro-GFP-TNF-α-ARE, with the TNF-α ARE inserted into the β-globin 3′ UTR (B); and pBABEpuro-GFP-TNF-α-II, containing region II of TNF-α inserted into the β-globin 3′ UTR (C). Subclones expressing detectable reporter mRNA levels were isolated and treated for 1 to 24 h with LPS (5 μg/ml) prior to RNA extraction and Northern blot analysis. The mRNA decay rates were determined by actinomycin D (act.D) chase experiments after 0, 2, and 6 h of LPS treatment and quantified as described in the legend to Fig. 1. (D) The expression of endogenous TNF-α and GM-CSF mRNAs was monitored in nontransfected RAW 264.7 cells in response to LPS stimulation. The mRNA decay rates were measured after 2 and 6 h of LPS treatment and quantified.
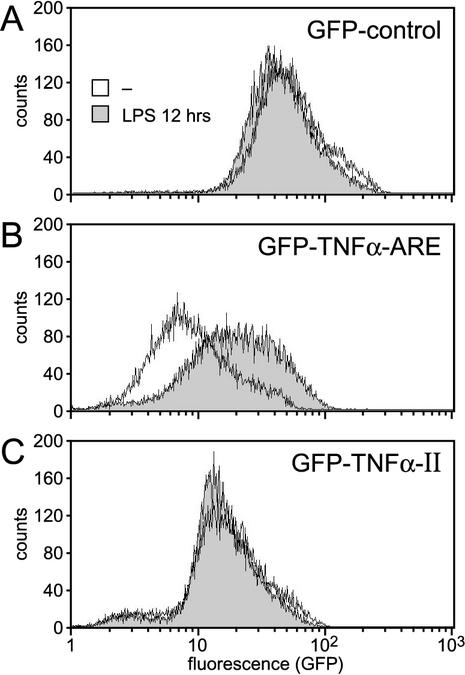
To investigate whether the differences observed at the level of mRNA expression would correlate with protein levels, we performed FACS analysis of GFP-transfected RAW 264.7 cells. Since endogenous TNF-α production reached its maximum after 12 h of LPS stimulation in RAW 264.7 cells (as determined by an enzyme-linked immunosorbent assay; data not shown), the transfected cells were treated for 12 h with LPS. No increase in the expression of the GFP control construct was observed (Fig. 3A). The GFP-TNF-α-ARE construct was expressed weakly in nonstimulated cells but responded to LPS treatment with a three- to fourfold increase in expression (Fig. 3B). In contrast, the expression of the GFP-TNF-α-II construct remained low in LPS-stimulated cells (Fig. 3C), confirming that the CDE acts in a constitutive fashion not subject to inhibition by LPS.
FIG. 3.
Analysis of protein expression under the control of the ARE or the CDE of TNF-α. GFP levels were measured by FACS analysis of RAW 264.7 macrophages stably expressing pBABEpuro-GFP-control (A), pBABEpuro-GFP-TNF-α-ARE (B), and pBABEpuro-GFP-TNF-α-II (C), which contains the CDE. Cells were treated for 12 h with LPS (5 μg/ml) prior to FACS analysis, and the expression profile was compared to that of nonstimulated cells.
Differential regulation of TNF-α and GM-CSF mRNAs.
We next compared the patterns of endogenous TNF-α mRNA expression in nontransfected RAW 264.7 cells. In nonstimulated cells, this mRNA was undetectable, and LPS treatment led to a transient induction, with maximum levels being attained after 4 h (Fig. 2D, left panel). Notably, TNF-α mRNA remained labile during the phase of LPS induction (Fig. 2D, middle and right panels), similar to the region II reporter mRNA (Fig. 2C). We also analyzed the expression of GM-CSF mRNA; like TNF-α mRNA, this mRNA contains a potent ARE that mediates decay in a TTP-dependent fashion (4, 31, 39). LPS caused a more delayed induction of GM-CSF mRNA, with maximum levels being attained after 8 h (Fig. 2D, lower panels). In sharp contrast to TNF-α mRNA, GM-CSF mRNA was stabilized by LPS treatment. Taken together, these results indicated that the decay of GM-CSF mRNA is primarily regulated through its ARE, whereas the CDE seems to allow for differential regulation of TNF-α mRNA by maintaining this transcript labile during the phase of LPS induction.
The CDE is unresponsive to upstream signals stabilizing ARE mRNA in NIH 3T3 cells.
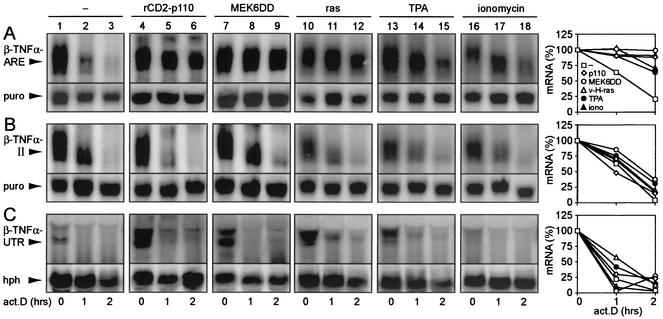
Activation of c-Jun N-terminal kinase, p38 mitogen-activated protein kinase, and phosphatidylinositol 3-kinase (PI3-kinase) pathways has been shown to inhibit ARE-dependent mRNA decay (7, 22, 23, 44). To determine whether decay mediated by region II would also be inhibited by these signal transduction pathways, we performed actinomycin D chase experiments upon transient transfection of NIH 3T3 B2A2 cells (47). Together with the β-globin reporter gene, cells were cotransfected with empty vector (Fig. 4, lanes 1 to 3); rCD2-p110, an activated form of the catalytic subunit of PI3-kinase (lanes 4 to 6); or MEK6DD, a constitutively active form of the p38 mitogen-activated protein kinase upstream activator MEK6 (lanes 7 to 9). Whereas the activation of the pathways resulted in marked stabilization of the β-TNF-α-ARE transcript (Fig. 4A), decay of the β-TNF-α-II and β-TNF-α-UTR mRNAs was not affected (Fig. 4B and C, respectively). As the activation of more than one signal transduction pathway may be required to antagonize CDE-mediated decay, we tested cotransfected v-H-ras, which is known to activate several signal transduction pathways (21, 46). v-H-ras had a stabilizing effect on the β-TNF-α-ARE mRNA but not on the β-TNF-α-II and β-TNF-α-UTR transcripts (Fig. 4, lanes 10 to 12). Treatment of the cells with TPA or ionomycin could stabilize the β-TNF-α-ARE mRNA but had again no effect on the decay of the β-TNF-α-II and β-TNF-α-UTR transcripts (Fig. 4, lanes 13 to 18). In conclusion, the CDE was unresponsive to all of the signal activation procedures that we tested. As the β-TNF-α-UTR transcript also did not respond to these signals, the CDE appears to impose constitutive decay on the mRNA even in the presence of an ARE.
FIG. 4.
Comparison of ARE- and CDE-mediated mRNA decay in response to the activation of different signal transduction pathways. NIH 3T3 B2A2 cells were transiently transfected with the following β-globin reporter constructs: puroMXβ-TNF-α-ARE (A), puroMXβ-TNF-α-II (B), and Mxh-β-TNF-α-UTR (C). Cells either were cotransfected with vector alone (lanes 1 to 3), rCD2-p110 (lanes 4 to 6), SRα3-MEK6DD (lanes 7 to 9), or v-H-ras (lanes 10 to 12) or were treated for 30 min with TPA (10 ng/ml; lanes 13 to 14) or ionomycin (5 μM; lanes 16 to 18) prior to the addition of actinomycin D (act.D). For each time point, total cytoplasmic RNA was extracted from a 10-cm dish of transfected cells and subjected to Northern blot analysis. Reporter mRNA was detected by using a radiolabeled SP6 probe specific for β-globin. To control for transfection efficiency and loading, the membranes were rehybridized with a probe against puromycin N-acetyltransferase (puro) or the hph gene product (hph), which is encoded on a reporter plasmid. For quantification of mRNA decay rates, the signal intensities of β-globin were normalized to that of puro or hph and plotted as a percentage of the initial value against time. iono, ionomycin.
Deletion mapping of the CDE.
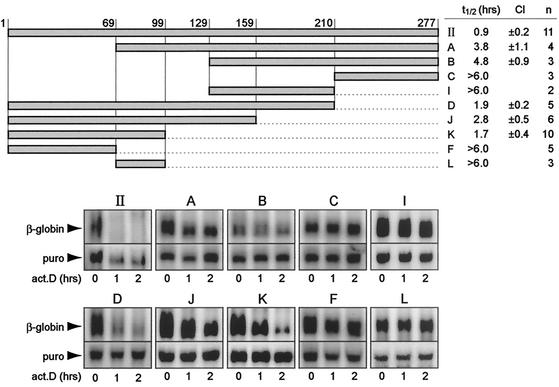
For precise mapping of the CDE, deletions were created from both ends of 277-nt region II. The fragments were inserted into the β-globin reporter gene and transiently transfected into NIH 3T3 B2A2 cells. As shown in Fig. 5, the reporter transcript carrying entire region II decayed rapidly. By linear regression of the normalized signal intensities, the half-life was calculated to be 0.9 h (number of repeat experiments and 95% confidence interval [CI] are given in Fig. 5). Deletion of 69 nt from the 5′ end reduced decay activity substantially (fragment A, with an mRNA half-life of 3.8 h), and deletion of 129 nt or more resulted in a further loss of decay activity (fragments B, C, and I, with mRNA half-lives of 4.8, >6.0, and >6.0 h, respectively). In contrast, deletion of up to 178 nt from the 3′ end caused only a weak reduction of decay activity (fragments D, J, and K, with mRNA half-lives of 1.9, 2.8, and 1.7 h, respectively). Further deletion of 30 nt, however, led to a complete loss of decay activity (fragment F, with an mRNA half-life of >6.0 h). The deleted 30-nt fragment on its own was not active either (fragment L, with an mRNA half-life of >6.0 h). Thus, fragment K was the shortest sequence within region II that could induce mRNA degradation.
FIG. 5.
Deletion mapping of the CDE within region II of the TNF-α 3′ UTR. Fragments A, B, C, I, D, J, K, F, and L, schematically represented in the upper panel, were inserted into the 3′ UTR of the β-globin reporter gene in plasmid puroMXβglobin. Upon transient transfection into NIH 3T3 B2A2 cells, the stability of the reporter mRNAs was determined by actinomycin D (act.D) chase experiments. Northern blot analysis was carried out as described in the legend to Fig. 4. Quantification of the signal intensities allowed the determination of first-order decay kinetics. The average mRNA half-lives (t1/2) were calculated from several (n) independent transfection experiments; the 95% CI is also shown. puro, puromycin N-acetyltransferase.
CDE sequence conservation.
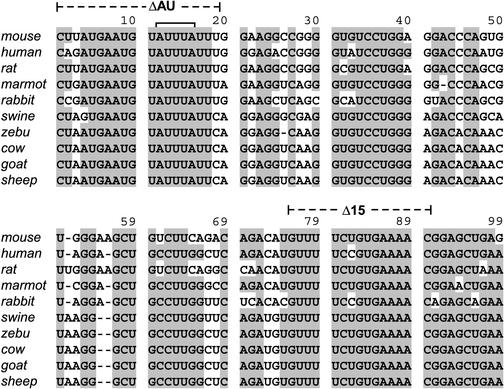
In a search for conserved motifs that may determine CDE function, TNF-α 3′ UTR sequences corresponding to fragment K from 10 different mammalian species were retrieved from GenBank and manually aligned (Fig. 6). A high degree of overall sequence conservation was observed, a result which seems to be in agreement with the notion that besides the ARE, the TNF-α 3′ UTR harbors additional sequences important for posttranscriptional regulation. The two sequences within fragment K showing the highest homology were (i) the U-rich 5′-terminal sequence (nt 1 to 18) including a canonical AUUUA motif characteristic of AREs and (ii) the 3′-terminal portion from nt 75 to 99.
FIG. 6.
Sequence conservation of the CDE. TNF-α 3′ UTR sequences for the following 10 mammalian species were derived from GenBank: mouse (Mus musculus; accession number X02611), human (Homo sapiens; accession number X02910), rat (Rattus norvegicus; accession number D00475), marmot (Marmota monax; accession number AF096268), rabbit (Oryctolagus cuniculus; accession number M12845), swine (Sus scrofa; accession number X54859), zebu (Bos indicus; accession number AF011927), cow (Bos taurus; accession number AF011926), goat (Capra hircus; accession number X14828), and sheep (Ovis aries; accession number X56756). Sequences corresponding to fragment K (Fig. 5) were aligned manually and edited with the help of Seqapp software. Grey shading indicates nucleotides that are conserved in ≥8 of 10 sequences.
An essential region of the CDE.
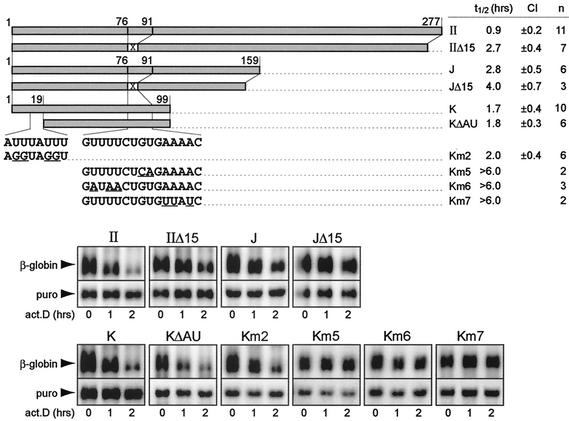
To test the functional significance of the two conserved sequences within fragment K, we performed mutational analysis. As shown in Fig. 7, deletion of 19 nt from the 5′ terminus including the AUUUA pentamer (ΔAU; Fig. 6) had no significant effect on the mRNA decay rate (fragment KΔAU, with an mRNA half-life of 1.8 h) compared to fragment K (with an mRNA half-life of 1.7 h). Likewise, transversion of uridine residues of the AUUUA motif to guanosine, a mutation that efficiently abolishes the decay activity of an ARE (37, 42), did not significantly alter the mRNA decay rate of fragment K (fragment Km2, with an mRNA half-life of 2.0 h). In conclusion, the 5′-terminal AU-rich sequence did not appear to be essential for the CDE, providing further evidence that the CDE recruits a non-ARE decay pathway. In fact, the first 19 nt of fragment K may still belong to the ARE, which is situated ultimately 5′ of the CDE.
FIG. 7.
Mutational analysis of two conserved sequences within fragment K. A conserved sequence of 15 nt (76 to 91) close to the 3′ terminus of fragment K was deleted and replaced by an XbaI site in the β-globin reporter constructs puroMXβ-TNF-α-IIΔ15 and puroMXβ-TNF-α-JΔ15, schematically represented in the upper panel. Point mutations were introduced into the same sequence in constructs puroMXβ-TNF-α-Km5, puroMXβ-TNF-α-Km6, and puroMXβ-TNF-α-Km7. An AU-rich sequence at the 5′ terminus of fragment K (nt 1 to 19) was either deleted in reporter construct puroMXβ-TNF-α-KΔAU or mutated in puroMXβ-TNF-α-Km2. Underlining in the sequences indicates point mutations. Following transient transfection into NIH 3T3 B2A2 cells, the rate of decay of the reporter transcripts was determined by actinomycin D (act.D) chase experiments. Northern blot analysis and quantification were carried out as described in the legend to Fig. 5. The average mRNA half-lives (t1/2) were calculated from several (n) independent transfection experiments; the 95% CI is also shown. puro, puromycin N-acetyltransferase.
We then tested the activity of the second conserved sequence within fragment K by deleting nt 76 to 90 (Δ15; Fig. 6) from entire region II. A threefold decrease in the mRNA decay rate was observed (fragment IIΔ15, with mRNA half-life of 2.7 h) compared to that seen with region II (with an mRNA half-life of 0.9 h) (Fig. 7). Removal of the same 15 nt from fragment J confirmed the stabilizing effect of this deletion (fragment JΔ15, with an mRNA half-life of 4.0 h, compared to fragment J, with an mRNA half-life of 2.8 h). In the context of fragment K, the effect of mutagenesis was more dramatic, as all point mutations between nt 77 and 89 completely abolished decay activity (fragments Km5, Km6, and Km7, with an mRNA half-life of >6.0 h). The pronounced effect of these mutations clearly indicates that the conserved sequence between nt 76 and 91 is essential for the function of the CDE.
DISCUSSION
In this report, we describe a novel determinant of mRNA decay which is located in the 3′ UTR of TNF-α mRNA and has regulatory properties that are very different from those of the well-characterized ARE. Like the ARE, it targets transcripts for rapid degradation, but unlike the activity of the ARE, its activity is not subject to inhibition by upstream signaling events; the latter fact prompted us to call it CDE. Evidence for the CDE was obtained from studies with three different cell culture models: human HT1080 fibrosarcoma cells, mouse macrophage line RAW 264.7, and mouse NIH 3T3 cells. Basic to discovery of the CDE were two HT1080-derived mutant cell lines (slowA and slowC) which are defective in ARE-mediated mRNA decay. Reporter transcripts carrying the entire 3′ UTR of TNF-α were still labile in the mutant background, suggesting the presence of an additional decay element (Fig. 1). By dividing the 3′ UTR into three portions, we could assign the CDE to the region downstream of the ARE (region II).
Mapping experiments with transiently transfected NIH 3T3 cells (Fig. 5 and 7) restricted the activity to an 80-nt fragment (KΔAU) which is adjacent to the ARE. Mutational analysis identified a highly conserved 15-nt sequence (nt 76 to 90; Fig. 6) that is required but not sufficient for CDE-mediated mRNA decay. UV cross-linking experiments with whole-cell extracts revealed a 69-kDa complex that binds to the CDE (data not shown). The stabilizing point mutation (Km5) within the critical 15-nt sequence, however, had no effect on the formation of this complex, indicating that it may not contain the protein that mediates CDE-mRNA decay. The use of an affinity step and a suitable purification strategy should help to resolve this issue. Identification of the relevant CDE-binding protein(s) should help to provide an understanding of why CDE-mediated mRNA decay is not subject to regulation. Another interesting task will be to investigate whether the CDE triggers 3′-5′ degradation through the exosome, as demonstrated for the ARE (8), or whether it recruits an alternative decay pathway.
Opposing effects of the ARE and the CDE on reporter mRNA decay were observed in NIH 3T3 cells that were cotransfected with activated forms of PI3-kinase, MEK6, and H-ras or stimulated with TPA and ionomycin (Fig. 4). Whereas all of these signals inhibited ARE-mediated mRNA decay, CDE-mediated decay was not affected. The β-TNF-α-UTR transcript, which contains both the ARE and the CDE, was not stabilized by these stimuli either, indicating that the CDE retains its decay-promoting activity even when the ARE-dependent decay pathway is inhibited.
Several transcripts, including c-myc, c-fos, and granulocyte colony-stimulating factor transcripts, have also been shown to harbor additional destabilizing elements (3, 32, 45), and it has been suggested that the presence of more than one decay element should allow for differential regulation under various physiological conditions. Differential regulation of ARE- and CDE-dependent decay pathways became particularly obvious when reporter transcripts carrying the individual elements were analyzed in LPS-stimulated RAW 264.7 macrophages. Both the ARE and the CDE were able to effectively suppress mRNA expression in nonstimulated cells (compare the first lanes in Fig. 2B and C), where the reporter mRNA transcribed from the strong MMLV long terminal repeat promoter was hardly detectable. Treatment with LPS led to sustained induction and permanent stabilization of GFP-TNF-α-ARE mRNA. In contrast, the CDE remained a strong suppressor of mRNA expression throughout the entire LPS time course experiment, allowing only for a transient and much weaker induction of GFP-TNF-α-II mRNA. Importantly, these transcripts remained labile in LPS-treated cells, demonstrating the constitutive nature of CDE-dependent mRNA decay. When the expression profiles for ARE and region II mRNAs were compared to the pattern for endogenous TNF-α, it was striking that the regulation of TNF-α mRNA resembled that of region II mRNA rather than that of ARE mRNA, as it also remained labile 2 and 6 h after LPS treatment. These results indicated that in LPS-stimulated cells, the CDE exerts powerful negative control by maintaining TNF-α mRNA short-lived and thereby antagonizing ARE-directed stabilizing signals. In contrast to TNF-α mRNA, GM-CSF mRNA, which also carries a strong ARE in its 3′ UTR (31, 39) but lacks evidence for a second decay element, was stabilized by LPS treatment. Thus, TNF-α and GM-CSF provide telling examples for two ARE-containing messages whose turnover is differentially regulated.
In accordance with our findings, other investigators have also observed that endogenous TNF-α mRNA as well as TNF-α reporter transcripts remain labile in LPS-stimulated macrophages (14, 28, 29). Particularly relevant to our study is work by Kontoyiannis et al. (17). In their mouse model, the ARE was specifically deleted from the TNF-α locus (TNFΔARE), including the first 18 nt of fragment K (Fig. 6), whereas the CDE (KΔAU fragment) was retained in the 3′ UTR. Primary macrophages derived from these mice showed excessive TNF-α mRNA and protein levels upon LPS stimulation. As estimated from the net mRNA levels during the LPS time course, the TNFΔARE transcript was reported to be essentially stable, a result which seems to preclude the existence of a second, ARE-independent decay pathway. Nevertheless, TNFΔARE mRNA declined to baseline levels in the 6 h following peak accumulation. It is important to readdress the issue of mRNA half-lives in these cells by using more accurate half-life measurements with transcription inhibitors. In another mouse model, the ARE-dependent decay function was inhibited in trans by knocking out the ARE-binding protein TTP (TTP−/−) (4, 5). In the TTP−/− mice, the TNF-α mRNA half-lives increased from 39 to 85 min in LPS-treated macrophages and from 35 to 90 min in LPS-treated bone marrow stromal cells. The fact that TNF-α mRNA was still fairly labile in the TTP−/− cells could be explained either by the activity of TTP-related proteins, such as BRF1 and BRF2 (19, 36), or by a second, ARE-indpendent decay pathway. Since GM-CSF mRNA was entirely stable over a 160-min time course in the TTP−/− bone marrow stromal cells compared to a half-life of 99 min in wt cells, TTP appears to be the main activity targeting cytokine AREs in these cells. Thus, the results of the TTP knockout model are fully consistent with a second decay element contributing to TNF-α mRNA turnover.
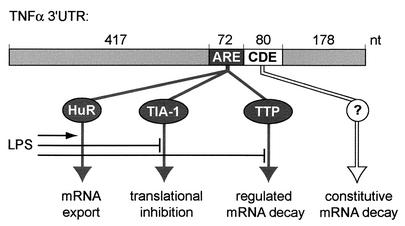
Based on our results and on the work of other investigators (2, 5, 8, 10-12, 17, 18, 28, 29), we propose the following model for the posttranscriptional regulation of TNF-α mRNA (Fig. 8). The ARE is a multifunctional negative control element involved in (i) mRNA export; (ii) cytoplasmic degradation of the mRNA through binding of TTP, which targets the mRNA to the exosome; and (iii) translational inhibition by binding of TIA-1. The CDE, through an as-yet-undefined mechanism, contributes to rapid degradation of the mRNA in resting cells. Stimulation by LPS interferes with the inhibitory functions of the ARE: Export of the mRNA is induced by a Tpl2- and ERK-dependent signal and may involve, by analogy to the export of c-fos mRNA, the ARE-binding protein HuR. A MAPKAP kinase 2-dependent signal allows for efficient translation, and a p38 kinase-dependent signal also inhibits ARE-mediated mRNA decay. These kinases are likely to act by affecting, directly or indirectly, ARE-binding proteins, which in turn may favor competitive binding of the positive regulator HuR. This LPS-induced switch will primarily result in the efficient translation of TNF-α. The net amount of mRNA available for translation remains, even in the presence of LPS, under the control of the CDE, ensuring that transcripts are continuously cleared from the cytoplasm by being targeted to a constitutive decay pathway. As a “safeguard” element, the CDE is likely to play a role in maintaining stringent control over TNF-α production and thereby limiting TNF-α-mediated pathologic processes.
FIG. 8.
Model of posttranscriptional mechanisms regulating TNF-α mRNA expression in LPS-stimulated macrophages. The ARE is involved in (i) export of the mRNA from the nucleus to the cytoplasm, (ii) translational inhibition through binding of TIA-1, and (iii) targeting of the mRNA for rapid decay through binding of TTP. LPS stimulates export of the mRNA, perhaps through binding of HuR; allows for efficient protein translation; and inhibits ARE-mediated decay of the mRNA. The CDE, through an as-yet-unidentified mechanism, ensures that transcripts are continuously cleared from the cytoplasm by targeting them to a constitutive decay pathway.
Acknowledgments
We are grateful to Brigitte Gross for help with transfections, Sebastien Viatte for generating the TNF-α probe, and Witold Filipowicz for advice on sequence analysis. We also thank Hans H. Hirsch, Helena Côrte-Real, and Paul Anderson for helpful comments on the manuscript.
This work was supported by grant 31-57065.99 from the Swiss National Science Foundation to C.M.
REFERENCES
- 1.Anderson, P. 2000. Post-transcriptional regulation of tumour necrosis factor alpha production. Ann. Rheum. Dis. 59(Suppl. 1):i3-i5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brook, M., G. Sully, A. R. Clark, and J. Saklatvala. 2000. Regulation of tumour necrosis factor alpha mRNA stability by the mitogen-activated protein kinase p38 signalling cascade. FEBS Lett. 483:57-61. [DOI] [PubMed] [Google Scholar]
- 3.Brown, C. Y., C. A. Lagnado, and G. J. Goodall. 1996. A cytokine mRNA-destabilizing element that is structurally and functionally distinct from A+U-rich elements. Proc. Natl. Acad. Sci. USA 93:13721-13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carballo, E., W. S. Lai, and P. J. Blackshear. 2000. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95:1891-1899. [PubMed] [Google Scholar]
- 5.Carballo, E., W. S. Lai, and P. J. Blackshear. 1998. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281:1001-1005. [DOI] [PubMed] [Google Scholar]
- 6.Chen, C. Y., and A. B. Shyu. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465-470. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. Y., F. Del Gatto-Konczak, Z. Wu, and M. Karin. 1998. Stabilization of interleukin-2 mRNA by the c-Jun NH2-terminal kinase pathway. Science 280:1945-1949. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. Y., R. Gherzi, S. E. Ong, E. L. Chan, R. Raijmakers, G. J. Pruijn, G. Stoecklin, C. Moroni, M. Mann, and M. Karin. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107:451-464. [DOI] [PubMed] [Google Scholar]
- 9.Collart, M. A., P. Baeuerle, and P. Vassalli. 1990. Regulation of tumor necrosis factor alpha transcription in macrophages: involvement of four κB-like motifs and of constitutive and inducible forms of NF-κB. Mol. Cell. Biol. 10:1498-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dean, J. L., R. Wait, K. R. Mahtani, G. Sully, A. R. Clark, and J. Saklatvala. 2001. The 3′ untranslated region of tumor necrosis factor alpha mRNA is a target of the mRNA-stabilizing factor HuR. Mol. Cell. Biol. 21:721-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumitru, C. D., J. D. Ceci, C. Tsatsanis, D. Kontoyiannis, K. Stamatakis, J. H. Lin, C. Patriotis, N. A. Jenkins, N. G. Copeland, G. Kollias, and P. N. Tsichlis. 2000. TNF-alpha induction by LPS is regulated posttranscriptionally via a Tpl2/ERK-dependent pathway. Cell 103:1071-1083. [DOI] [PubMed] [Google Scholar]
- 12.Gallouzi, I. E., and J. A. Steitz. 2001. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 294:1895-1901. [DOI] [PubMed] [Google Scholar]
- 13.Han, J., T. Brown, and B. Beutler. 1990. Endotoxin-responsive sequences control cachectin/tumor necrosis factor biosynthesis at the translational level. J. Exp. Med. 171:465-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han, J. H., B. Beutler, and G. Huez. 1991. Complex regulation of tumor necrosis factor mRNA turnover in lipopolysaccharide-activated macrophages. Biochim. Biophys. Acta 1090:22-28. [DOI] [PubMed] [Google Scholar]
- 15.Jarrous, N., F. Osman, and R. Kaempfer. 1996. 2-Aminopurine selectively inhibits splicing of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 16:2814-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karima, R., S. Matsumoto, H. Higashi, and K. Matsushima. 1999. The molecular pathogenesis of endotoxic shock and organ failure. Mol. Med. Today 5:123-132. [DOI] [PubMed] [Google Scholar]
- 17.Kontoyiannis, D., M. Pasparakis, T. T. Pizarro, F. Cominelli, and G. Kollias. 1999. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10:387-398. [DOI] [PubMed] [Google Scholar]
- 18.Kotlyarov, A., A. Neininger, C. Schubert, R. Eckert, C. Birchmeier, H. D. Volk, and M. Gaestel. 1999. MAPKAP kinase 2 is essential for LPS-induced TNF-alpha biosynthesis. Nat. Cell Biol. 1:94-97. [DOI] [PubMed] [Google Scholar]
- 19.Lai, W. S., E. Carballo, J. M. Thorn, E. A. Kennington, and P. J. Blackshear. 2000. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to Au-rich elements and destabilization of mRNA. J. Biol. Chem. 275:17827-17837. [DOI] [PubMed] [Google Scholar]
- 20.Lindsten, T., C. H. June, J. A. Ledbetter, G. Stella, and G. B. Thompson. 1989. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science 244:339-343. [DOI] [PubMed] [Google Scholar]
- 21.Marshall, C. J. 1996. Ras effectors. Curr. Opin. Cell Biol. 8:197-204. [DOI] [PubMed] [Google Scholar]
- 22.Ming, X. F., M. Kaiser, and C. Moroni. 1998. c-jun N-terminal kinase is involved in AUUUA-mediated interleukin-3 mRNA turnover in mast cells. EMBO J. 17:6039-6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ming, X. F., G. Stoecklin, M. Lu, R. Looser, and C. Moroni. 2001. Parallel and independent regulation of interleukin-3 mRNA turnover by phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase. Mol. Cell. Biol. 21:5778-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myokai, F., S. Takashiba, R. Lebo, and S. Amar. 1999. A novel lipopolysaccharide-induced transcription factor regulating tumor necrosis factor alpha gene expression: molecular cloning, sequencing, characterization, and chromosomal assignment. Proc. Natl. Acad. Sci. USA 96:4518-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nair, A. P., I. D. Diamantis, J. F. Conscience, V. Kindler, P. Hofer, and C. Moroni. 1989. A v-H-ras-dependent hemopoietic tumor model involving progression from a clonal stage of transformation competence to autocrine interleukin 3 production. Mol. Cell. Biol. 9:1183-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osman, F., N. Jarrous, Y. Ben-Asouli, and R. Kaempfer. 1999. A cis-acting element in the 3′-untranslated region of human TNF-alpha mRNA renders splicing dependent on the activation of protein kinase PKR. Genes Dev. 13:3280-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piecyk, M., S. Wax, A. R. Beck, N. Kedersha, M. Gupta, B. Maritim, S. Chen, C. Gueydan, V. Kruys, M. Streuli, and P. Anderson. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 19:4154-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raabe, T., M. Bukrinsky, and R. A. Currie. 1998. Relative contribution of transcription and translation to the induction of tumor necrosis factor-alpha by lipopolysaccharide. J. Biol. Chem. 273:974-980. [DOI] [PubMed] [Google Scholar]
- 30.Reif, K., S. Lucas, and D. Cantrell. 1997. A negative role for phosphoinositide 3-kinase in T-cell antigen receptor function. Curr. Biol. 7:285-293. [DOI] [PubMed] [Google Scholar]
- 31.Shaw, G., and R. Kamen. 1986. A conserved AU sequence from the 3′untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46:659-667. [DOI] [PubMed] [Google Scholar]
- 32.Shyu, A.-B., J. G. Belasco, and M. E. Greenberg. 1991. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 5:221-231. [DOI] [PubMed] [Google Scholar]
- 33.Shyu, A.-B., M. E. Greenberg, and J. G. Belasco. 1989. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 3:60-72. [DOI] [PubMed] [Google Scholar]
- 34.Shyu, A. B., and M. F. Wilkinson. 2000. The double lives of shuttling mRNA binding proteins. Cell 102:135-138. [DOI] [PubMed] [Google Scholar]
- 35.Steer, J. H., K. M. Kroeger, L. J. Abraham, and D. A. Joyce. 2000. Glucocorticoids suppress tumor necrosis factor-alpha expression by human monocytic THP-1 cells by suppressing transactivation through adjacent NF-kappa B and c-Jun-activating transcription factor-2 binding sites in the promoter. J. Biol. Chem. 275:18432-18440. [DOI] [PubMed] [Google Scholar]
- 36.Stoecklin, G., M. Colombi, I. Raineri, S. Leuenberger, M. Mallaun, M. Schmidlin, B. Gross, M. Lu, T. Kitamura, and C. Moroni. 2002. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 21:4709-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoecklin, G., S. Hahn, and C. Moroni. 1994. Functional hierarchy of AUUUA motifs in mediating rapid interleukin-3 mRNA decay. J. Biol. Chem. 269:28591-28597. [PubMed] [Google Scholar]
- 38.Stoecklin, G., X. F. Ming, R. Looser, and C. Moroni. 2000. Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway. Mol. Cell. Biol. 20:3753-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoecklin, G., P. Stoeckle, M. Lu, O. Muehlemann, and C. Moroni. 2001. Cellular mutants define a common mRNA degradation pathway targeting cytokine AU-rich elements. RNA 7:1578-1588. [PMC free article] [PubMed] [Google Scholar]
- 40.Tracey, K. J., and A. Cerami. 1994. Tumor necrosis factor: a pleiotropic cytokine and therapeutic target. Annu. Rev. Med. 45:491-503. [DOI] [PubMed] [Google Scholar]
- 41.Trede, N. S., A. V. Tsytsykova, T. Chatila, A. E. Goldfeld, and R. S. Geha. 1995. Transcriptional activation of the human TNF-alpha promoter by superantigen in human monocytic cells: role of NF-kappa B. J. Immunol. 155:902-908. [PubMed] [Google Scholar]
- 42.Vakalopoulou, E., J. Schaack, and T. Shenk. 1991. A 32-kilodalton protein binds to AU-rich domains in the 3′ untranslated regions of rapidly degraded mRNAs. Mol. Cell. Biol. 11:3355-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilusz, C. J., M. Wormington, and S. W. Peltz. 2001. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell. Biol. 2:237-246. [DOI] [PubMed] [Google Scholar]
- 44.Winzen, R., M. Kracht, B. Ritter, A. Wilhelm, C. Y. Chen, A. B. Shyu, M. Muller, M. Gaestel, K. Resch, and H. Holtmann. 1999. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 18:4969-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wisdom, R., and R. Lee. 1991. The protein-coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev. 5:232-243. [DOI] [PubMed] [Google Scholar]
- 46.Wolthuis, R. M., and J. L. Bos. 1999. Ras caught in another affair: the exchange factors for Ral. Curr. Opin. Genet. Dev. 9:112-117. [DOI] [PubMed] [Google Scholar]
- 47.Xu, N., P. Loflin, C. Y. Chen, and A. B. Shyu. 1998. A broader role for AU-rich element-mediated mRNA turnover revealed by a new transcriptional pulse strategy. Nucleic Acids Res. 26:558-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang, Y., J. F. Chang, J. R. Parnes, and C. G. Fathman. 1998. T cell receptor (TCR) engagement leads to activation-induced splicing of tumor necrosis factor (TNF) nuclear pre-mRNA. J. Exp. Med. 188:247-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao, J., N. Mackman, T. S. Edgington, and S. T. Fan. 1997. Lipopolysaccharide induction of the tumor necrosis factor-alpha promoter in human monocytic cells. Regulation by Egr-1, c-Jun, and NF-kappaB transcription factors. J. Biol. Chem. 272:17795-17801. [DOI] [PubMed] [Google Scholar]
- 50.Zuckerman, S. H., G. F. Evans, Y. M. Snyder, and W. D. Roeder. 1989. Endotoxin-macrophage interaction: post-translational regulation of tumor necrosis factor expression. J. Immunol. 143:1223-1227. [PubMed] [Google Scholar]