Abstract
p220NPAT is a substrate of cyclin E/Cdk2 that localizes in nuclear organelles called Cajal bodies in a cell cycle-regulated manner. In normal diploid fibroblasts, p220 is concentrated in two Cajal bodies tethered to histone gene clusters at chromosome 6p21 during G1, S, and G2 phases and two additional Cajal bodies tethered to histone genes at 1q21 during S, and G2 phases. Overexpression of p220 in U2OS cells can promote the G1/S transition and can also promote transcription from histone H2B and H4 luciferase reporter constructs. How p220 expression induces these activities and whether the two activities are related are unknown. In this study, we developed a “lox-scanning” mutagenesis approach to identify functional domains in p220. We identified two distinct functional regions of p220. The C-terminal half of the protein contains multiple elements that are required for its ability to induce S phase in transfected cells. In contrast, sequences at the N terminus appear to be critical for activation of histone H4 and H2B reporter constructs. We identified an ∼30-amino-acid motif at the N terminus of p220 that has the characteristics of a LisH motif. LisH motifs are found in a large number of proteins in the database but are of unknown function. Point mutations in conserved residues in the LisH motif of p220 block histone H4 transcriptional activity without affecting localization in Cajal bodies or phosphorylation on Cdk2 phosphorylation sites. These studies indicate that the ability of p220 to promote S phase is independent of its ability to promote histone H4 transcription and suggests that p220 may link cyclin E/Cdk2 to multiple independent downstream functions.
Cell cycle transitions are regulated by cyclin-dependent kinases, a family of enzymes composed of a catalytic Cdk subunit and a regulatory cyclin subunit (19). Cdk proteins act to promote specific cell cycle events, such as DNA replication and entry into mitosis, by phosphorylating particular substrates. E-, D-, and A-type cyclins function together with Cdk4 and Cdk2 to promote entry into and completion of DNA synthesis (19). Important targets for these kinases include a family of transcriptional repressors typified by the retinoblastoma protein. Phosphorylation of Rb leads to derepression of E2F transcription factors, which then activate a transcriptional program designed to promote S phase (reviewed in references 14 and 26).
DNA synthesis is accompanied by additional transcriptional programs that promote completion of replication. One of these is the activation of core histone synthesis that occurs as cells enter S phase. Histones are the building blocks of nucleosomes and are required for packaging of newly synthesized DNA into chromatin. Histone synthesis is complex and is regulated by multiple mechanisms which together provide an ∼35-fold increase in the quantity of histone mRNA during S phase (20; for a review, see reference 21). Histone gene transcription is activated ∼5-fold as cells enter S phase (20). Linker histone (H1) and core histones (H2A, H2B, H3, and H4) are relatively unusual in that the cell requires a large number of gene copies to support adequate nucleosome formation during DNA replication, and it appears that gene clustering (3) provides a means by which to coordinate the expression of different classes of histone genes. There are both common and specific cis-acting regulatory elements in replication-dependent histone genes (21). Both subtype-specific consensus elements (SSCEs) and the common YY1 element located in the structural gene contribute to cell cycle-regulated transcription, and in some cases, trans-acting factors that interact with SSCEs have been identified (Oct1 in the case of H2B and IFR2 in the case of histone H4) (4, 5, 11, 12, 24, 29, 33-35). In addition, histone mRNA is stabilized during S phase. This process involves recognition of a stem-loop element in the 3′ untranslated region of histone mRNAs by the stem-loop binding protein (SLBP1) followed by RNA processing and stabilization in the cytoplasm (13, 30). One level of cell cycle control comes from the fact that SLBP1 is translated only during S phase and is rapidly destroyed as cells complete DNA synthesis (36).
Histone synthesis is thought to be linked to the basic cell cycle machinery by the cyclin E/Cdk2 substrate p220NPAT. p220 was identified in screens for cyclin E substrates (22, 39) and demonstrated to promote S-phase entry from G1 when overexpressed (39). Like many other Cdk substrates (1, 22, 31, 41), p220 interacts with cyclin E through a cyclin-binding RXL motif located between residues 1062 and 1066 (Y. Wei and J. W. Harper, unpublished data). Insight into p220 function came from the finding that p220 is localized in discrete nuclear organelles called Cajal bodies in a cell cycle-regulated manner in normal diploid fibroblasts (23, 40). Although Cajal bodies were initially described in 1903 (8), their precise functions are still emerging (18, 27). Studies of both Xenopus and mammalian systems suggest that they function, at least in part, as sites of assembly of transcription and splicing complexes (17; reviewed in references 18 and 27). In particular, there is strong evidence that Cajal bodies are responsible for maturation of snRNPs and nuclear RNPs and may also be the sites of assembly of RNA polymerase complexes (18, 25, 27). In G1 normal diploid fibroblast cells, p220 is localized in two Cajal bodies, and these are physically tethered to histone gene clusters located at chromosome 6p21 (23, 40). As these cells enter S phase, two additional p220-containing Cajal bodies appear and are tethered to histone gene clusters at 1q21. Both sets of p220-positive Cajal bodies are maintained until the metaphase-anaphase transition, when Cajal bodies are disassembled (23). As cells exit mitosis and reenter G1, p220-containing Cajal bodies reform and colocalize at 6p21. Additional Cajal bodies that lack p220 are also seen in some cell types and are sometimes associated with other chromosomal loci, such as snRNA genes on chromosome 17 (16). Phospho-specific antibodies were used to demonstrate that p220 becomes phosphorylated at the G1/S transition as cyclin E/Cdk2 accumulates in Cajal bodies and that this phosphorylation is maintained until the metaphase-anaphase transition (23). In transformed cells, the number of Cajal bodies appears to be more variable. The mechanism underlying this is not clear, but the facts that additional p220-containing Cajal bodies exist in particular cell types and that these are also linked to histone gene clusters (40) suggest that aneuploidy may be responsible for some of the heterogeneity observed.
Insight into a role for p220 in histone transcription came from the finding that ectopic expression of p220 can activate the expression of histone 2B (H2B) and H4 promoter constructs (40). This activity is potentiated by coexpression of cyclin E/Cdk2 (40) and is inhibited by mutations in Cdk2 sites in p220 (23). These data have led to a model wherein cyclin E/Cdk2 links the S-phase cell cycle machinery with the histone gene transcription program that is critical for completion of DNA replication. Precisely how p220 activates histone transcription is unknown. Although p220 can be cross-linked with histone promoters (40) and its ability to activate transcription requires known cell-cycle regulated promoter elements (23, 40), there is no evidence that it directly interacts with DNA sequences. In addition, the major location of p220, Cajal bodies, is devoid of DNA, indicating that if p220 functions directly in transcription through interaction with chromatin, then this occurs outside Cajal bodies. An alternative mechanism might involve a function for p220 within the Cajal body, such as the assembly or activation of transcription complexes that function to activate histone genes during S phase (23).
A major limitation in the analysis of p220 function is the fact that it lacks obvious functional motifs that would link it to histone transcription or cell proliferation. To begin to understand p220 function, we have performed an extensive mutagenic analysis of the protein using a recombination-based mutagenesis approach we refer to as “lox-scanning” mutagenesis. These studies reveal that the two known activities of p220—G1 acceleration and activation of histone gene transcription—primarily involve two independent functional elements in p220. Sequences at the extreme N terminus are required for activation of histone H2B and H4 transcription. This region displays similarities with an ∼33-amino-acid motif referred to as a lissencephaly type 1-like homology (LisH) motif. This motif was originally identified in a number of proteins in the database (15), including the protein lissencephaly 1 (Lis-1), which is required for neuronal migration. The precise function of LisH motifs in these proteins is not known, but several proteins containing LisH domains have been linked to microtubule and kinetochore function (15). We found that point mutations in conserved residues in the LisH-like motif of p220 abolish histone H4 transcriptional activation without affecting the localization in Cajal bodies or the ability of the protein to promote S-phase entry. In contrast, sequences in the C terminus are required for efficient G1 acceleration but not histone transcription. The identification of separation of function mutations in p220 indicates that its ability to activate transcription is not linked with its ability to promote G1 progression and suggests the possibility that activation of p220 by cyclin E/Cdk2 links upstream cell cycle signals with independent cellular outputs that both promote completion of S phase.
MATERIALS AND METHODS
loxP-scanning mutagenesis.
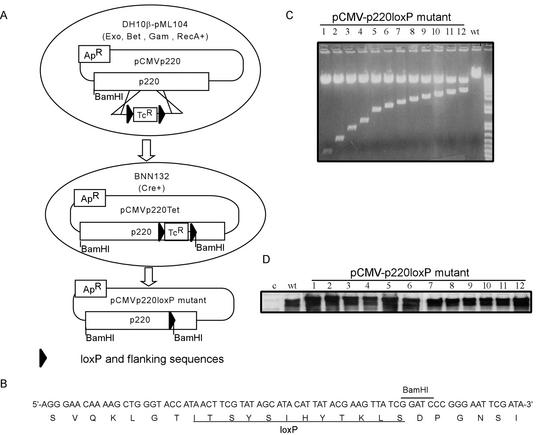
Tcr cassettes flanked with two loxP sites and a total of 120 bp of p220 sequences for homologous recombination were obtained by two rounds of PCR using plasmid pKOEZ81 (38) as the template. The sequences of oligonucleotides employed for PCR are provided in Table 1. PCR products were gel purified and then electroporated into Escherichia coli strain DH10β(pML104) containing pCMV-p220 (40) prior to selection on medium containing tetracycline (15 μg/ml) and ampicillin (50 μg/ml). Plasmids were screened for recombination by digestion with BamHI. pCMV-p220-Tet plasmids were then transformed into BNN132 cells expressing Cre recombinase to excise the Tcr cassette. The resulting plasmids were digested with BamHI and sequenced.
TABLE 1.
Oligonucleotides used to create p220 lox-scanning mutants
| p220 mutant | Primer description | Primer sequence (5′-3′)a |
|---|---|---|
| 1 (LoxP121-145) | 1st round, forward | GCC CGA ACG AGA ACT GGA ATT GCA GAA ATC AGG GAA CAA AAG CTG GGT ACC ATA ACT TCG |
| 1st round, reverse | TGT ACC TGT GGA AGG AGG AGT GGT AAA CTG TAT CGA ATT CCC GGG ATC CGA TAA CTT CG | |
| 2nd round, forward | TCC CCA AGG TTT GCT GGC AGT CAG AGA GCC CGA ACG AGA ACT GGA ATT GCA GAA ATC | |
| 2nd round, reverse | AAT TTG GCC ACT TGG TCG AGT AAC CTG TGT ACC TGT GGA AGG AGG AGT GGT AAA CTG | |
| 2 (LoxP225-249) | 1st round, forward | TCT CAG AGA AAA AGT ACC ACT TTG TCT GGC AGG GAA CAA AAG CTG GGT ACC ATA ACT TCG |
| 1st round, reverse | TTG AAG AGA TTT GTT GCT TAG TAT TTT TTC TCG TAT CGA ATT CCC GGG ATC CGA TAA CTT CG | |
| 2nd round, forward | TCT CCC GGT AGA CGC AAA AGT GAA TCT CAG AGA AAA AGT ACC ACT TTG TCT GGC | |
| 2nd round, reverse | TTT ATT TAT GTT TTC TGC TAG CTT TTC TTG AAG AGA TTT GTT GCT TAG TAT TTT TTC TCG | |
| 3 (LoxP325-349) | 1st round, forward | TTG GAA CAG ACA GAA TCA GAC CCA GCA TTT AGG GAA CAA AAG CTG GGT ACC ATA ACT TCG |
| 1st round, reverse | GAC TAT ACT GGG ATT GGA TTC CAT AGG TTG TAT CGA ATT CCC GGG ATC CGA TAA CTT CG | |
| 2nd round, forward | ATG TCT GAA GAA GCT ATA CAG GAC ATA TTG GAA CAG ACA GAA TCA GAC CCA GCA TTT | |
| 2nd round, reverse | AAC TGC TAG ATT AGT TTC ATC TGC TAA GAC TAT ACT GGG ATT GGA TTC CAT AGG TTG | |
| 4 (LoxP422-446) | 1st round, forward | GAA GAC CAG GAA AAT TTT TCC CAA ATA AGT ACC AGG GAA CAA AAG CTG GGT ACC ATA ACT TCG |
| 1st round, reverse | CCC TCT TTG GTT AAA GTC ATT CAA ATT AGG CAC TAT CGA ATT CCC GGG ATC CGA TAA CTT CG | |
| 2nd round, forward | AGC AAC AAC CAT GAT GTG CTT AGA CAA GAA GAC CAG GAA AAT TTT TCC CAA ATA AGT ACC | |
| 2nd round, reverse | ATG TGG ATT ACA TTC AGC ATT AGA ATT CCC TCT TTG GTT AAA GTC ATT CAA ATT AGG CAC | |
| 5 (LoxP629-653) | 1st round, forward | GGA CAA GTA GAA ATT CAT CTT GGA GAT TCG AGG GAA CAA AAG CTG GGT ACC ATA A TCG |
| 1st round, reverse | AGG CTC CTG TGA ATT CTC AGA CTT GGA TAT CGA ATT CCC GGG ATC CGA TAA CTT CG | |
| 2nd round, forward | GTT GAA AGT TCA CAT TTA AAT GTA TCT GGA CAA GTA GAA ATT CAT CTT GGA GAT TCG | |
| 2nd round, reverse | ATT CTC TTC TTT TAC AGA AGA TGA AGG CTC CTG TGA ATT CTC AGA CTT GGA | |
| 6 (LoxP727-752) | 1st round, forward | TCC CAA AAT ACT GAT GAT AAA CCT TCT AGC AGG GAA CAA AAG CTG GGT ACC ATA ACT TCG |
| 1st round, reverse | ACT AGA AAC AGC ACT GGT AAG TTC AGT ATC TAT CGA ATT CCC GGG ATC CGA TAA CTT CG | |
| 2nd round, forward | TCT TCA GTG GGA GAT TCT CAC CCT GAG TCC CAA AAT ACT GAT GAT AAA CCT TCT AGC | |
| 2nd round, reverse | TAT AGT TGG CAG GTT TTC TCC ATT AAT ACT AGA AAC AGC ACT GGT AAT TTC AGT ATC | |
| 7 (LoxP828-853) | 1st round, forward | TCT GAA GAC TCT GCA GTA AAC AAT ACT CAG AGG GAA CAA AAG CTG GGT ACC ATA ACT TCG |
| 1st round, reverse | TGA ATT GCC AAA AGC TGT GCT TGT GGC TGG TAT CGA ATT CCC GGG ATC CGA TAA CTT CG | |
| 2nd round, forward | ATG GAA CAG AGT CTT TTA ACA TTC AAA TCT GAA GAC TCT GCA GTA AAC AAT ACT CAG | |
| 2nd round, reverse | CAC ACA GGT AGC TAT CAG AAT GTT ATT TGA ATT GCC AAA AGC TGT GCT TGT GGC TGG | |
| 8 (LoxP926-950) | 1st round, forward | TTT GCT GTC AAC CAA GCT GTG TCA CCA AAC AGG GAA CAA AAG CTG GGT ACC ATA ACT TCG |
| 1st round, reverse | GTT ATT TCC ATT CTG TCC AAC CAC AGA TAC TAT CGA ATT CCC GGG ATC CGA TAA CTT CG | |
| 2nd round, forward | CAG ACA CCA CCA AGG TCA AAC AGT GTA TTT GCT GTC AAC CAA GCT GTG TCA CCA AAC | |
| 2nd round, reverse | AAG AAC CTG CCG AGG AGG AGT AGA AAA GTT ATT TCC ATT CTG TCC AAC CAC AGA TAC | |
| 9 (LoxP1039-1054) | 1st round, forward | TGT CAT GCA CAA AAA ACT GAA GTT TCT GAC AGG GAA CAA AAG CTG GGT ACC ATA ACT TCG |
| 1st round, reverse | TAC ACG TCT GTG GCA TGG TTT AGC AGC TAT CGA ATT CCC GGG ATC CGA TAA CTT CG | |
| 2nd round, forward | GTG GAT TCG TCA GGT CAT TCA GTT GGA TGT CAT GCA CAA AAA ACT GAA GTT TCT GAC | |
| 2nd round, reverse | CAC AGG AGC AGT AGT GCT GTC GAA ACA GAG TAC ACG TCT GTG GCA TGG TTT AGC AGC | |
| 10 (LoxP1125-1149) | 1st round, forward | ATC AAA AGA GAG AAA GAG AAG CCT CCT CTG AGG GAA CAA AAG CTG GGT ACC ATA ACT TCG |
| 1st round, reverse | AGA TTC AAG CAC AAT TTC TGT TGG TGA AAC TAT CGA ATT CCC GGG ATC CGA TAA CTT CG | |
| 2nd round, forward | ACC TTA AAA CCC CCT TCT AAT AAT GCT ATC AAA AGA GAG AAA GAG AAG CCT CCT CTG | |
| 2nd round, reverse | CTC CTT ATT AGC TGT TGC TTT ATG GAA AGA TTC AAG CAC AAT TTC TGT TGG TGA AAC | |
| 11 (LoxP1228-1252) | 1st round, forward | AAA AAT GTA CTT TCA GTA GGT ACA GCT GTG AGG GAA CAA AAG CTG GGT ACC ATA ACT TCG |
| 1st round, reverse | ACT ATC AGC AAG CCT ACT TAC TGA GCT GTG TAT CGA ATT CCC GGG ATC CGA TAA CTT CG | |
| 2nd round, forward | AAA AAA CAA GGC ACA TCT TCA AAC AAT AAA AAT GTA CTT TCA GTA GGT ACA GCT GTG | |
| 2nd round, reverse | AGG TGT CCG GGG CAC AGG TAA ATC ACT ACT ATC AGC AAG CCT ACT TAC TGA GCT GTG | |
| 12 (LoxP1325-1349) | 1st round, forward | AGC CCT GCC AGT GAA ACA GGA AGT AGG GAA CAA AAG CTG GGT ACC ATA ACT TCG |
| 1st round, reverse | AAA CTG TTG TGT GTT ATC TTT CAG AGG AGT TAT CGA ATT CCC GGG ATC CGA TAA CTT CG | |
| 2nd round, forward | CCT GTC ACC CCA GAC TTG CCT GCC TGC AGC CCT GCC AGT GAA ACA GGA AGT | |
| 2nd round, reverse | TTT TGT GGT GCT CCT TGA AGA TGC TCT AAA CTG TTG TGT GTT ATC TTT CAG AGG AGT |
Nucleotides in italics correspond to sequences homologous to the Tcr gene used for selection.
PCR mutagenesis.
Mutations in the LisH motif were generated by PCR using the following forward primers: for pCMV-p220ΔLisH, 5′-GGA TCC GCC ACC ATG TTG TTA CCC CAG ACT TTT ATT TTG GAA AGT TCA G-3′; for pCMV-p220VLVLE/A, 5′-GGA TCC GCC ACC ATG TTG TTA CCC TCG GAC GCA GCC CGG GCT GCA TTG GGT TAC GCA CAGCAAGCAAAC CTC ATT TCT AC-3′; for pCMV-p220-F/A, 5′-GGA TCC GCC ACC ATG TTG TTA CCC TCG GAC GTA GCC CGG CTT GTA TTG GGT TAC TTA CAG CAA GAA AAC CTC ATT TCT ACC TGC CAG ACT GCT ATT TTG GAA AGT TCA GAT TTA AAA G-3′; for pCMV-p220-FE/A, 5′-GGA TCC GCC ACC ATG TTG TTA CCC TCG GAC GTA GCC CGG CTT GTA TTG GGT TAC TTA CAG CAA GAA AAC CTC ATT TCT ACC TGC CAG ACT GCT ATT TTG GCA AGT TCA GAT TTA AAA G-3′. In all cases, the reverse primer was 5′-GTT TTC TGC TAG CTT TTC TTG-3′. PCR products were ligated into pCR2.1 (Invitrogen), and mutations were verified by sequence analysis. Mutant fragments were released from pCR2.1 with BamHI/NheI and subcloned into pCMV-p220 to generate the appropriate expression construct.
Cell culture and cell cycle analysis.
Human U2OS cells and rodent Rat1 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37°C with 0.5% CO2. For transfection, U2OS cells were seeded in 60-mm dishes and cultured overnight to a confluence of 60 to 70%. pCMV-p220 or mutant plasmids (10 μg) and 0.3 μg of pCMV-CD20 plasmid were cotransfected with FuGENE 6 (Roche). The medium was changed 8 h later. Thirty-six hours after transfection, cells were washed with phosphate-buffered saline (PBS) and treated with PBS-0.1% EDTA at room temperature for 10 min. Cells were then collected and washed with PBS prior to incubation with fluorescein isothiocyanate-conjugated anti-CD20 (Becton Dickinson) in PBS on ice for 15 min. Cells washed with PBS-0.1% serum and then fixed with 80% ethanol-PBS at 4°C for at least 1 h. Fixed cells were washed with PBS once and incubated in PBS with 10 μg of propidium iodide/ml and 40 μg of RNase A/ml at 37°C for 30 min. The DNA content of CD20-positive cells was determined by flow cytometry using an EPICS XL-MCL (Beckman-Coulter) instrument.
Luciferase assay.
U2OS cells were seeded in 12-well tissue culture dishes and cultured overnight to a confluence of 60 to 70%. pCMV-p220 or mutant plasmids (4 μg), pCMV-LacZ (0.1 μg), and pGL2-histone H4 promoter luciferase plasmids (0.1 μg) (40) were cotransfected by using calcium phosphate. In some experiments, a pGL2-histone H2B luciferase reporter containing the H2BFR promoter from chromosome 6 was used (23). The medium was changed 8 h later. Thirty-six hours after transfection, cells were collected and luciferase assays were performed with a luciferase assay kit (Promega). A β-galactosidase assay was used to normalize the luciferase activities.
Antibodies and immunofluorescence assays.
Generation of polyclonal antibodies against p220 and two phosphorylation sites (T1270 and T1350) were described previously (23). U2OS or Rat1 cells on coverslips were transfected with 4 μg of pCMV-p220 or mutant plasmids. In some cases, 0.5 μg of pCMV-EYFP-coilin/ml (28) was used included in the transfection. Cells were fixed with 4% paraformaldehyde for 10 min and then treated with 0.5% Triton X-100 in PBS for 10 min. p220 was detected with an antibody raised against the C terminus (23) For detection of p220, cells were stained with an anti-C-terminus antibody (37°C for 1 h). Detection was done with Cy3-labeled anti-rabbit antibodies (Amersham). Nuclear DNA was revealed with DAPI (4′,6′-diamidino-2-phenylindole) staining. To determine the phosphorylation status of p220, Triton X-100-treated cells were incubated with 3% H2O2 in PBS for 10 min and subsequently incubated with antigen retrieval solution (Dako) at 95°C for 30 min. Cells were incubated with antibodies against P-T1270 or P-T1350 in PBS for 1 h and then with horseradish peroxidase-conjugated anti-rabbit antibody (Roche) in PBS for 1 h. A tyramide signal amplification system (NEN) was used to detect the secondary antibodies. Cells were then fixed with 4% paraformaldehyde in PBS for 10 min prior to staining with anti-p220 C-terminus antibodies as described above. DNA was revealed with DAPI staining. Microscopic analysis was performed with an Olympus BX-60 microscope fitted with a Hamamatsu camera. Images were captured with a 100× objective lens using single-pass filters and merged by using NIH Image Pro software or a Deltavision deconvolution microscope.
RESULTS
Development of a lox-scanning mutagenesis approach to identify functional domains in p220NPAT.
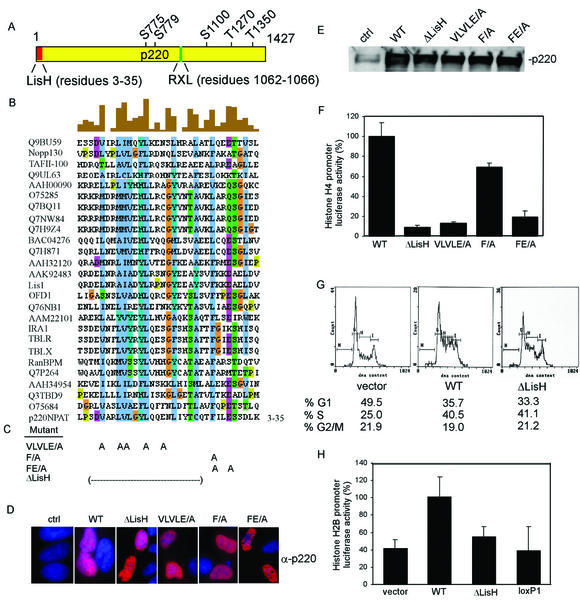
The large size of p220, coupled with the absence of obvious domains that can be linked with function, make the identification of biologically active sequence elements difficult. Therefore, we developed an approach we call lox-scanning mutagenesis, which allows systematic screening for potential functional domains in large proteins (Fig. 1). In this approach, homologous recombination is used to precisely replace 25-amino-acid segments at ∼100-amino-acid intervals along the entire length of p220 with a 25-amino-acid sequence encoded by a loxP cassette and flanking sequences (Fig. 1B). Replacement of functional elements contained within independent folded domains would be expected to alter the biological activity of p220, which is measured subsequently in cell-based assays. The approach takes advantage of recently developed methods that allow efficient homologous recombination of linear DNA fragments with circular plasmids in E. coli (38). PCR was used to generate recombination substrates containing a total of 120 bp of p220 sequences (corresponding to sequences 5′ and 3′ of the segment to be replaced) flanking a loxP-Tcr-loxP cassette encoding resistance to tetracycline. This linear template was transformed into recombination-proficient DH10β cells harboring both pML104 (Spr; encoding λ exonuclease and bet and gam recombination functions as well as RecA) (38) and the recombination target, a pCMV-p220 plasmid (Apr) (Fig. 1A). Recombination events were selected for resistance to tetracycline and ampicillin. Plasmids were then recovered and transformed into pBNN132 cells expressing Cre recombinase in order to excise the Tcr cassette. The identification of appropriate clones was aided by the inclusion of a BamHI restriction site in the loxP cassette (Fig. 1A to C), and mutations were verified by sequence analysis. The resulting set of 12 plasmids encode p220 mutants in which desired sequence elements along the length of p220 have been replaced precisely by residues encoded by the loxP cassette (Fig. 2). Mutant p220 plasmids were tested for expression by immunoblotting after transfection into U2OS cells. All 12 p220 mutant proteins were expressed at levels equal to or higher than that found with wild-type p220 expressed in parallel (Fig. 1D).
FIG. 1.
Strategy for lox-scanning mutagenesis and its application to p220. (A) Schematic representation of the approach used to generate lox-scanning mutants in p220. See the text for details. (B) Nucleotide and translated peptide sequences of the loxP cassette used to generate lox-scanning mutants. The position of an engineered BamHI site used to aid in the identification of appropriate recombinants is indicated. (C) Restriction enzyme analysis of pCMV-p220 plasmids generated by lox-scanning mutagenesis. Plasmids were digested with BamHI and analyzed by agarose gel electrophoresis. BamHI cuts at the 5′ end of the p220 cassette as well as in the recombined loxP site to generate a family of restriction fragments. Wild-type (wt) p220 was employed as a control. (D) pCMV-p220loxP mutant expression plasmids were transfected into U2OS cells, and the expression of mutant proteins was detected by immunoblotting with anti-p220 antibodies.
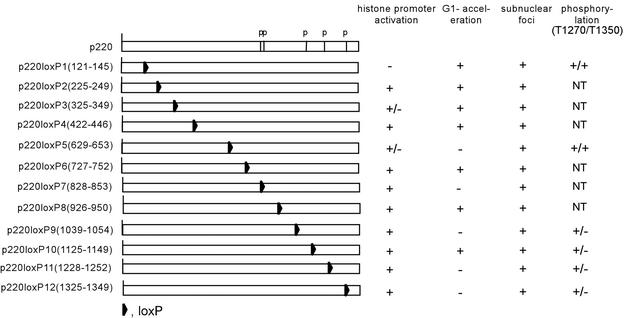
FIG. 2.
Functional analysis of lox-scanning mutant p220 proteins. The relative positions of loxP cassettes in 12 p220 mutants are shown along with the positions of five known phosphorylation sites (p). The activities of these proteins in cell based assays are summarized on the right.
Independent functional domains in p220NPAT control its G1 acceleration and histone transcription functions.
Previous studies have identified two activities for p220. First, overexpression of p220 in U2OS cells leads to a substantial increase in the population of S-phase cells at the expense of G1 cells (39). This activity does not appear to reflect aberrant accumulation in S phase because p220-overexpressing cells accumulate in G2/M in the presence of nocodazole (40; Wei and Harper, unpublished), indicating that p220 primarily promotes progression from G1 to S phase. Second, expression of p220 activates expression from histone H4 and H2B luciferase reporter constructs in U2OS cells (23, 40). It is not clear whether these two activities are linked.
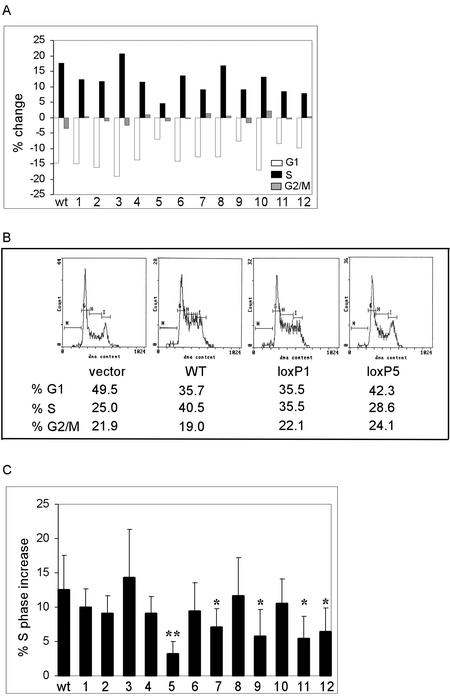
We tested the activity of our p220 lox-scanning mutants in both of these assays. The data are summarized in Fig. 2. Consistent with previous results (39), expression of p220 led to an ∼15% increase in S-phase cells from ∼25 to ∼40%, with only a small (2%) increase in the percentage of G2/M cells, as determined by using a CD20 cotransfection flow cytometry assay (Fig. 3A and B). The majority of lox-scanning mutants, including mutants 1, 2, 3, 4, 6, 8, and 10, were also found to elevate the percentage of S-phase cells to an extent comparable to that seen with wild-type p220 (8 to 14% increase), as measured over five replicate experiments (Fig. 3A to C). However, five mutants—5 (LoxP629-653), 7 (LoxP828-853), 9 (LoxP1039-1054), 11 (LoxP1128-1252), and 12 (LoxP1325-1349)—displayed substantially reduced activity in this assay. Mutant 5 showed the most dramatic reduction, as it increased S phase by only 3% over five independent experiments (P < 0.01), while mutants 7, 9, 11, and 12 displayed 5 to 7% increases in S phase (P < 0.05) (Fig. 3A to C). These data indicate that much of the C terminus of p220 is critical to its ability to promote S phase when overexpressed.
FIG. 3.
Multiple C-terminal elements in p220 are required for its ability to promote G1 acceleration. The indicated pCMV-p220 plasmids (10 μg) were cotransfected with pCMV-CD20 (300 ng), and the DNA content of CD20-positive cells was determined by flow cytometry after 36 h (see Materials and Methods). (A) Representative results showing the change in G1, S, and G2/M relative to a control employing pcDNA3. (B) Representative flow diagrams for pCMV-p220, pCMV-p220loxP1, and pCMV-p220loxP5. The percentages of G1, S, and G2/M cells are shown below each flow diagram. (C) Summary of the S-phase increase for five replicate transfection experiments. ∗∗, P < 0.01; ∗, P < 0.05.
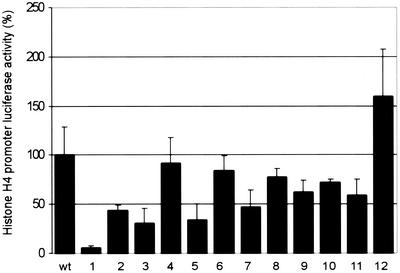
To examine the effect of these mutations on histone transcription activity, we employed a histone H4-luciferase reporter construct (Fig. 4). This reporter contains nucleotides −113 to +8 of the pHu4A (H4/e) promoter derived from the chromosome 6 histone gene cluster, and mutation of the SSCE in this promoter abolishes activation by p220 (40). In typical cotransfection studies, p220 expression led to a 50-fold increase in the level of luciferase activity relative to empty vector used as a negative control. The majority of p220 lox-scanning mutants, especially those affecting the C-terminal half of p220, displayed activities within twofold of that seen with wild-type p220 (Fig. 2 and 4). In contrast, mutant 1 (LoxP121-145) was essentially devoid of histone H4 transcriptional activity. In replicate assays, the activity was reduced by >20-fold (Fig. 4). As noted above, mutant 1 was essentially fully active in the G1 acceleration assay. These data suggest that the histone H4 transcriptional activation function of p220 is genetically separable from its ability to promote S-phase entry. Two additional lox-scanning mutants representing residues 325 to 349 and 629 to 653 reduced activity threefold, indicating that these regions may also contribute to histone H4 transcriptional activity (Fig. 4).
FIG. 4.
The N terminus of p220 is required for its ability to activate histone H4 promoter luciferase expression. U2OS cells were transfected with 4 μg of pCMV-p220, 100 ng of pCMV-lacZ, and 100 ng of pGL3-histone H4-luciferase. After 36 h, cell extracts were generated, and luciferase and LacZ assays were performed. Relative activities for three independent experiments, each done in triplicate, are shown as averages with standard deviations.
p220NPAT lox-scanning mutants are not defective in Cajal body localization.
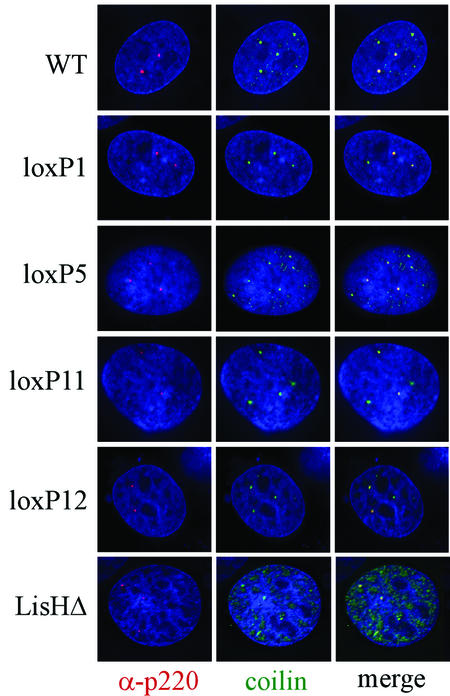
p220 is normally localized in Cajal bodies, and therefore, we considered the possibility that loss of transcriptional or cell cycle activities was due to improper localization. Transient expression of wild-type p220 in U2OS cells leads to accumulation through the nucleoplasm (see Fig. 7), making it difficult to assess the ability of p220 to localize properly in Cajal bodies. Therefore, we employed Rat1 cells to examine localization. This approach has two advantages. First, our p220 antibodies are specific for the human protein, and therefore, immunofluorescence can be used to directly examine the localization of the transiently expressed human p220 protein in Rat1 cells. Second, the level of expression achieved in Rat1 cells is markedly lower than that observed with U2OS cells, allowing visualization of p220 in nuclear foci. In order to simultaneously visualize Cajal bodies, we cotransfected cells with pCMV-EYFP-coilin. Although EYFP (enhanced yellow fluorescent protein)-coilin is found throughout the nucleoplasm when transiently expressed, it is concentrated in Cajal bodies (28, 32). Using this system, we examined the localization of all of the lox-scanning mutants described previously (Fig. 2 and 5). We found that all of these mutant proteins localized in small nuclear foci that are coincident with foci generated with EYFP-coilin. Thus, the absence of transcriptional or cell cycle activity found with particular lox-scanning mutants is not a reflection of defects in localization within Cajal bodies.
FIG. 7.
Differential effects of cyclin E/Cdk2 coexpression on p220 mutant protein activity and phosphorylation. (A) LisH mutants are not activated by cotransfection with cyclin E/Cdk2. The indicated plasmids were used in histone H4 reporter assays as described in Fig. 4, using 0.5 μg of pCMV-cyclin E and pCMV-Cdk2, as indicated. The results are means and standard deviations for triplicate assays. (B) Phosphorylation of p220 in U2OS cells. The indicated p220 expression plasmid (4 μg) was transfected into U2OS cells and subjected to immunofluorescence using the indicated antibodies. ΔCdk (23) is a mutant of p220 in which 5 of 17 phosphorylation sites (S775, S779, S1100, T1270, and T1350) are replaced by alanine. This protein has reduced transcriptional activation activity relative to the wild-type protein using both H2B and H4-luciferase reporter constructs (reference 23 and data not shown). p220 is red, T1270- and T1350-phosphorylated p220 are green, and DNA stained with DAPI is blue.
FIG. 5.
p220 lox-scanning and LisH mutants localize in Cajal bodies. The indicated pCMV-p220 plasmids (4 μg) and pCMV-EYFP-coilin (0.5 μg) were transfected into Rat1 cells. After 36 h, cells were fixed and subjected to immunofluorescence using anti-p220 antibodies as described in Materials and Methods. p220 is red, coilin is green, and DNA stained with DAPI is blue. Images were generated by deconvolution microscopy.
p220NPAT contains an N-terminal LisH-like motif that is required for histone transcription.
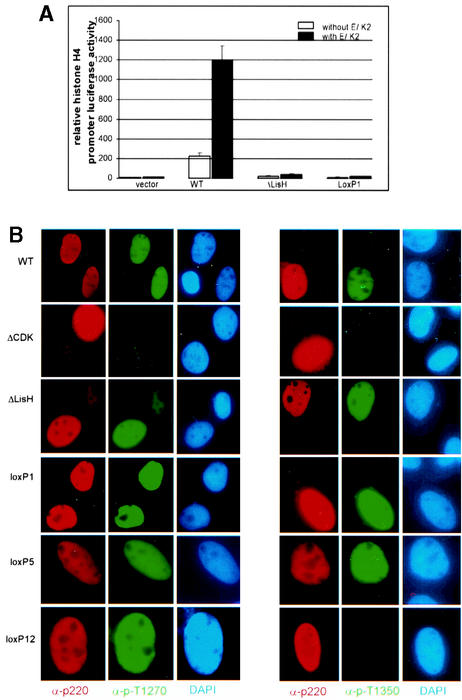
The finding that the N terminus of p220 is required for histone transcriptional activity with reporter constructs (Fig. 4) led us to examine this region in greater detail. Sequence analysis revealed that the extreme N terminus of p220 (Fig. 6A) has characteristics of a small motif—the LisH motif (15)—found in a number of other proteins in the database, including Lis1. The LisH motif contains approximately 33 amino acids and is characterized by hydrophobic residues at positions 5, 8, 9, 10, 12, 13, 18, 25, and 31, which are highly conserved in 26 human LisH-motif-containing proteins (Fig. 6B). The biochemical functions of LisH motifs are not clear. Given the proximity of this motif to sequences found to be important for histone H4 reporter activation (i.e., residues 121 to 145), we generated specific mutations in the LisH-like motif (Fig. 6C). These included deletion of residues 4 to 24 and compound point mutations in residues conserved in the LisH motif. Each of these mutant proteins was found to be expressed at levels similar to that observed with wild-type p220 when transfected into U2OS cells (Fig. 6D and E). These mutants were then tested for G1 acceleration and histone H4 promoter luciferase activity. Deletion of the LisH motif led to a 20-fold decrease in transcriptional activity (Fig. 6F). Moreover, mutation of conserved hydrophobic residues (VLVLE/A mutant) led to a 10-fold decrease in activity (Fig. 6F). This mutant includes replacement of the highly conserved leucine residue at position 15 in p220 (Fig. 6B). The analogous residue (Leu-16) was found to be mutated in the LisH motif of the Drosophila Ebi protein, which is involved in neuronal differentiation (6). We also examined Phe-27 in p220, which corresponds to Phe-31 in human Lis1, and this residue is replaced by serine in patients with lissencephaly (9). Mutation of Phe-27 in p220 to alanine led to a modest decrease in transcriptional activity (∼30%), but when it was combined with a mutation in the highly conserved Glu-30 residue, activity was reduced to 20% (Fig. 6F). In stark contrast, the ΔLisH p220 mutant displayed essentially wild-type activity in the G1 acceleration assay (Fig. 6G). Moreover, all of the LisH mutant p220 proteins localized in Cajal bodies when expressed in Rat1 cells (Fig. 5 and data not shown). These data strongly argue that the LisH-like motif at the N terminus of p220 is critical to its ability to activate histone H4 reporter constructs.
FIG. 6.
An N-terminal LisH domain in p220 is required for activation of the histone H4 promoter. (A) Schematic diagram of p220 showing the position of the LisH motif, the cyclin interaction motif, and known Cdk phosphorylation sites. (B) Clustal analysis of LisH motifs from 26 human proteins. Highly conserved residues are shown in blue. Alignments were generated by using Clustal W (http://www.ebi.ac.uk/clustalw/) and displayed by using Jalview (http://www2.ebi.ac.uk/∼michele/jalview/contents.html). (C) Positions of mutations generated in the LisH domain of p220. (D and E) p220 LisH mutants are expressed at levels indistinguishable from that of wild-type p220 in U2OS cells. The indicated pCMV-p220 expression plasmids were transfected into U2OS cells, and expression was examined after 36 h by immunofluorescence with anti-p220 antibodies (D) or immunoblotting (E). p220 is red; DNA is blue. (F) p220 LisH mutants are defective in activation of the histone H4 promoter reporter. U2OS cells were transfected with 4 μg of pCMV-p220 plasmid, 100 ng of pCMV-lacZ, and 100 ng of pGL3-histone H4-luciferase. After 36 h, cell extracts were generated, and luciferase and LacZ assays were performed. Relative activities from triplicate assays are shown. (G) A p220 ΔLisH mutant is not defective in G1 acceleration activity. Cell cycle acceleration assays using the indicated p220 expression plasmids were performed as described in Fig. 3. (H) Activation of a histone H2B-luciferase reporter by mutant p220 proteins. Plasmids expressing the indicated p220 mutants were transfected together with the pGL2-histone H2B-luciferase reporter as described in Materials and Methods. The results of duplicate assays, each done in triplicate, are shown as averages and standard deviations.
Previous studies indicate that p220 can promote transcription of not only the H4 promoter but also an H2B promoter, although the extent of H2B activation is significantly lower than seen with the H4 reporter. To address whether the N-terminal region of p220 is also involved in activation of the H2B promoter, the activities of ΔLisH and lox-scanning mutant 1 were compared with that of wild-type p220. Consistent with previous results (23, 40), expression of p220 led to an ∼3-fold increase in reporter activity (Fig. 6H). In contrast, lox-scanning mutant 1 displayed reporter activity comparable to that found in the absence of p220. The ΔLisH mutant displayed intermediate activity, consistent with the results found with the H4 reporter (Fig. 6H). These data indicate that the N terminus of p220 also contributes to activation of the H2B promoter.
LisH mutants in p220NPAT are not defective in Cdk-mediated phosphorylation.
Previous studies have demonstrated that phosphorylation of p220 is important for its ability to activate histone transcription and to promote S-phase entry in U2OS cells. Overexpression of cyclin E/Cdk2 potentiates both the activation of histone H4 luciferase reporter constructs and the accumulation of S-phase cells by p220 (39, 40), while mutation of five Cdk2 phosphorylation sites reduces activity in histone H4 reporter assays (23). Thus, we considered the possibility that defects in p220 function observed with LisH and lox-scanning mutants might reflect defects in Cdk2-mediated phosphorylation which could possibly be rescued by coexpression of cyclin E/Cdk2. To begin to address this question, we transfected pCMV-p220 expression plasmids with and without plasmids expressing cyclin E and Cdk2 into U2OS cells, and histone H4-luciferase activity was then measured (Fig. 7A). While the luciferase activity of wild-type p220 was stimulated ∼6-fold by coexpressed cyclin E/Cdk2, the activities of the LisH motif mutants were increased less than 2-fold. In empty vector control experiments (Fig. 7A), cyclin E/Cdk2 cotransfection also led to a 2-fold increase in background luciferase activity. These data indicate that defects in p220 LisH mutant protein function cannot be substantially enhanced by high levels of cyclin E/Cdk2.
Consistent with these data, we found that all p220 LisH mutant proteins retain the capability of being phosphorylated on Cdk sites. We performed immunofluorescence assays using anti-phospho-T1270 and anti-phospho-T1350 antibodies after transfection of p220 mutants in U2OS cells (Fig. 7B). These antibodies have been demonstrated to react with p220 in Cajal bodies during S phase in normal diploid fibroblasts (23). Cells expressing ectopic wild-type or mutant p220 proteins display anti-p220 staining throughout the nucleoplasm (Fig. 7B), as expected (16). All of the LisH mutant proteins were found to be phosphorylated on both T1270 and T1350, while p220ΔCdk (23), used as a control for phospho-antibody specificity, was devoid of staining with either phosphorylation site antibody (Fig. 7B). In contrast, certain lox-scanning mutants, in particular, mutants 9 to 12, were found to be defective in T1350 phosphorylation but not in phosphorylation of T1270 (Fig. 2 and 7B). In the case of mutant 12, the loxP replacement cassette is located adjacent to T1350 and could interfere directly in kinase recognition. However, mutants 9 to 11 are located some distance from T1350 in the linear sequence. Interestingly, mutant 10 was not phosphorylated on T1350 but was active in both the G1 acceleration assay and the histone transcription assay, suggesting that phosphorylation at this site is not critical to either of these activities. In addition, p220(LoxP629-653), which is the most severely affected with respect to G1 acceleration activity was found to be phosphorylated on both T1270 and T1350, suggesting that defects in phosphorylation are not responsible for the absence of S-phase-promoting activity with this p220 mutant.
DISCUSSION
Available data indicate that p220NPAT, a cyclin E/Cdk2 target and constituent of Cajal bodies, serves to link the basic cell cycle machinery with activation of histone gene expression during S phase. p220 expression in U2OS and 293T cells can activate expression of histone H4 and H2B reporter constructs and this requires known cell cycle regulatory sequences within these promoters (23, 40). This activity is stimulated upon phosphorylation of p220 by cyclin E/Cdk2. In addition, p220 has been shown to be closely associated with histone promoters based on chromatin immunoprecipitation assays (40). Currently, there is no evidence that p220 directly binds to histone promoters and it lacks motifs of known transcriptional activators. Other studies indicate that p220 can, in some instances, be rate limiting for S-phase entry. For example, expression of p220 can force G1 cells into S phase, and this activity is also enhanced by cyclin E/Cdk2 (39). Cells overexpressing p220 complete S phase and accumulate in mitosis in the presence of nocodazole, indicating that accumulation of S-phase cells does not reflect cell cycle arrest at this stage (reference 39 and data not shown). It is not clear whether these two activities of p220 are mechanistically related.
In this study, we have begun to explore how p220 functions in histone transcription and cell proliferation through the identification of functional domains. p220 contains a RXL-like motif (residues 1062 to 1066) (Fig. 6) that in other Cdk substrates is known to facilitate interaction with the cyclin (1, 31, 41). We found that replacement of the RVL sequence by AVA leads to a loss of cyclin E/Cdk2 binding in vitro (data not shown). In addition, short peptides containing wild-type (but not mutant) sequences from the E2F-1 RXL motif block association of p220 with cyclin E/Cdk2 in vitro. However, p220AVA is still phosphorylated by cyclin E/Cdk2 in vitro on at least a subset of sites, and it is not substantially defective in either S-phase-promoting activity or histone H4 reporter activation when transiently expressed (data not shown). Whether endogenous levels of p220 would require an intact RXL motif for function remains unknown.
Outside of this RXL motif, p220 lacks obvious functional domains that would link it mechanistically to known regulatory processes. Moreover, p220 is a large protein, making systematic analysis of functional domains difficult. We developed an approach we call lox-scanning mutagenesis (Fig. 1) as an approach to systematically analyze p220 sequences for functional activity. The approach is analogous to linker-scanning mutagenesis, which is frequently employed with smaller proteins, in that the LoxP replacement would be expected to disrupt the structure of folded domains and would therefore be expected to reveal regions of the protein important for function. Using this approach, we have identified multiple elements important for p220 function and have found that the ability of p220 to promote histone transcription can be genetically uncoupled from its ability to promote S phase.
The ability of p220 to promote the G1/S transition depends heavily upon sequences located in the C-terminal half of p220 (Fig. 2 and 3). lox-scanning mutants that have replacements of residues 629 to 653, 1228 to 1252, and 1325 to 1349 displayed reduced activity in cell cycle assays. Residues 629 to 653 appear to be most critical in this regard. This region is proximal to two previously identified Cdk sites, S775 and S779 (23). Whether this mutation affects phosphorylation at these sites is unknown, as phospho-specific antibodies against these sites are not available, but phosphorylation at T1270 and T1350 was not affected by this mutation. Two additional mutations that affect the G1 acceleration activity (LoxP1228-1252 and LoxP1325-1349) are located at the extreme C terminus of p220. Both of the mutant proteins are defective in phosphorylation of T1350 but not T1270. However, the fact that p220(LoxP1125-1149) is active in the G1 acceleration assay but is also not phosphorylated on T1350 indicates that this event is not required for S-phase promotion.
In contrast with the cell cycle function of p220, the histone H4 transcriptional activity appears to depend primarily on the N terminus (Fig. 2, 4, and 6). lox-scanning mutant 1, affecting residues 121 to 145, displayed the greatest reduction in activity toward the histone H4 promoter (20-fold). Importantly, this mutant displayed activity in cell cycle assays that was similar to that found with wild-type p220 (Fig. 3), indicating that transcriptional activity toward the H4 promoter is not required for the G1 acceleration function of p220. Additional lox-scanning mutants, particularly those affecting residues 325 to 349 (mutant 3) and 629 to 653 (mutant 5), also displayed significantly reduced activity (∼3-fold). Interestingly, mutant 5 was also defective in G1 acceleration. These data suggest that the region defined by mutant 5 may contribute to both cell cycle acceleration and histone transcription.
Sequence analysis of the extreme N terminus of p220 led to the identification of a LisH-like motif in p220 (Fig. 6). This ∼33 amino acid motif contains a series of conserved largely hydrophobic residues (9). To examine whether this motif might be involved in p220 function, we performed further mutagenic studies focusing on residues that are conserved in this motif. Deletion of this domain decreased activity toward histone H4 reporter constructs to an extent similar to that found with lox-scanning mutant 1. Additional point mutations in conserved residues reduced activity at least 10-fold. In contrast, none of the LisH mutations significantly affected the G1 acceleration activity of p220. These data indicate that this LisH domain is critical to the ability of p220 to activate histone transcription but is not required for its ability to promote S-phase entry. Consistent with this notion, we found that a p220 deletion mutant lacking residues 121 to 426 was fully active in promoting S phase and localized in Cajal bodies yet was defective in H4 reporter activation (data not shown). Given the close proximity of the LisH motif and sequences disrupted in lox-scanning mutant 1 (residues 121 to 145), it is possible that they act together to promote histone transcription. Alternatively, residues 121 to 145 may represent a second N-terminal domain that is involved in histone transcription independently of the LisH motif. Structural studies are required to determine if these two regions form a single functional domain.
p220 is localized in small nuclear organelles called Cajal bodies (23, 40). p220-containing Cajal bodies are associated with histone gene clusters on chromosome 6p21 during G1, S, and G2 phases of the cell cycle and with histone gene clusters on chromosome 1q21 during S and G2 phases. How p220 is targeted to these organelles and whether this localization is required for function is unknown. One potential explanation for the absence of activity of p220 mutants is that they do not localize properly to Cajal bodies. We tested each of the mutants examined here for their localization in Cajal bodies by transfection into Rat1 cells (Fig. 5). Surprisingly, all of the mutants examined localized specifically in Cajal bodies, as determined by colocalization with EYFP-coilin, analyzed in parallel. Thus, effects on transcription and cell cycle function do not appear to reflect an inability of the mutant proteins to localize in these organelles. We cannot, however, rule out the possibility that there are redundant sequences that target p220 to Cajal bodies and that within the Cajal body, the mutant p220 proteins are defective in association with other proteins that could independently serve to tether the p220 protein. Given the extent to which our panel of lox-scanning mutants explore p220 structure, it seems likely that a short linear sequence in p220 is involved in targeting it to Cajal bodies. The mechanisms responsible for localization of other Cajal body constituents is largely unknown, although it is clear that many Cajal body proteins display dynamic association with this organelle (reviewed in reference 27). Coilin itself requires its N terminus to associate with Cajal bodies, and this region is subject to specific arginine methylation (reviewed in reference 27). It is unclear whether p220 itself might be recruited to Cajal bodies in a coilin-dependent manner or whether it might require particular modifications for proper localization.
A major finding of this study is that p220 employs a LisH-like motif to promote histone transcription. LisH motifs are found in bacteria, fungi, and plant and animal proteins. Although LisH-containing proteins frequently have additional domains (15), such as WD40 repeats found in the human Lis1 and Drosophila Ebi proteins, there are a number of proteins in the database which, like p220, contain only LisH domains. Interestingly, p220 is not the only protein with a LisH domain that is involved in transcriptional regulation. Human TBLR1/IRA1 (Fig. 6A) and budding yeast Sif2p both contain an N-terminal LisH domain and C-terminal WD40 repeats. TBLR1/IRA1 is a component of an HDAC3 corepressor complex (37), while Sif2p is a component of the Sir4 histone deacetylase transcriptional silencing complex. Deletion of Sif2 in budding yeast leads to increased telomeric silencing (10). The precise functions of LisH motifs are unknown. In Lis1, the LisH motif is located on the N terminus of the protein and is adjacent to a coiled-coil domain, and this N-terminal fragment is known to be involved in Lis1 dimerization (2, 7). Given the known role of coiled-coil domains in dimerization, it seems likely that this motif is involved in the dimerization process, although a role for the LisH domain cannot be ruled out. It seems likely that LisH motifs function as protein interaction surfaces. Given the diverse array of proteins that contain this domain (Fig. 6B), it is probable that these different motifs confer binding to distinct targets in various settings. The identification of a functionally important LisH-like motif in p220 provides a starting point for the identification of proteins that collaborate with p220 to control histone gene transcription.
Acknowledgments
We thank S. Elledge (Baylor College of Medicine) for E. coli strain DH10β(pML104). We also thank A. Lamond (University of Dundee) and J. Zhao (University of Rochester) for plasmids.
This work was supported by grants from the National Institutes of Health (GM54137) and the Welch Foundation to J.W.H. Y.W. was supported by a Department of Defense predoctoral fellowship. J.J. was supported by a Department of Defense postdoctoral fellowship.
REFERENCES
- 1.Adams, P. D., W. R. Sellers, S. K. Sharma, A. D. Wu, C. M. Nalin, and W. G. Kaelin, Jr. 1996. Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol. Cell. Biol. 16:6623-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, C., and N. R. Morris. 2001. Nudf, a fungal homolog of the human LIS1 protein, functions as a dimer in vivo. J. Biol. Chem. 276:9903-9909. [DOI] [PubMed] [Google Scholar]
- 3.Albig, W., and D. Doenecke. 1997. The human histone gene cluster at the D6S105 locus. Hum. Genet. 101:284-294. [DOI] [PubMed] [Google Scholar]
- 4.Aziz, F., A. J. van Wijnen, J. L. Stein, and G. S. Stein. 1998. HiNF-D (CDP-cut/CDC2/cyclin A/pRB-complex) influences the timing of IRF-2-dependent cell cycle activation of human histone H4 gene transcription at the G1/S phase transition. J. Cell Physiol. 177:453-464. [DOI] [PubMed] [Google Scholar]
- 5.Aziz, F., A. J. van Wijnen, P. S. Vaughan, S. Wu, A. R. Shakoori, J. B. Lian, K. J. Soprano, J. L. Stein, and G. S. Stein. 1998. The integrated activities of IRF-2 (HiNF-M), CDP/cut (HiNF-D) and H4TF-2(HiNF-P) regulate transcription of a cell cycle controlled human histone H4 gene: mechanistic differences between distinct H4 genes. Mol. Biol. Rep. 25:1-12. [DOI] [PubMed] [Google Scholar]
- 6.Boulton, S. J., A. Brook, K. Staehling-Hampton, P. Heitzler, and N. Dyson. 2000. A role for Ebi in neuronal cell cycle control. EMBO J. 19:5376-5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cahana, A., T. Escamez, R. S. Nowakowski, N. L. Hayes, M. Giacobini, A. von Holst, O. Shmueli, T. Sapir, S. K. McConnell, W. Wurst, S. Martinez, and O. Reiner. 2001. Targeted mutagenesis of Lis1 disrupts cortical development and LIS1 homodimerization. Proc. Natl. Acad. Sci. USA 98:6429-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cajal, S. R. Y. 1903. Un sencillo metodo de coloracion seletiva del reticulo protoplasmatico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Trab. Lab. Investig. Biol. 2:129-221. [Google Scholar]
- 9.Cardoso, C., R. J. Leventer, N. Matsumoto, J. A. Kuc, M. B. Ramocki, S. K. Mewborn, L. L. Dudlicek, L. F. May, P. L. Mills, S. Das, D. T. Pilz, W. B. Dobyns, and D. H. Ledbetter. 2000. The location and type of mutation predict malformation severity in isolated lissencephaly caused by abnormalities within the LIS1 gene. Hum. Mol. Genet. 9:3019-3028. [DOI] [PubMed] [Google Scholar]
- 10.Cockell, M., H. Renauld, P. Watt, and S. M. Gasser. 1998. Sif2p interacts with Sir4p amino-terminal domain and antagonizes telomeric silencing in yeast. Curr. Biol. 8:787-790. [DOI] [PubMed] [Google Scholar]
- 11.Dailey, L., S. M. Hanly, R. G. Roeder, and N. Heintz. 1986. Distinct transcription factors bind specifically to two regions of the human histone H4 promoter. Proc. Natl. Acad. Sci. USA 83:7241-7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dailey, L., S. B. Roberts, and N. Heintz. 1988. Purification of the human histone H4 gene-specific transcription factors H4TF-1 and H4TF-2. Genes Dev. 2:1700-1712. [DOI] [PubMed] [Google Scholar]
- 13.Dominski, Z., L. X. Zheng, R. Sanchez, and W. F. Marzluff. 1999. Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol. Cell. Biol. 19:3561-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dyson, N. 1998. The regulation of E2F by pRb-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 15.Emes, R. D., and C. P. Ponting. 2001. A new sequence motif linking lissencephaly, Treacher Collins and oral-facial-digital type 1 syndromes, microtubule dynamics and cell migration. Hum. Mol. Gen. 10:2813-2820. [DOI] [PubMed] [Google Scholar]
- 16.Frey, M. R., and A. G. Matera. 1995. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc. Natl. Acad. Sci. USA 92:5915-5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gall, J. G., M. Bellini, Z. Wu, and C. Murphy. 1999. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol. Biol. Cell 10:4385-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gall, J. G. 2000. Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol. 16:273-300. [DOI] [PubMed] [Google Scholar]
- 19.Harper, J. W., and P. D. Adams. 2001. Cyclin-dependent kinases. Chem. Rev. 101:2511-2526. [DOI] [PubMed] [Google Scholar]
- 20.Harris, M. E., R. Bohni, M. H. Schneiderman, L. Ramamurthy, D. Schumperli, and W. F. Marzluff. 1991. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol. Cell. Biol. 11:2416-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heintz, N. 1991. The regulation of histone gene expression during the cell cycle. Biochim. Biophys. Acta 1088:327-339. [DOI] [PubMed] [Google Scholar]
- 22.Ma, T., N. Zou, B. Y. Lin, L. T. Chow, and J. W. Harper. 1999. Interaction between cyclin-dependent kinases and human papillomavirus replication-initiation protein E1 is required for efficient viral replication. Proc. Natl. Acad. Sci. USA 96:382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma, T., B. Van Tine, Y. Wei, M. Garrett, D. Nelson, P. D. Adams, J. Wang, J. Qin, L. T. Chow, and J. W. Harper. 2000. Cell-cycle regulated phosphorylation of p220NPAT by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 14:2298-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinelli, R., and N. Heintz. 1994. H1TF2A, the large subunit of a heterodimeric, glutamine-rich CCAAT-binding transcription factor involved in histone H1 cell cycle regulation. Mol. Cell. Biol. 14:8322-8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matera, A. G. 1999. RNA splicing: more clues from spinal muscular atrophy. Curr. Biol. 9:R140-R142. [DOI] [PubMed]
- 26.Nevins, J. R. 1998. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 9:585-593. [PubMed] [Google Scholar]
- 27.Ogg, S. C., and A. I. Lamond. 2002. Cajal bodies and coilin—moving towards function. J. Cell Biol. 159:17-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Platani, M., I. Goldberg, J. R. Swedlow, and A. I. Lamond. 2000. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J. Cell Biol. 151:1561-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramsey-Ewing, A., A. J. Van Wijnen, G. S. Stein, and J. L. Stein. 1994. Delineation of a human histone H4 cell cycle element in vivo: the master switch for H4 gene transcription. Proc. Natl. Acad. Sci. USA 91:4475-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez, R., and W. F. Marzluff. 2002. The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol. Cell. Biol. 22:7093-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schulman, B. A., D. L. Lindstrom, and E. Harlow. 1998. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc. Natl. Acad. Sci. USA 95:10453-10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shpargel, K. B., J. K. Ospina, K. E. Tucker, A. G. Matera, and M. D. Hebert. 2003. Control of Cajal body number is mediated by the coilin C-terminus. J. Cell Sci. 116:303-312. [DOI] [PubMed] [Google Scholar]
- 33.van der Meijden, C. M., P. S. Vaughan, A. Staal, W. Albig, D. Doenecke, J. L. Stein, G. S. Stein, and A. J. van Wijnen. 1998. Selective expression of specific histone H4 genes reflects distinctions in transcription factor interactions with divergent H4 promoter elements. Biochim. Biophys. Acta 1442:82-100. [DOI] [PubMed] [Google Scholar]
- 34.Vaughan, P. S., C. M. van der Meijden, F. Aziz, H. Harada, T. Taniguchi, A. J. van Wijnen, J. L. Stein, and G. S. Stein. 1998. Cell cycle regulation of histone H4 gene transcription requires the oncogenic factor IRF-2. J. Biol. Chem. 273:194-199. [DOI] [PubMed] [Google Scholar]
- 35.Vaughan, P. S., F. Aziz, A. J. van Wijnen, S. Wu, H. Harada, T. Taniguchi, K. J. Soprano, J. L. Stein, and G. S. Stein. 1995. Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature 377:362-365. [DOI] [PubMed] [Google Scholar]
- 36.Whitfield, M. L., L. X. Zheng, A. Baldwin, T. Ohta, M. M. Hurt, and W. F. Marzluff. 2000. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 20:4188-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, J., M. Kalkum, B. T. Chait, and R. G. Roeder. 2002. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 9:611-623. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, P., M. Z. Li, and S. J. Elledge. 2002. Towards genetic genome projects: genomic library screening and gene-targeting vector construction in a single step. Nat. Genet. 30:31-39. [DOI] [PubMed] [Google Scholar]
- 39.Zhao, J., B. D. Dynlacht, T. Imai, T. Hori, and E. Harlow. 1998. Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S-phase entry. Genes Dev. 12:456-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao, J., B. K. Kennedy, B. D. Lawrence, D. Barbie, A. G. Matera, J. A. Fletcher, and E. Harlow. 2000. NPAT links cyclin E-cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 14:2283-2297. [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu, L., E. Harlow, and B. D. Dynlacht. 1995. p107 uses a p21CIP1-related domain to bind cyclin/cdk2 and regulate interactions with E2F. Genes Dev. 9:1740-1752. [DOI] [PubMed] [Google Scholar]