Abstract
Chromatin structure is believed to exert a strong effect on replication origin function. We have studied the replication of the chicken β-globin locus, whose chromatin structure has been extensively characterized. This locus is delimited by hypersensitive sites (HSs) that mark the position of insulator elements. A stretch of condensed chromatin and another HS separate the β-globin domain from an adjacent folate receptor (FR) gene. We demonstrate here that in erythroid cells that express the FR but not the globin genes, replication initiates at four sites within the β-globin domain, one at the 5′ HS4 insulator and the other three near the ρ- and βA-globin genes. Three origins consist of G+C-rich sequences enriched in CpG dinucleotides. The fourth origin is A+T rich. Together with previous work, these data reveal that the insulator origin has unmethylated CpGs, hyperacetylated histones H3 and H4, and lysine 4-methylated histone H3. In contrast, opposite modifications are observed at the other G+C-rich origins. We also show that the whole region, including the stretch of condensed chromatin, replicates early in S phase in these cells. Therefore, different early-firing origins within the same locus may have opposite patterns of epigenetic modifications. The role of insulator elements in DNA replication is discussed.
The replicon model (35) hypothesized two essential components for replication initiation: the initiator, a trans-acting factor, and the replicator, a cis-acting element. The initiator would bind the replicator, and appropriate signals would direct DNA unwinding and recruitment of additional factors at a replication origin that, in the simplest case, would coincide with the replicator. This model has been confirmed in bacteria and viruses, which use a single origin to duplicate their genome. In the genomes of eukaryotic cells, replication initiates at multiple origins that have been more difficult to identify. Studies with Saccharomyces cerevisiae led to the identification of replicators, the so-called autonomously replicating sequences (ARSs) that share an essential 11-bp ARS consensus sequence (for reviews see references 49 and 61) and to a single initiator, the origin recognition complex (ORC) (8, 19). The discovery that ORC and additional factors required for initiation are highly conserved suggested that the replicon model might be directly applicable to all eukaryotes (7).
However, no element with properties similar to those of the yeast ARS consensus sequence has been identified to date in any other eukaryote. In several eukaryotic systems, any DNA sequence can function as a replicator (17, 29). In early embryos of Xenopus and Drosophila melanogaster, replication initiates with no regard for specific sequences, whereas later in development origins are progressively restricted to more specific sequences (33, 46, 58). In differentiated cells, nevertheless, there is evidence that defined cis-acting elements are required for origin function. For example, an 8-kb deletion encompassing the initiation region located between the δ- and β-globin genes abolishes initiation within the human β-globin locus (39). One interpretation of these variable-sequence requirements is that the use of specific replication origins is not needed to duplicate the genome but rather to coordinate replication with chromatin remodeling and gene expression. Indeed, ORC has been implicated in the establishment of transcriptionally repressed domains in S. cerevisiae (25, 48) and in heterochromatin formation or maintenance in Drosophila (50).
Another feature of metazoan but not yeast origins is that initiation at a specific sequence sometimes depends on far-removed sequences (2, 15, 37). For example, initiation at the human β-globin origin is abolished in the Hispanic deletion, which removes sequences located >50 kb upstream (2). This deletion also results in a closed chromatin structure and abolition of transcription of the β-globin locus in erythroid cells (24). A potential role for these distal sequences is to prevent inhibition of initiation by a repressive chromatin structure. In fact, initiation from the human β-globin origin introduced into ectopic sites does not require these distal elements (3).
Insulator elements, first described to protect gene expression from inappropriate signals (64), may play a role in DNA replication. Insulators can block the action of an enhancer on a promoter if placed between (but not outside) them (enhancer-blocking effect) or can prevent the advance of nearby condensed chromatin that might otherwise silence gene expression (protection against position effects, or barrier effect). Interestingly, studies of Drosophila chorion gene amplification by P-element-mediated transformation have shown that flanking the chorion gene replication initiation zone with exogenous insulators protects replication initiation from position effects (43, 44).
To address the role of insulators in replication in their natural context, we analyzed the replication of the chicken folate receptor/β-globin region, a 52-kb chromosomal region containing three independent domains separated by well-defined boundaries marked by DNase I-hypersensitive sites (HSs) (Fig. 1C) (41, 42, 53). The 30-kb chicken β-globin gene cluster contains four globin genes that are developmentally regulated and expressed in erythroid cells (23). The β-globin gene cluster is separated by an upstream (5′ HS4) and a downstream (3′ HS) insulator element from an upstream 16-kb region of constitutively condensed chromatin and a downstream olfactory receptor gene (14, 57). The HSs 5′ HS4 and 3′ HS mark the positions of binding sites for the ubiquitous DNA-binding protein CTCF that are necessary and sufficient for the enhancer-blocking activity of these insulators (6). In contrast, the CTCF binding site appears unnecessary for the barrier activity of 5′ HS4 (54). Further upstream of the 16-kb condensed chromatin region is a folate receptor (FR) gene which is expressed only in immature erythroid cells before the globin genes are active. Another hypersensitive site, HSA, sits at the boundary between the FR gene and the condensed chromatin region (53). In previous work, several types of chicken cells had been used to study the correlation between transcription, chromatin structure, and histone modifications throughout the FR gene, condensed chromatin stretch, and β-globin cluster (41, 42, 53). These detailed analyses have made the chicken β-globin locus very attractive for replication studies.
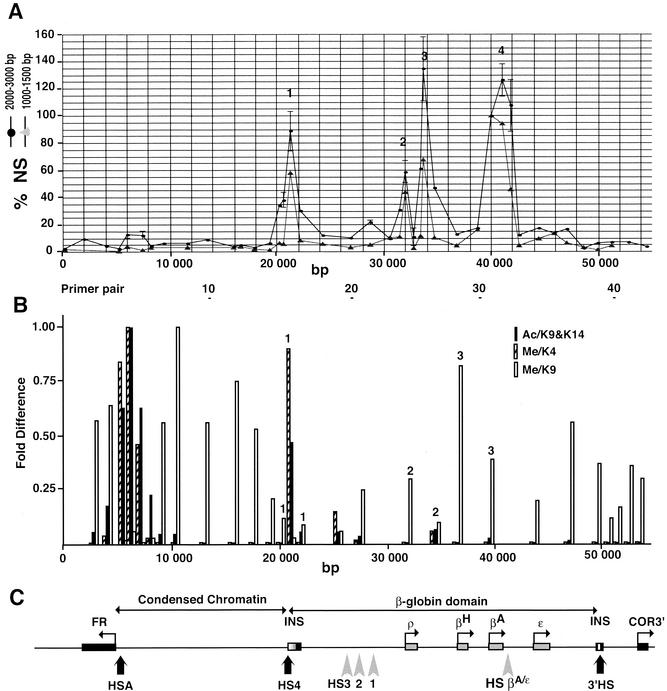
FIG. 1.
Replication origin mapping of the chicken β-globin locus. (A) Relative enrichments of 1.0- to 1.5-kb and 2.0- to 3.0-kb nascent strands in the 52-kb FR/β-globin region reveal four sites of initiation of replication in the β-globin locus. Two different preparations of nascent strands (1.0 to 1.5 kb, gray triangles, and 2.0 to 3.0 kb, black circles) were used to determine the abundance of short RNA-primed DNA chains along the chromosome by real-time PCR with 36 and 42 different primer pairs, respectively. Each quantification was repeated twice except in the case of the 2.0- to 3.0-kb fraction (four times for primer pairs 5, 6, 16, 21, 24, 27, 32, and 33 and six times for primer pairs 17 and 25). The scale corresponds to a relative nascent-strand abundance (NS) normalized with respect to primer pair 31 (arbitrarily set as 100%). The abscissa scale is map position (nucleotide number). The positions of primer pairs 10, 20, 30, and 40 are shown below. (B) Map of histone H3 modifications at the chicken β-globin/FR locus in 6C2 cells (adapted from reference 41). Normalized data of H3 diacetylation at K9 and K14 (Ac/K9&K14), H3 methylation at K4 (Me/K4), and H3 methylation at K9 (Me/K9). Note the correlation between diacetylation at K9 and K14 and methylation at K4 and anticorrelation with methylation at K9. Numbers 1, 2, and 3 above vertical bars correspond to probes reported in Fig. 2, 3, and 4, respectively. (C) Map of the FR gene, condensed chromatin region, and β-globin domain. DNase I HSs are indicated (HSA, HS1 to -4, HS βA/ɛ, 3′ HS). HSA is located 1 kb upstream of the FR transcription start site. HS4 maps within the upstream insulator element (INS) of the β-globin locus. The 16-kb region between HSA and HS4 consists of micrococcal nuclease-resistant chromatin. The embryonic (ρ- and ɛ-), hatching (βH-), and adult (βA-) globin genes are indicated. The β-globin locus is controlled by an LCR consisting of HS1, HS2, HS3, and the strong enhancer HS βA/ɛ. The HS1 and HS2 sites are not detectable in 6C2 cells. The 3′ end of the locus is marked by the 3′ HS insulator. Beyond the 3′ HS insulator (INS) a gene coding for an olfactory receptor (COR3′) is indicated.
The 6C2 cell line mimics the CFU-E stage of differentiation, which precedes the final differentiation step in erythrocytes. At this stage, the FR gene is expressed but the globin genes are not. In this work, we identify at least four replication origins within the β-globin cluster in 6C2 cells. Replication initiates at the 5′ HS4 insulator as well as 5′ and 3′ of the silent ρ-globin gene and over a broader 1.5-kb region that covers the βA-globin gene and its promoter. We also show that the entire 52-kb region is replicated early in S phase, and therefore the four origins fire early in S phase. The initiation site located 3′ of the ρ-globin gene is A+T rich, but the three other replication origins are G+C-rich, CpG-enriched sequences. The CpGs are totally unmethylated at the insulator but are partially methylated at the two other origins. The 5′ HS4 insulator is known to be associated with hyperacetylated histones, but the origin located 5′ of the ρ-globin gene is associated with hypoacetylated histones. Whereas the data confirm that CpG- and A+T-enriched sequences may define a subset of metazoan DNA replication origins, we conclude that different active origins within the same locus can show different patterns of epigenetic modifications. We discuss the possible role of insulator elements in DNA replication.
MATERIALS AND METHODS
Cell culture.
6C2 is a colony-forming unit-erythrocyte (CFU-E)-stage erythroid precursor cell line obtained by transformation of bone marrow with wild-type avian erythroblastosis virus. 6C2 cells were grown as previously described (53) and were harvested in exponential phase.
Preparation of short nascent strands.
Total genomic DNA was extracted from 108 asynchronously growing 6C2 cells with DNAzol reagent (Gibco-BRL) according to the manufacturer's instructions. DNA was resuspended in a solution containing 10 mM Tris-HCl (pH 8.0), 1 mM EDTA in the presence of 1 U of RNase inhibitors (Promega)/μl, denatured by heating for 5 min at 95°C, and size fractionated on a sucrose gradient as described previously (28). Fractions were analyzed by electrophoresis on an alkaline 1.2% agarose gel and staining with SYBR-Gold (Molecular Probe). Fractions containing fragments from 1 to 1.5 kb, 2 to 3 kb, and 4 to 5 kb were pooled, and the DNA was precipitated and treated with polynucleotide kinase and λ-exonuclease as described previously (1).
Quantitative real-time PCR.
Quantitative real-time PCR was performed by using the LightCycler (Roche) detection system with a QuantiTect SYBR Green PCR kit (Qiagen). Primers were synthesized for each of the 45 target sites to be amplified (Table 1). For each reaction, amplification of the purified nascent strands was performed in duplicate. Five fourfold dilutions of BamHI-digested total genomic DNA and a reaction mixture without template DNA were used as controls. Amplification was run for 50 cycles in 20 μl with 1 μl of nascent-strand preparation (1/100 of the 1.0- to 1.5-kb nascent-strand preparation or 1/200 of the 2.0- to 3.0-kb and 4.0- to 5.0-kb nascent-strand preparation), 10 μl of 2× QuantiTect SYBR Green PCR Master Mix, 8 μl of H2O, and 1 μl of primers (10 μM each). Primer pair 31 was used as a standard in each PCR experiment. Subsequent to amplification, a melting curve analysis was performed to analyze the products and to control the specificity of the reaction. Quantitations were made by using the second derivative maximum method described in the LightCycler Software.
TABLE 1.
List of primer sequences
| Primer pair | Sequence (5′ to 3′) | Map position |
|---|---|---|
| 1 | TGAGCTGCTGACTCTGCCCA | 80 |
| CATGTCCTCATGGCCAACAC | 382 | |
| 2 | CAATGCAGCTCTGGCAGTCT | 1904 |
| GTTACTCGTGTCCCTGCATG | 2075 | |
| 3 | CAGCAAGATGGTGTTGAAGG | 4016 |
| CAGTGGTGTGGGTAATTGGA | 4322 | |
| 4 | GCCTCCTGACACAGCAGAGC | 5217 |
| GCAGGTTTGAGGCAAGTTAG | 5512 | |
| 5 | CTGAATGGTCTTTGTGGTCCC | 6064 |
| CTTGCTGTGGGAGACCTGCT | 6348 | |
| 6 | TGGGAGATAATTGTTAGCCAC | 7499 |
| TATGCTGGGAACAGCTATAC | 7651 | |
| 7 | GCCATACCCATTCACTCCAT | 8166 |
| AATGGGACACGCAGTCCCAG | 8391 | |
| 8 | GCATCCTTCATCCAGCTG | 9380 |
| CAGTGAGCAGACTGTGAAGC | 9533 | |
| 9 | CTGTGGTCTGGTTTGTTTGG | 11649 |
| CAGCCTCTCCACTCATGCAC | 11956 | |
| 10 | CTGAGCTGTGACAGTGCC | 13489 |
| AATGGTCTTTGCCTGGGTCA | 13652 | |
| 11 | TGGCACCGATGGGATCACAT | 15895 |
| CACTGATACTGCAGCCTCTT | 16189 | |
| 12 | GGTGTTCAATAATCAGACTGTC | 16535 |
| TCACATTTCAGCTGGCGCTC | 16824 | |
| 13 | TCTCACTGTGCAGATGATGG | 17905 |
| CATGGTGTCATCACAGCATC | 18159 | |
| 14 | GGATCCTGAAGCTGTTTGGC | 19230 |
| CCTGCCTCGTGTCTTCTGG | 19488 | |
| 15 | ACGGGATGGTGAAGGCACAG | 20165 |
| TTAGCTCCCATTCCCCCACA | 20472 | |
| 16 | CCTTGTTGCCAGCTGTTAGCA | 20649 |
| ACTGTTAGACAAACATCTCCGTG | 20800 | |
| 17 | CCAAAGTGAAATCATGAAGGC | 21298 |
| GACCTTCCAGGAAAGCCTG | 21538 | |
| 18 | GGACGTGGACATGCAGGTG | 22200 |
| CCAGCCTTCATGATTTGACG | 22508 | |
| 19 | TGGGCAGTTTTCTAAGGGAA | 24240 |
| GTGCTGGCAGGGCATTCCAA | 24554 | |
| 20 | CAGCTCTGAGCACAGCACTG | 26910 |
| CAGGGAATTCCTTCTGGGG | 27216 | |
| 21 | CTGTCTGAATATCCTGGCTC | 28642 |
| GTGATTCAATGTCAGGCACT | 29021 | |
| 22 | GTGAGAGGGGCACTCCAGG | 30441 |
| GCAGTGCTCCGATAATGCC | 30739 | |
| 23 | TGCACAGGGGCACCATTTTG | 31444 |
| CCTTGCATAAGGACAGCAGG | 31610 | |
| 24 | GAAGGGTGAGGGAAGTGCC | 31941 |
| TCAGTGTGCACAAGGTGTGG | 32211 | |
| 25 (external) | GAACACCTACGCCAAGCTGT | 32738 |
| GTCAGTTCCATGTCTGCCTG | 33060 | |
| 5′ half | TGCTGCACCTTGTTCCACAC | 32925 |
| 3′ half | CAGCAGATGAAGGAGGGAAG | 32919 |
| 26 | TTACCCCATTGCTCCCCTTC | 33317 |
| ACAGCCCTGAGCCCTCTTTC | 33600 | |
| 27 | GACGGTCAGGTTTGCCAAAG | 33620 |
| TCCTGAGGATACGTTTTTCAG | 33887 | |
| 28 | CTGGGAGCAAAGACACTGAC | 34701 |
| TGGTCACTCTGATTGCAGCA | 34869 | |
| 29 | GATACGCACTGAGCTCTCGT | 36700 |
| CACCCATGATCTCGTAGCCA | 37000 | |
| 30 | GAACAAGTCATTGCACAACGG | 38690 |
| GGCAGTGAAACCAAGTGCTC | 38994 | |
| 31 | TGGTGTGGCCACGGATCTG | 40040 |
| GTGATGAGCTGCTTCTCCTC | 40333 | |
| 32 | CATAGAGCAAGGGACGGTG | 41070 |
| TACTGTGGGAAGAGTAGCTC | 41231 | |
| 33 | AGCGCTTTGTGCTCAGTGG | 41835 |
| GTTTATCTGCAACCTGCCCCC | 42112 | |
| 34 | TCCCCTGACTCACTGCTGG | 42570 |
| TACCCTTTTCCTTCCGGCTG | 42857 | |
| 35 | GCAGCTCCGCTCCAAGCTCT | 44490 |
| GGCTGGAGAGGTTCCCAAAG | 44810 | |
| 36 | TCCAAGCAGCACTAACCCTG | 45900 |
| ACGTTGACCAGCTTCTGCCA | 46039 | |
| 37 | GCTGAATGCTGTGCCTCTGG | 47020 |
| CCATCAACCTGCTAGAGAAG | 47318 | |
| 38 | CCCAGATTTGCTTCATCAGGAG | 48560 |
| GAGATGTGATAACAGACTGCCA | 48758 | |
| 39 | CACTTCTGCTCTACAAGGCC | 49950 |
| ACTGACATGGAAACACACGG | 50103 | |
| 40 | GCAGTGCAGACCCATCCCT | 51170 |
| TCTTTCTGTGCGTCATCAGC | 51455 | |
| 41 | CCAAGCAAGTCCAGACAGAG | 52817 |
| GCAAGCCTACATTCCTCCC | 52996 | |
| 42 | TCATGAACTCCCAGTACCAG | 54424 |
| TGCACACTGATCAGTAGGTC | 54567 | |
| LYS | CGGGTATCATTAGTGCCGAG | |
| CTGCCAGTATATCCTGGCAAA | ||
| MIT | CATCCCATGCATAACTCCTG | |
| GTAGTCCAGGCTTCACTTGA | ||
| AM | AGGCTTCTCTCCCGTAAATG | |
| CAAGCTGCAAGAAAGAGCTC |
Replication timing analysis.
Approximately 107 exponentially growing cells were labeled with 50 μM BrdU (Roche) for 1 h. Cells were harvested, washed twice in phosphate-buffered saline, fixed in 75% ethyl alcohol, and stored at 4°C overnight. For fluorescent-activated cell sorting, fixed cells were rehydrated at a concentration of 2.106 cells/ml in cold phosphate-buffered saline with 0.05% NP-40, propidium iodide (50 μg/ml), and RNase A (0.5 mg/ml). By using an Elite Beckman Counter cells were separated into seven compartments of the cell cycle based on propidium iodide staining intensity (DNA content), corresponding to G1 and S1 to S6. Ten thousand cells from each compartment were sorted into tubes containing 100 μl of lysis buffer (50 mM Tris-HCl [pH 8], 10 mM EDTA, 100 mM NaCl, 0.5% sodium dodecyl sulfate, 0.2 mg of proteinase K/ml). DNA was then purified, sonicated to a size range of 1 kb, and denatured by boiling. Immunopurification of BrdU-labeled DNA was performed as described previously (16). From each cell cycle fraction DNA equivalent to 500 sorted cells was used as a template for either multiplex PCR or real-time PCR. Fifty-microliter reaction mixtures were made with 2.5 U of AmpliTaq Gold Taq polymerase (Applied Biosystems), which was activated at 95°C for 6 min before PCR was started. Each cycle consisted of denaturation for 30 s at 95°C, annealing for 1 min at 55°C, and extension for 15 s at 72°C. Aliquots (10 μl) were taken after 25, 30, and 35 cycles. For multiplex PCR experiments, amplified products were separated on a 2.5% MetaPhor agarose, transferred on nylon membranes, and probed with fragments radiolabeled by random priming. The blots were exposed to storage phospho-screens and were visualized by using an FLA-3000 phospho-fluoroimager (Fuji). Quantitative real-time PCRs were performed as described above.
DNA methylation analysis.
Five micrograms of genomic DNA extracted with DNAzol from exponentially growing 6C2 cells was digested with 100 U of either MspI or HpaII for 2 h at 37°C. The digested DNA was extracted with phenol-chloroform and was precipitated with ethanol. Fifty nanograms of either undigested or MspI-digested or HpaII-digested DNA was used as a template for PCR with various primer pairs. The PCR protocol was the same as that for replication timing analysis for 30 cycles. Amplification products were visualized by gel electrophoresis and ethidium bromide staining. Quantitation by real-time PCR of MspI- or HpaII-digested DNA was performed as described for nascent strands. Primer pair 25 amplifies a PCR fragment lacking a CCGG site and was used as a control for input DNA.
RESULTS
Initiation of replication along the chicken FR/β-globin region is restricted to specific sites.
We mapped replication origins of the chicken FR/β-globin region by quantifying the abundance of short nascent strands at various positions along this region. Genomic DNA from proliferating 6C2 cells was heat denatured and size fractionated by sucrose gradient centrifugation. DNA in the 1.0- to 1.5-kb and 2.0- to 3.0-kb fractions were collected and treated with λ-exonuclease after phosphorylation by T4 polynucleotide kinase. This digestion eliminates nicked DNA but not short nascent strands, which are protected against λ-exonuclease by their RNA primers (27). Sequences close to replication origins are expected to be enriched in the 1.0- to 1.5-kb nascent-strand preparation. The 2.0- to 3.0-kb nascent-strand preparation is also expected to be enriched in sequences located 1.0 to 1.5 kb away from origins. The relative abundance of specific sequences within two nascent-strand preparations of different sizes was quantified by real-time PCR by using 42 different primer pairs distributed at 1.0- to 2.0-kb intervals along the 52-kb region (Fig. 1A). We also used as an exogenous negative control a primer pair that amplifies a fragment located 4 kb upstream of the 5′ matrix attachment region of the lysozyme locus and reported to lack initiation activity (51).
The results of all the quantitations are summarized in Fig. 1A. The strongest abundance in the 1.0- to 1.5-kb nascent-strand preparations (arbitrarily set as 100%) was detected with a primer pair located just upstream of the βA-globin gene (Fig. 1A, primer pair 31). The relative nascent-strand abundance for the other primer pairs was therefore normalized with respect to primer pair 31. This primer pair was used as a standard in each PCR experiment. For each primer pair two quantitations were performed, and the average is reported on the graph. The abundance profile of the 2- to 3-kb nascent-strand preparation was identically performed, except that six supplementary primer pairs were used (primer pairs 2, 3, 8, 10, 41, and 42) and more than two quantitations were made with several primer pairs (see the legend to Fig. 1). The two profiles showed four discrete peaks indicative of replication initiation sites located inside the β-globin locus (Fig. 1A). One corresponded to the 5′ insulator HS4 (primer pair 17). Two initiation sites were observed upstream and downstream of the ρ-globin gene (primer pairs 24 and 27). Finally, a fourth initiation region was observed over the βA-globin gene and promoter region (primer pairs 31, 32, and 33). The relative abundance of 1.0- to 1.5-kb nascent strands was <4% over the condensed chromatin region and the FR gene, <6% over the locus control region (LCR), and 3% at the negative control fragment from the lysozyme locus (primer pair LYS), arguing against initiation in these regions. Taking the relative abundance of primer pair 1 as background, the enrichment in nascent-strand abundance observed at the 5′ HS4 insulator, the ρ-globin gene and the βA-globin promoter are 30-, 35-, and 50-fold, respectively. These values are quite high compared to those of similar studies at other loci and suggest that origins located in the chicken β-globin locus may be active during a large fraction of the cell cycle. The different abundance of nascent strands at the four origins may be due to differences in origin efficiency or in fork speed or both.
Our results are reminiscent of the human (39) and murine (4) β-globin loci inasmuch as in all cases initiation was detected within or close to the globin genes but not at the LCR. The distribution of initiation sites in the chicken locus is somewhat intermediary between the sharply defined sites found in the human locus and the dispersed initiation zone found for the murine locus. The location of an initiation region over the chicken βA gene was reminiscent of the location of the initiation site between the human δ- and β-globin genes. However, initiation in the chicken locus also occurs at the 5′ HS4 insulator, whose functional equivalents in the mouse and human loci (22) were not reported to show origin activity.
The replication origins of the chicken β-globin locus are associated with either hyper- or hypoacetylated histones.
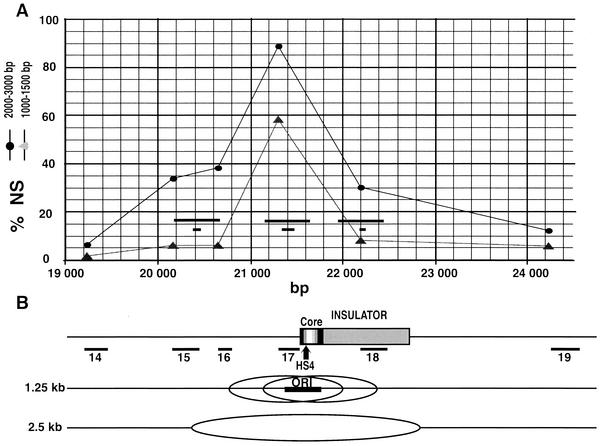
A close comparison of the abundance profiles of nascent strands of different sizes allowed us to refine the mapping of replication initiation sites. An enlargement of the region containing the 5′ HS4 insulator and quantitations of 1.0- to 1.5-kb and 2.0- to 3.0-kb nascent strands are shown in Fig. 2. With 1.0- to 1.5-kb nascent strands (gray triangles), only primer pair 17 showed a significant enrichment above the background level. Thus, bubbles up to 1.5 kb in length do not significantly overlap primer pairs 16 and 18. We have drawn on panel B the leftmost and rightmost possible positions of 1.25-kb bubbles that would not completely encompass probes 16 and 18. The centers of these two bubbles define the boundaries of a 400-bp segment where the origin has to be located. This region is centered on the 5′ end of the HS4 core insulator region. The analysis of the longer 2.0- to 3.0-kb nascent strands (black circles) confirmed this positioning. As expected, primer pairs located less than 1.5 kb away on either side of this potential zone of initiation showed significant enrichments (probes 15, 16, and 18), whereas more distal probes (14 and 19) were not enriched. The amplified probes used in previous studies (41, 42; see also Fig. 1B) to characterize the modifications of histone tails in this region are reported on the graph (small black bars). In these studies, the authors immunoprecipitated mono- and dinucleosomes and thus probed a region of about 500 bp centered on the primer pair used. These regions are represented above the probes (long black bars). The site of initiation coincides with a region containing histone H3 hyperacetylated on K9 and K14 and methylated on K4 (Fig. 1B, bars numbered 1) as well as hyperacetylated histone H4 (41, 42).
FIG. 2.
The leftmost initiation site colocalizes with the 5′ boundary of the β-globin locus. (A) A blow-up of the graph shown in Fig. 1A around the 5′ boundary of the chicken β-globin locus. The short and long horizontal black bars indicate the amplified PCR products used previously (41) to characterize histone tail modifications and the corresponding detection zones (3 nucleosomes), respectively. (B) The map shows the 1.2-kb insulator, the 250-bp core insulator region, the position of HS4, and PCR fragments used to quantify nascent strands shown in panel A. An interpretation of the profiles obtained with the two nascent-strand sizes is drawn below. The leftmost and rightmost possible positions of 1.25-kb bubbles that do not span probes 16 and 18 are indicated. The centers of these two bubbles define the ends of a 400-bp segment (black horizontal bar) where the origin has to be located. The predicted extent of bidirectional progression of the replication forks from the center of this segment up to a bubble size of 2.5 kb is indicated on the bottom line.
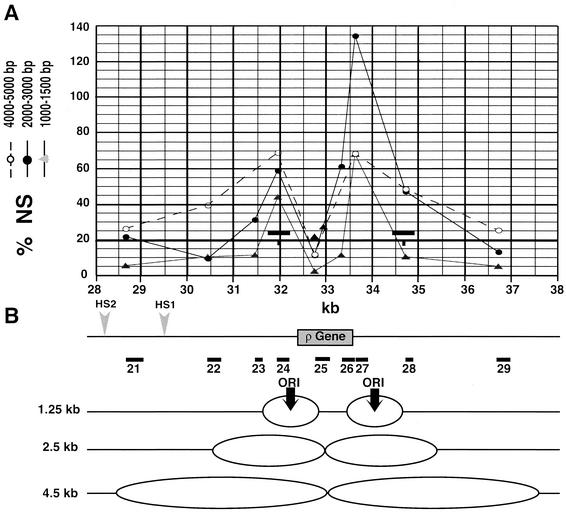
The region surrounding the ρ-globin gene and the two peaks of nascent-strand abundance detected 5′ and 3′ of the gene with both nascent-strand preparations are shown in Fig. 3. The narrowness of the peaks found with the 1.0- to 1.5-kb nascent-strand preparation allowed us to very precisely define two sites of initiation (Fig. 3B, black vertical arrows). The profile obtained with the 2.0- to 3.0-kb nascent strands revealed a progression of replication forks 5′ of the leftmost initiation site (primer pair 23) and on both sides of the rightmost initiation site (primer pairs 26 and 28). Surprisingly, only a faint increase in the amount of 2.0- to 3.0-kb nascent strands was observed with primer pair 25. This suggested either that none of the forks had sufficiently progressed to replicate this region or that the two small bubbles initiated 5′ and 3′ of the ρ-globin gene fused at the time they reached this region, so that the smallest nascent strands containing this probe were already larger than 2.0 to 3.0 kb. To resolve this issue, we quantified the abundance of 4.0- to 5.0-kb nascent strands (Fig. 3, white circles). Again, only a very low abundance of nascent strands was detected with primer pair 25. This result suggested that replication forks oriented in both directions pause or progress very slowly in this region. To map the pausing region more precisely, we quantified the abundance of the 4.0- to 5.0-kb nascent strands with two primer pairs that amplified the 5′ and 3′ halves, respectively, of the fragment amplified by primer pair 25. Both of them also gave a low enrichment (Fig. 3, black triangles), suggesting that forks can pause at several positions between primer pairs 24 and 26.
FIG. 3.
Two sites of initiation surround the ρ-globin gene. (A) A blow-up of the graph shown in Fig. 1A around the ρ-globin gene. In addition to the 1.0- to 1.5-kb (gray triangles) and 2.0- to 3.0-kb (black circles) nascent strands (NS), quantitations of a 4.0- to 5.0-kb nascent-strand preparation are shown (white circles, dotted line). The two black triangles show quantitations of the same 4.0- to 5.0-kb nascent-strand preparation with two PCR primer pairs that amplify the 5′ and 3′ halves of fragment 25. The short and long horizontal black bars indicate the amplified PCR products used previously (41) to characterize histone tail modifications and the corresponding detection zones (3 nucleosomes). (B) Map of the ρ-globin gene, upstream HS1, HS2, and the PCR fragments used in panel A. An interpretation of the profiles obtained is drawn at the bottom. Two small bubbles are initiated at position 32.2 and 34.2 kb either on the same molecule of DNA or on different molecules. The progression of replication forks inside the ρ-globin gene is slowed or blocked so that the region amplified by primer pair 25 is underrepresented in short (<5 kb) nascent strands.
The small and long black bars reported on the graph again indicate the PCR probes used to characterize the modifications of histone tails and the corresponding 500-bp detection zones, respectively (41). One detection zone includes probe 24 and the associated replication initiation site. No enrichment was detected in this region after immunoprecipitation of mono- and dinucleosomes with antibodies directed against acetylated H3-K9, H3-K14, methylated H3-K4 (Fig. 1B, bars numbered 2), or acetylated H4-K5, H4-K8, and H4-K12 (41, 42). In contrast, some enrichment in methylated H3-K9 (Fig. 1B, bars numbered 2), a modification characteristic of condensed chromatin, was detected. We conclude that the replication initiation site located upstream of the ρ-globin gene is associated with hypoacetylated histones. The downstream initiation site is also located very close to a region containing hypoacetylated histones (Fig. 1B and 3).
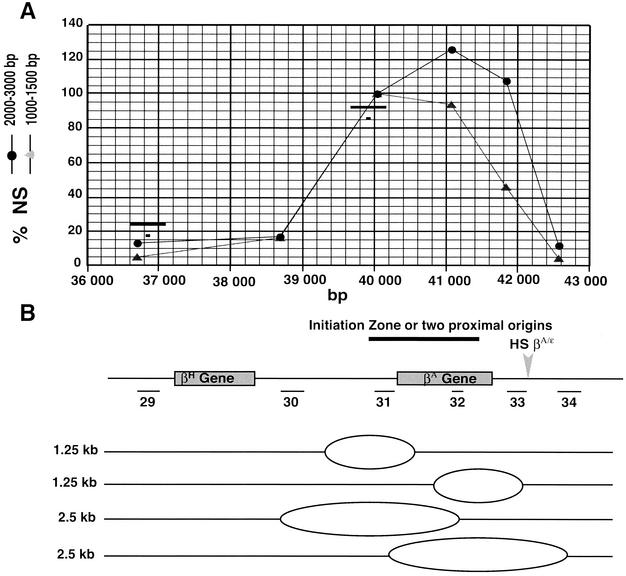
Finally, the region surrounding the βA-globin gene and containing the fourth initiation region is represented in Fig. 4. An enrichment of both the 1.0- to 1.5-kb and the 2.0- to 3.0-kb nascent strands was detected with primer pairs 31, 32, and 33 but not with primer pairs 29, 30, and 34 (Fig. 4). We conclude that the 1.5-kb region located between map positions 40.0 and 41.5 kb contains at least two closely spaced initiation sites. In contrast with the observation made with the two origins surrounding the ρ-globin gene, the strong enrichment found with primer pair 32 favored a model in which replication forks progress bidirectionally in this region. The closest region mapped for histone modifications is centered around map position 39.9 kb (Fig. 1B, bars numbered 3, and Fig. 4). Again, this chromosomal region has properties of condensed chromatin.
FIG. 4.
Dispersive initiation within the βA-globin gene. (A) A blow-up of the graph shown in Fig. 1A around the βA-globin gene. The short and long horizontal black bars indicate the amplified PCR products used previously (41) to characterize histone tail modifications and the corresponding detection zones (3 nucleosomes), respectively. (B) Map of the position of the βH- and βA-globin genes, HS βA/ɛ, and PCR fragments used for quantitations of nascent strands (NS) shown in panel A. Below the map an interpretation of the graph is drawn. Small bubbles initiate at multiple sites inside a region located between 40.0 and 41.5 kb and progress bidirectionally. The strong abundance of 2.0- to 3.0-kb nascent strands observed with primer pair 32 result from the simultaneous detection of the 5′-most and 3′-most bubbles with this probe.
Origins colocalize with either methylated or unmethylated CpG-rich sequences.
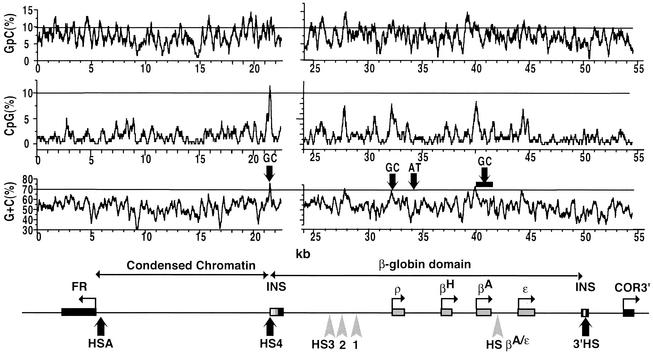
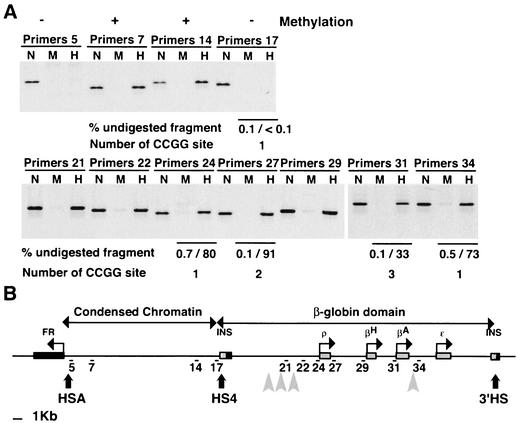
We have analyzed the G+C richness and the density of CpG and GpC dinucleotides over the entire region mapped for origin activity (Fig. 5). The core insulator origin, the βA origin, and the 5′ ρ origin are located in regions that are 77, 74, and 69% G+C rich, respectively. The nucleotide sequence of the core insulator is enriched in CpG dinucleotides and has been shown previously to lack CpG methylation. Although these properties are typical of CpG islands, the lack of detectable promoter activity and nearby transcripts suggest it is not a standard CpG island, because these structures are normally associated with genes (13). The promoter regions of the βA- and ρ-globin genes also contain a high density of CpGs. No other comparably G+C-rich region of the locus shows a similar CpG enrichment. This is reminiscent of the suggestion that G+C-rich, CpG-enriched sequences define a subset of mammalian DNA replication origins (5). A+T-rich sequences have also been proposed to be important functional components of replication origins in various species (11). In the chicken β-globin locus, initiation sites 1, 2, and 4 are not themselves A+T rich, but A+T-rich sequences can be found 1 to 2 kb upstream or downstream. Interestingly, initiation site 3 is located in an A+T-rich sequence (Fig. 5, black arrow labeled AT). Previous studies with chicken erythrocytes as well as with the 6C2 cell line showed that CpGs within and just upstream of the core insulator region are unmethylated (13). We further explored the methylation status of the chicken FR/β-globin region and in particular its replication origins. Genomic DNA extracted from 6C2 cells was digested with either MspI or HpaII (Fig. 6A). Both enzymes recognize the CCGG sequence, but HpaII is inhibited when the internal cytosine is methylated, whereas MspI is insensitive to methylation. Undigested, MspI-digested, or HpaII-digested genomic DNA was used as template for amplification of fragments containing at least one CCGG site. Methylated sites are protected against HpaII digestion, and thus only genomic fragments that are methylated at CCGG sites can be amplified by PCR. The upper part of Fig. 6A is a control experiment confirming the status of sites previously shown to be methylated (primer pairs 7 and 14, condensed chromatin region) or unmethylated (primer pair 5, HSA, and 17, core insulator) in the 6C2 cell line (53). Analysis of the same samples with primer pairs located throughout the β-globin locus showed that this region contains mainly methylated sites. In particular, the use of primer pairs 24 and 31 showed that the two GC-rich origins of replication contain methylated CpGs. Precise quantitations of undigested CCGG sites were made by real-time PCR with primer pairs 17, 24, 27, 31, and 34 and are indicated by numbers below the gels in Fig. 6A. Overall these results show that the extent of CpG methylation at CpG-enriched origins can vary from unmethylated to highly methylated.
FIG. 5.
Three out of the four replication origins are located at CpG-enriched sequences. The DNA sequence of the entire FR/β-globin region (with the exception of 1 kb of unsequenced DNA upstream of the β-globin locus) was analyzed by using a Genetics Computer Group program with a window of 200 nucleotides and a shift of 1 nucleotide. Shown are the percentages of G+C (bottom graph), CpG dinucleotides (middle), and GpC dinucleotides (top). The horizontal lines mark a G+C content of 70% and a CpG or GpC content of 10%. Black arrows on the top of the lowest graph indicate the position of either G+C- or A+T-rich replication origins. The βA-globin gene initiation zone is delimited by a horizontal black bar. The map of the region is shown at the bottom.
FIG. 6.
The G+C-rich, CpG-enriched replication origins contain either methylated or unmethylated CpGs. (A) Genomic DNA extracted from 6C2 cells was digested either with MspI or HpaII. Undigested (N), MspI-digested (M), or HpaII-digested DNA (H) was amplified by PCR with the indicated primer pairs. The upper gels are controls analyzing genomic fragments whose methylation status of CCGG sites is already known (pairs 5, 7, 14, and 17). Quantitation by real-time PCR of MspI- or HpaII-digested DNA was also made with primer pairs 17, 24, 27, 31, and 34. Primer pair 25 amplifies a PCR fragment lacking a CCGG site and was used as a control for input DNA. The real-time PCR results and the number of CCGG sites in the amplified fragment are indicated below the corresponding lanes. Each experiment was repeated at least twice with similar results. (B) The map shows the positions of fragments amplified by primer pairs used for panel A. INS, insulator.
The whole FR/β-globin region is replicated early in S phase.
Studies with animal cells have suggested a striking correlation between replication timing and gene expression (reference 59 and references therein). In general, loci that are expressed in a tissue-specific manner replicate early in S phase in expressing cells but later in S phase in nonexpressing cells (30). In the 6C2 cell line, which mimics the CFU-E stage of erythropoiesis, the FR gene is transcribed and the β-globin locus is silent. Moreover, the two loci are separated by a 16-kb region of condensed chromatin enriched in histone modifications typical of inactive, late-replicating chromatin.
To analyze the replication timing of this region in 6C2 cells, we used an assay that determines the relative abundance of specific sequences in nascent DNA synthesized during different windows of the cell cycle. An exponentially growing culture was labeled with BrdU for 1 h, and cells were sorted by fluorescence-activated cell sorting on the basis of DNA content into six different compartments within S phase and one in G1 as a control. The cell cycle profile of the culture and the sort gates corresponding to G1 and S1 to S6 are shown in Fig. 7A. DNA was prepared from equal numbers of cells from each cell cycle compartment, and BrdU-labeled DNA was purified by immunoaffinity chromatography with anti-BrdU antibodies and was used as a template for multiplex PCRs with primer pairs specific for different parts of the 52-kb region or the lysosyme locus and primer pair 14 as a reference. After 25 amplification cycles (conditions shown by comparison with 30 and 35 cycles to be well below the plateau phase), PCR products were resolved on agarose gels and were analyzed by Southern blot and hybridization with various pairs of probes (Fig. 7C). Whenever coamplification resulted in a strong advantage of one primer pair, the Southern blot was probed sequentially rather than simultaneously with the two probes (Fig. 7C, rightmost blots).
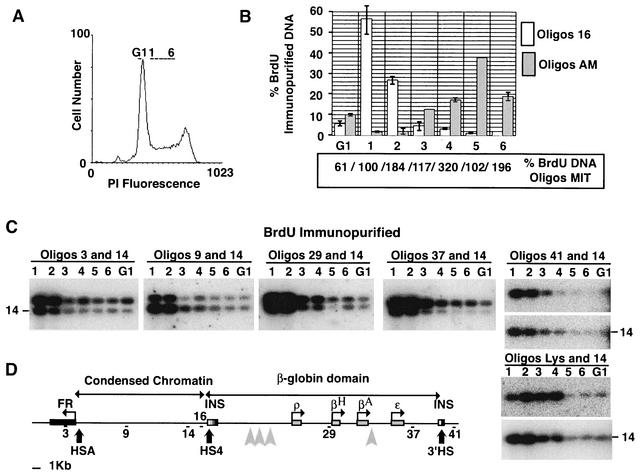
FIG. 7.
The entire FR/β-globin region is replicated early in S phase. (A) Histogram of propidium iodide (PI) staining intensity of a population of asynchronous cells pulse labeled with BrdU and sorted for timing analysis. The gates used to sort cells into a G1 compartment and six different compartments of S phase are labeled G1 and 1 to 6, respectively. (B) Quantitative real-time PCR of DNA extracted from different compartments of the cell cycle and immunoprecipitated with anti-BrdU antibodies. Numerical values at the bottom of the graph indicate the relative amounts of immunoprecipitated DNA detected with a primer pair specific for the D-loop region of the chicken mitochondrion (Oligos MIT). One hundred was arbitrarily set for fraction 1. Since mitochondrial genomes are replicated throughout the cell cycle, we used these data to control for variations in DNA recovery and PCR amplification efficiency. The graph shows data (normalized with respect to mitochondrial DNA abundance) obtained with the same fractions with either a chicken β-globin locus (Oligos 16) or a chicken α-amylase (Oligos AM)-specific primer pair. Each quantification was repeated twice. Error bars indicate the range of variation between the two values. The absence of an error bar indicates that these values are equal. (C) Multiplex PCRs made with different primer pairs on DNA extracted from different compartments of the cell cycle and immunoprecipitated with anti-BrdU antibodies. The PCR products were analyzed by Southern blotting and hybridization with the two cognate probes simultaneously (leftmost four gels) or consecutively (rightmost two gels). The experiment was performed twice with identical results. (D) The map shows the positions of fragments amplified by primer pairs used in the experiments depicted in panels B and C. INS, insulator.
The analysis was performed with primer pairs located in the FR gene (pair 3), in the condensed chromatin region (pairs 9 and 14), between the two major β-globin cluster origins (pair 29), or downstream of the β-globin cluster (pairs 37 and 41). In each case the strongest signal was clearly in fractions 1 and 2, suggesting that the entire region (including sequences downstream of the 3′ HS boundary) is replicated early in S phase (Fig. 7C). We believe that the much weaker signal in fractions S3 to S6 represents background rather than late replication of the locus in a minor fraction of the cell cycles, because a comparable signal was observed in G1 cells. In 6C2 cells S phase lasts for about 6 h, and thus each fraction of S phase represents approximately 1 h. These results imply that the replication origins of this locus fire most of the time within the first 2 h in S phase.
A control was made with the LYS primer pair, which amplifies a region reported to replicate 2 to 3 h after the beginning of S phase in cell lines that either express or do not express the lysozyme (51). The LYS pair showed a strong signal in fractions 2, 3, and 4, consistent with its reported replication timing. Pair 14 showed a strong signal in the first two fractions, consistent with the earlier replication timing of the FR/β-globin region.
A further control was made to ascertain that the data reflect replication timing and not a variation in the efficiency of immunoprecipitation and/or amplification between the different S-phase fractions. The abundance of mitochondrial DNA in each fraction was measured by quantitative real-time PCR by using a specific primer pair (MIT) alongside β-globin (primer pair 16) and α-amylase gene sequences (primer pair AM). Mitochondrial genomes are replicated throughout the cell cycle and are therefore expected to show uniform abundance in all fractions. Indeed, we only observed minor variations in mitochondrial DNA abundance between fractions (Fig. 7B, numerical values at the bottom of the graph). The data for mitochondrial DNA were then used to normalize the data for primer pairs 16 and AM (Fig. 7B, graph). The results clearly confirmed that the chicken β-globin locus replicates within the first two hours of S phase in the 6C2 cell line. In contrast, the chicken α-amylase locus was found to replicate late in S phase in the same cells, as previously reported for the homologous locus in nonexpressing human cells (15). Along with the results for the lysozyme locus, this control further validates that the assay can distinguish between replication in early, mid-, and late S phase.
The fact that the origin located near 5′ HS4 is highly enriched in acetylated histones (42) raises the possibility that histone hyperacetylation causes early firing of some replication origins in chicken cells, as recently reported for S. cerevisiae origins (63). However, the ρ and βA origins are either unacetylated or located near unacetylated histones yet replicate early in most cells. We therefore suggest that unacetylated origins can also fire early in S phase in chicken cells.
DISCUSSION
In this study we find that the replication of the chicken FR/β-globin region in the 6C2 cell line initiates early in S phase at three G+C-rich, CpG-enriched discrete sites localized close to the 5′ HS4 insulator and the promoters of the ρ- and the βA-globin genes. A fourth, A+T-rich initiation site is also found 3′ of the ρ-globin gene. The CpG dinucleotides are totally unmethylated at the insulator origin but are partially methylated at the ρ and the βA origins. The nucleosomes are methylated at H3-K4 and are acetylated on histones H3 and H4 at the insulator origin but not at the other origins. Methylation at H3-K9 is very low at the insulator origin but is more abundant and typical of bulk DNA at the other origins (41, 42).
G+C-rich sequences and replication origins.
Despite intensive study, no obvious consensus sequence for metazoan DNA replication origins has been found to date, although there is evidence that cis-acting elements influence replication initiation in differentiated cells (29). The results reported here show that G+C-rich, CpG-enriched sequences are found at three of the four origins in the chicken β-globin locus. No A+T-rich region is associated with these three sites, in contrast with the requirement for A+T-rich sequences at either S. cerevisiae or Saccharomyces pombe origins. These observations are consistent with the finding of a replication origin within a CpG island at the 3′ end of the chicken lysozyme gene (52). Several studies with human cells also suggested a role for CpG islands in DNA replication initiation. Unmethylated CpG islands are enriched in a library of short nascent strands prepared from human erythroleukemic cells (18). More recently, it has been found that half of ORC-bound human DNA sequences isolated after protein-DNA cross-linking and immunoprecipitation have a high G+C content with many CpG dinucleotides (40). Finally, replication origins located at the promoters of the c-MYC gene, the hsp70 gene, and the PPV-1 gene also map close to or within CpG islands (5). The only origin that is not G+C rich in the chicken β-globin locus is A+T rich. A characterization of additional initiation sites would be required to determine if these distinct sequence features reflect the existence of two distinct classes of replication origins in higher eukaryotes.
CpG methylation and replication origin activity.
A study of the dihydrofolate reductase (DHFR) locus of Chinese hamster cells has suggested a potential role for CpG methylation in the control of mammalian DNA replication origins. Replication of this locus can initiate at a large number of sites in a 55-kb zone downstream of the DHFR gene, with two subregions (ori-β and ori-γ) somewhat preferred (reference 20 and references therein). ori-β, although not particularly G+C rich, was found to contain a cluster of methylated CpGs, and similar sequence features were reported for another origin located at the 5′ end of the RPS14 gene in the same cells (56). Cells treated with 5-azacytidine no longer exhibit initiation at ori-β, suggesting that DNA methylation promotes the initiation process at this origin. On the other hand, the CpG cluster associated with the replication origin of the c-MYC gene is not methylated (55), and other mammalian origins are neither G+C rich nor CpG enriched (1, 39, 65). Furthermore, a targeted deletion of ori-β does not prevent initiation in the rest of the DHFR locus initiation zone (37). The role of CpG methylation in regulating replication initiation is therefore not completely clear. In the case of the chicken β-globin locus, the lack of methylation of the insulator origin as opposed to the methylation of the ρ and the βA origins confirms that DNA methylation is not a universal component of replication origins. Different epigenetic modifications are probably involved in replication origin control. DNA methylation may be just one of them but may be dispensable, as illustrated by its association with late replication, although organisms that lack DNA methylation nevertheless have a replication timing program.
The chromatin of unmethylated CpG islands has been found to contain highly acetylated histones H3 and H4 (62), just as the chicken β-globin insulator does (42). On the other hand, methylated DNA can be specifically bound by proteins, such as MeCP2, which in turn recruits histone deacetylase to repress transcription (36). It remains to be seen whether ORC, or a factor that facilitates ORC binding, preferentially binds CpG clusters or G+C-rich sequences and whether DNA methylation can influence this interaction.
Histone modifications and replication origin specification.
Numerous studies suggest that the packaging of the genome into nucleosomes and higher-order structures affects the selection and activation of DNA replication origins (26). Although histone acetylation destabilizes nucleosomes and is known to favor the interaction of transcription factors with their target site (10), the influence of histone acetylation on the binding of replication initiation factors is unknown. Interestingly, three of the four origins identified here are located within the underacetylated β-globin locus, whereas the fourth origin maps at the hyperacetylated 5′ HS4 insulator. Histone acetylation, therefore, is not a systematic mark of replication origins. It should be noted that two replication initiation factors, ORC1 and MCM2, have been shown to interact with the histone acetyltransferase HBO1, suggesting a direct role for histone acetyltransferase activity in the process of DNA replication (12, 34). Whether HBO1 or some other histone acetyltransferase is recruited by ORC/MCM complexes at the insulator origin and participates in histone acetylation of this region or conversely recruits prereplication complexes remains to be determined.
In metazoa, initiation can occur at specific sites or within broad zones depending on the locus (1, 20). One interesting point raised by our study is that replication initiates at precise sites within a globally closed chromatin region. We and others (21, 32, 45) have suggested that broad initiation zones may arise from the spreading of multiple MCM complexes away from a single ORC complex during prereplication complex formation. It is possible that a condensed chromatin structure restrains initiation to sharply specific sites either by suppressing weak binding sites for ORC or by limiting the spreading of the MCM complexes away from ORC. It will be interesting to determine whether initiation within the β-globin locus becomes more dispersive at later differentiation stages, when the globin genes start to decondense and to be transcribed.
Histone modifications and replication timing.
A recent study has shown that the firing time of S. cerevisiae origins can be artificially advanced either by global hyperacetylation of the genome due to the deletion of a histone deacetylase or by local recruitment of a histone acetyltransferase, suggesting that histone acetylation is a direct determinant of origin firing time (63). However, we found that the four origins of the chicken β-globin locus, whether associated with hyperacetylated or hypoacetylated histones H3 and H4, fire early in S phase. Although these results do not exclude that histone acetylation can induce early firing of some origins, they clearly indicate that the status of histone acetylation is neither the sole nor a necessary determinant of origin firing time in chicken cells. However, it is possible that one of the four origins is a master origin that triggers initiation at the entire cluster. For example, the hyperacetylated 5′ HS4 origin could be programmed to fire early, and once poised to replicate it could set up the other origins to fire simultaneously or shortly following its initiation. Although speculative, this scenario is consistent with the long-standing evidence for synchronous replicon clusters in higher eukaryotes (reviewed in reference 9).
Studies of the human β-globin locus indicate that the LCR is sufficient for directing replication timing in transgenic mice (60), although it is not necessary for the ability of this locus to switch to early replication in erythroid cell hybrids (15), and that early replication is correlated with an open chromatin conformation rather than with globin gene expression itself. Conversely, inactivation of the X chromosome in early embryogenesis is characterized by a transition from early to late replication that is concomitant with H3-K9 deacetylation and methylation, whereas other epigenetic modifications occur subsequently (31, 47).
Except for the active FR gene and the 5′ HS4 insulator, the overall structure of the FR/β-globin region in 6C2 cells is typical of condensed chromatin. In particular, the high level of H3-K9 methylation and nuclease resistance observed over the 16-kb stretch that separates the FR gene domain from the β-globin domain appear typical of heterochromatin, which usually replicates in late S phase. Nevertheless, the whole FR/β-globin region replicates early in S phase. This result adds to the recent evidence that heterochromatin does not necessarily replicate late in S phase in D. melanogaster (59) and in S. pombe (37a, 38). Further work is required to establish whether a specific combination of epigenetic modifications is responsible for the replication timing of the chicken FR/β-globin domains.
Role of insulators in replication initiation.
One replication origin of the FR/β-globin region is located at the chicken 5′ HS4 β-globin insulator. Might origin activity be required for insulator function or vice versa, or is this a chance colocalization? The lack of replication initiation at the 3′ HS boundary suggests that a CTCF binding site is not sufficient to define a replication origin and that enhancer-blocking activity does not require an origin of replication. The barrier activity of the 5′ HS4 insulator was proposed to result from high levels of H3-K9 acetylation preventing H3-K9 methylation and encroachment of condensed chromatin into the β-globin gene cluster (41). In this regard, the ability of ORC and MCM proteins to interact with the HBO1 acetyltransferase (12, 34) is intriguing and may be implied in the recruitment of HAT activity at the 5′ HS4 insulator.
Studies of Drosophila chorion gene amplification (a developmentally regulated process that results from repeated rounds of replication initiation within the chorion genes) by using P-element-mediated transformation have revealed that amplification is highly subject to position effects but can be protected by a powerful transcriptional insulator, the suppressor of the Hairy-wing protein-binding site (SHWBS) from the gypsy transposon (43). By using transgenic constructs surrounded with two SHWBSs, two DNA elements of the chorion locus (ACE3 and ori-β) were found to be necessary and sufficient for amplification (44). Only ori-β has origin activity, suggesting that ACE3 function is to activate initiation at ori-β. The SHWBS insulators showed no origin activity on their own. Moreover, the SHWBS insulator was found to inhibit amplification if located between ACE3 and ori-β. These findings led to a model in which ACE3 and additional replicators activate in cis multiple origins within an initiation zone, ori-β being the most active. Thus, insulators may both protect initiation zones from position effects and prevent activation of cryptic origins outside the zone they delimit.
The initiation sites we have mapped in the chicken FR/condensed chromatin/β-globin region are restricted to within the β-globin gene cluster. Therefore, they may be controlled by replicators whose range of action is limited by the 5′ HS4 and the 3′ HS insulators. One way to test this model would be to examine the effect of removing these elements on the replication of this locus.
Acknowledgments
We thank Céline Hauzy for participating in these investigations during a stage de Magistère and Edith Heard and Kathrin Marheineke for critical reading of the manuscript.
This work was supported by the ATIPE-CNRS, the Ligue Nationale Française Contre le Cancer (Comité de Paris), the Association pour la Recherche sur le Cancer, and the Association Française contre les Myopathies.
REFERENCES
- 1.Abdurashidova, G., M. Deganuto, R. Klima, S. Riva, G. Biamonti, M. Giacca, and A. Falaschi. 2000. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287:2023-2026. [DOI] [PubMed] [Google Scholar]
- 2.Aladjem, M. I., M. Groudine, L. L. Brody, E. S. Dieken, R. E. Fournier, G. M. Wahl, and E. M. Epner. 1995. Participation of the human beta-globin locus control region in initiation of DNA replication. Science 270:815-819. [DOI] [PubMed] [Google Scholar]
- 3.Aladjem, M. I., L. W. Rodewald, J. L. Kolman, and G. M. Wahl. 1998. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science 281:1005-1009. [DOI] [PubMed] [Google Scholar]
- 4.Aladjem, M. I., L. W. Rodewald, C. M. Lin, S. Bowman, D. M. Cimbora, L. L. Brody, E. M. Epner, M. Groudine, and G. M. Wahl. 2002. Replication initiation patterns in the beta-globin loci of totipotent and differentiated murine cells: evidence for multiple initiation regions. Mol. Cell. Biol. 22:442-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antequera, F., and A. Bird. 1999. CpG islands as genomic footprints of promoters that are associated with replication origins. Curr. Biol. 9:R661-R667. [DOI] [PubMed]
- 6.Bell, A. C., A. G. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387-396. [DOI] [PubMed] [Google Scholar]
- 7.Bell, S. P. 2002. The origin recognition complex: from simple origins to complex functions. Genes Dev. 16:659-672. [DOI] [PubMed] [Google Scholar]
- 8.Bell, S. P., and B. Stillman. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357:128-134. [DOI] [PubMed] [Google Scholar]
- 9.Berezney, R., D. D. Dubey, and J. A. Huberman. 2000. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108:471-484. [DOI] [PubMed] [Google Scholar]
- 10.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 11.Bogan, J. A., D. A. Natale, and M. L. Depamphilis. 2000. Initiation of eukaryotic DNA replication: conservative or liberal? J. Cell Physiol. 184:139-150. [DOI] [PubMed] [Google Scholar]
- 12.Burke, T. W., J. G. Cook, M. Asano, and J. R. Nevins. 2001. Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J. Biol. Chem. 276:15397-15408. [DOI] [PubMed] [Google Scholar]
- 13.Chung, J. H., A. C. Bell, and G. Felsenfeld. 1997. Characterization of the chicken beta-globin insulator. Proc. Natl. Acad. Sci. USA 94:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung, J. H., M. Whiteley, and G. Felsenfeld. 1993. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74:505-514. [DOI] [PubMed] [Google Scholar]
- 15.Cimbora, D. M., D. Schubeler, A. Reik, J. Hamilton, C. Francastel, E. M. Epner, and M. Groudine. 2000. Long-distance control of origin choice and replication timing in the human beta-globin locus are independent of the locus control region. Mol. Cell. Biol. 20:5581-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Contreas, G., M. Giacca, and A. Falaschi. 1992. Purification of BrdUrd-substituted DNA by immunoaffinity chromatography with anti-BrdUrd antibodies. BioTechniques 12:824-826. [PubMed] [Google Scholar]
- 17.Coverley, D., and R. A. Laskey. 1994. Regulation of eukaryotic DNA replication. Annu. Rev. Biochem. 63:745-776. [DOI] [PubMed] [Google Scholar]
- 18.Delgado, S., M. Gomez, A. Bird, and F. Antequera. 1998. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J. 17:2426-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diffley, J. F., and J. H. Cocker. 1992. Protein-DNA interactions at a yeast replication origin. Nature 357:169-172. [DOI] [PubMed] [Google Scholar]
- 20.Dijkwel, P. A., S. Wang, and J. L. Hamlin. 2002. Initiation sites are distributed at frequent intervals in the chinese hamster dihydrofolate reductase origin of replication but are used with very different efficiencies. Mol. Cell. Biol. 22:3053-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards, M. C., A. V. Tutter, C. Cvetic, C. H. Gilbert, T. A. Prokhorova, and J. C. Walter. 2002. MCM2-7 complexes bind chromatin in a distributed pattern surrounding the origin recognition complex in Xenopus egg extracts. J. Biol. Chem. 277:33049-33057. [DOI] [PubMed] [Google Scholar]
- 22.Farrell, C. M., A. G. West, and G. Felsenfeld. 2002. Conserved CTCF insulator elements flank the mouse and human β-globin loci. Mol. Cell. Biol. 22:3820-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felsenfeld, G. 1993. Chromatin structure and the expression of globin-encoding genes. Gene 135:119-124. [DOI] [PubMed] [Google Scholar]
- 24.Forrester, W. C., E. Epner, M. C. Driscoll, T. Enver, M. Brice, T. Papayannopoulou, and M. Groudine. 1990. A deletion of the human beta-globin locus activation region causes a major alteration in chromatin structure and replication across the entire beta-globin locus. Genes Dev. 4:1637-1649. [DOI] [PubMed] [Google Scholar]
- 25.Foss, M., F. J. McNally, P. Laurenson, and J. Rine. 1993. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae. Science 262:1838-1844. [DOI] [PubMed] [Google Scholar]
- 26.Gerbi, S. A., and A. K. Bielinsky. 2002. DNA replication and chromatin. Curr. Opin. Genet. Dev. 12:243-248. [DOI] [PubMed] [Google Scholar]
- 27.Gerbi, S. A., and A. K. Bielinsky. 1997. Replication initiation point mapping. Methods 13:271-280. [DOI] [PubMed] [Google Scholar]
- 28.Giacca, M., C. Pelizon, and A. Falaschi. 1997. Mapping replication origins by quantifying relative abundance of nascent DNA strands using competitive polymerase chain reaction. Methods 13:301-312. [DOI] [PubMed] [Google Scholar]
- 29.Gilbert, D. M. 2001. Making sense of eukaryotic DNA replication origins. Science 294:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilbert, D. M. 2002. Replication timing and transcriptional control: beyond cause and effect. Curr. Opin. Cell Biol. 14:377-383. [DOI] [PubMed] [Google Scholar]
- 31.Heard, E., C. Rougeulle, D. Arnaud, P. Avner, C. D. Allis, and D. L. Spector. 2001. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell 107:727-738. [DOI] [PubMed] [Google Scholar]
- 32.Hyrien, O., K. Marheineke, and A. Goldar. Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. BioEssays, in press. [DOI] [PubMed]
- 33.Hyrien, O., C. Maric, and M. Méchali. 1995. Transition in specification of embryonic metazoan DNA replication origins. Science 270:994-997. [DOI] [PubMed] [Google Scholar]
- 34.Iizuka, M., and B. Stillman. 1999. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J. Biol. Chem. 274:23027-23034. [DOI] [PubMed] [Google Scholar]
- 35.Jacob, F., S. Brenner, and F. Cuzin. 1964. On the regulation of DNA replication in bacteria. Cold Spring Harbor Symp. Quant. Biol. 288:329-348. [Google Scholar]
- 36.Jones, P. L., G. J. Veenstra, P. A. Wade, D. Vermaak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19:187-191. [DOI] [PubMed] [Google Scholar]
- 37.Kalejta, R. F., X. Li, L. D. Mesner, P. A. Dijkwel, H. B. Lin, and J. L. Hamlin. 1998. Distal sequences, but not ori-beta/OBR-1, are essential for initiation of DNA replication in the Chinese hamster DHFR origin. Mol. Cell 2:797-806. [DOI] [PubMed] [Google Scholar]
- 37a.Kim, S. M., D. D. Dubey, and J. A. Huberman. 2003. Early replicating chromatin. Genes Dev. 17:330-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim, S. M., and J. A. Huberman. 2001. Regulation of replication timing in fission yeast. EMBO J. 20:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kitsberg, D., S. Selig, I. Keshet, and H. Cedar. 1993. Replication structure of the human beta-globin gene domain. Nature 366:588-590. [DOI] [PubMed] [Google Scholar]
- 40.Ladenburger, E. M., C. Keller, and R. Knippers. 2002. Identification of a binding region for human origin recognition complex proteins 1 and 2 that coincides with an origin of DNA replication. Mol. Cell. Biol. 22:1036-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Litt, M. D., M. Simpson, M. Gaszner, C. D. Allis, and G. Felsenfeld. 2001. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science 293:2453-2455. [DOI] [PubMed] [Google Scholar]
- 42.Litt, M. D., M. Simpson, F. Recillas-Targa, M. N. Prioleau, and G. Felsenfeld. 2001. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 20:2224-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu, L., and J. Tower. 1997. A transcriptional insulator element, the su(Hw) binding site, protects a chromosomal DNA replication origin from position effects. Mol. Cell. Biol. 17:2202-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu, L., H. Zhang, and J. Tower. 2001. Functionally distinct, sequence-specific replicator and origin elements are required for Drosophila chorion gene amplification. Genes Dev. 15:134-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lucas, I., M. Chevrier-Miller, J. M. Sogo, and O. Hyrien. 2000. Mechanisms ensuring rapid and complete DNA replication despite random initiation in Xenopus early embryos. J. Mol. Biol. 296:769-786. [DOI] [PubMed] [Google Scholar]
- 46.Maric, C., B. Levacher, and O. Hyrien. 1999. Developmental regulation of replication fork pausing in Xenopus laevis ribosomal RNA genes. J. Mol. Biol. 291:775-788. [DOI] [PubMed] [Google Scholar]
- 47.Mermoud, J. E., B. Popova, A. H. Peters, T. Jenuwein, and N. Brockdorff. 2002. Histone H3 lysine 9 methylation occurs rapidly at the onset of random X chromosome inactivation. Curr. Biol. 12:247-251. [DOI] [PubMed] [Google Scholar]
- 48.Micklem, G., A. Rowley, J. Harwood, K. Nasmyth, and J. F. Diffley. 1993. Yeast origin recognition complex is involved in DNA replication and transcriptional silencing. Nature 366:87-89. [DOI] [PubMed] [Google Scholar]
- 49.Newlon, C. S., and J. F. Theis. 1993. The structure and function of yeast ARS elements. Curr. Opin. Genet. Dev. 3:752-758. [DOI] [PubMed] [Google Scholar]
- 50.Pak, D. T., M. Pflumm, I. Chesnokov, D. W. Huang, R. Kellum, J. Marr, P. Romanowski, and M. R. Botchan. 1997. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91:311-323. [DOI] [PubMed] [Google Scholar]
- 51.Phi-Van, L., and W. H. Stratling. 1988. The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO J. 7:655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phi-van, L., and W. H. Stratling. 1999. An origin of bidirectional DNA replication is located within a CpG island at the 3′ end of the chicken lysozyme gene. Nucleic Acids Res. 27:3009-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prioleau, M. N., P. Nony, M. Simpson, and G. Felsenfeld. 1999. An insulator element and condensed chromatin region separate the chicken beta-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J. 18:4035-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Recillas-Targa, F., M. J. Pikaart, B. Burgess-Beusse, A. C. Bell, M. D. Litt, A. G. West, M. Gaszner, and G. Felsenfeld. 2002. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc. Natl. Acad. Sci. USA 99:6883-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rein, T., T. Kobayashi, M. Malott, M. Leffak, and M. L. DePamphilis. 1999. DNA methylation at mammalian replication origins. J. Biol. Chem. 274:25792-25800. [DOI] [PubMed] [Google Scholar]
- 56.Rein, T., H. Zorbas, and M. L. DePamphilis. 1997. Active mammalian replication origins are associated with a high-density cluster of mCpG dinucleotides. Mol. Cell. Biol. 17:416-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saitoh, N., A. C. Bell, F. Recillas-Targa, A. G. West, M. Simpson, M. Pikaart, and G. Felsenfeld. 2000. Structural and functional conservation at the boundaries of the chicken beta-globin domain. EMBO J. 19:2315-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sasaki, T., T. Sawado, M. Yamaguchi, and T. Shinomiya. 1999. Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolα-dE2F locus of Drosophila melanogaster. Mol. Cell. Biol. 19:547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schubeler, D., D. Scalzo, C. Kooperberg, B. van Steensel, J. Delrow, and M. Groudine. 2002. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat. Genet. 32:438-442. [DOI] [PubMed] [Google Scholar]
- 60.Simon, I., T. Tenzen, R. Mostoslavsky, E. Fibach, L. Lande, E. Milot, J. Gribnau, F. Grosveld, P. Fraser, and H. Cedar. 2001. Developmental regulation of DNA replication timing at the human beta globin locus. EMBO J. 20:6150-6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stillman, B. 2001. DNA replication. Genomic views of genome duplication. Science 294:2301-2304. [DOI] [PubMed] [Google Scholar]
- 62.Tazi, J., and A. Bird. 1990. Alternative chromatin structure at CpG islands. Cell 60:909-920. [DOI] [PubMed] [Google Scholar]
- 63.Vogelauer, M., L. Rubbi, I. Lucas, B. J. Brewer, and M. Grunstein. 2002. Histone acetylation regulates the time of replication origin firing. Mol. Cell 10:1223-1233. [DOI] [PubMed] [Google Scholar]
- 64.West, A. G., M. Gaszner, and G. Felsenfeld. 2002. Insulators: many functions, many mechanisms. Genes Dev. 16:271-288. [DOI] [PubMed] [Google Scholar]
- 65.Zhao, Y., S. Miyagi, T. Kikawada, and K. Tsutsumi. 1997. Sequence requirement for replication initiation at the rat aldolase B locus implicated in its functional correlation with transcriptional regulation. Biochem. Biophys. Res. Commun. 237:707-713. [DOI] [PubMed] [Google Scholar]