Abstract
In the absence of MEIS family proteins, two mechanisms are known to restrict the PBX family of homeodomain (HD) transcription factors to the cytoplasm. First, PBX is actively exported from the nucleus via a CRM1-dependent pathway. Second, nuclear localization signals (NLSs) within the PBX HD are masked by intramolecular contacts. In a screen to identify additional proteins directing PBX subcellular localization, we identified a fragment of murine nonmuscle myosin II heavy chain B (NMHCB). The interaction of NMHCB with PBX was verified by coimmunoprecipitation, and immunofluorescence staining revealed colocalization of NMHCB with cytoplasmic PBX in the mouse embryo distal limb bud. The interaction domain in PBX mapped to a conserved PBC-B region harboring a potential coiled-coil structure. In support of the cytoplasmic retention function, the NMHCB fragment competes with MEIS1A to redirect PBX, and the fly PBX homologue EXD, to the cytoplasm of mammalian and insect cells. Interestingly, MEIS1A also localizes to the cytoplasm in the presence of the NMHCB fragment. These activities are largely independent of nuclear export. We show further that the subcellular localization of EXD is deregulated in Drosophila zipper mutants that are depleted of nonmuscle myosin heavy chain. This study reveals a novel and evolutionarily conserved mechanism controlling the subcellular distribution of PBX and EXD proteins.
The Hox family genes encode homeodomain (HD)-containing transcription factors that play important roles in axial patterning during embryonic development (23). HOX proteins execute their functions by regulating downstream targets to specify regional identity (20). The weak DNA-binding affinity and specificity of HOX proteins are partially compensated for by DNA-binding partners (16, 27). The most studied of these partners are PBC and MEINOX proteins belonging to the TALE class of homeoproteins. The PBC proteins include EXD (EXTRADENTICLE) in Drosophila and the PBX (pre-B-cell homeobox) proteins of vertebrates. The MEINOX proteins include the fly HTH (HOMOTHORAX) and vertebrate MEIS (myeloid ecotropic viral integration site) and PREP proteins (7-9).
Additional controls of HOX function have recently been brought to light. The subcellular localization of EXD is regulated throughout Drosophila embryonic development (4, 26). HTH, the MEIS homologue in flies, is required for the nuclear translocation of EXD (21, 31, 36). For example, EXD is expressed in both proximal and distal regions of the Drosophila leg imaginal disk; however, nuclear EXD is found only in the proximal region, where HTH is coexpressed (1, 19). Mammalian MEIS1, and the related PREP1, can substitute for fly HTH to induce EXD nuclear translocation in both cell culture and Drosophila embryos (2, 6, 21, 36, 37). We and others have shown that PBX1 is localized in the cytoplasm when transfected into Drosophila Schneider 2 (S2) cells, whereas cotransfection of MEIS1 or PREP1 induces PBX1 nuclear translocation (6, 37). Consistent with this, PBX is localized to the nucleus only in the proximal mouse limb bud, where Meis genes are expressed, and not in the distal portion, where Meis transcripts cannot be detected (12, 19, 28, 30, 37). These findings suggest that in Drosophila, as well as in vertebrates, the access of HOX to its cofactors is controlled by the nuclear localization of PBX/EXD, a process regulated by MEIS/HTH family proteins.
Interaction between PBX/EXD and MEIS/HTH maps to the N-terminal PBC-A and HM1/HM2 domains, respectively (14, 22). The PBX/EXD N terminus also directs their cytoplasmic localization in the absence of MEIS/HTH, via the activity of CRM1-mediated nuclear export. CRM-1 was identified as a receptor mediating the nuclear export of proteins containing leucine-rich nuclear export sequences (NESs) (17). It has been shown that the cytotoxin leptomycin B (LMB) specifically inhibits CRM-1 activity by blocking its binding to its target proteins (24). Two studies characterized a nonconsensus NES in EXD and PBX; however, nonoverlapping domains were proposed (2, 6). The NES localizes to the EXD PBC-B domain (residues 178 to 220) and the PBX1 PBC-A region (residues 1 to 96). In any case, the interaction between PBX/EXD and nuclear export receptors must be directly or indirectly disrupted in the presence of MEIS or HTH.
Previous results showed that PBX1 possesses two nuclear localization signals (NLSs) within its HD that are masked by intramolecular interaction with the N terminus (37). PBX-MEIS interaction induces a conformational change in PBX, which in turn shifts the balance between nuclear import and export. However, this does not explain all the observations of PBX subcellular distribution in vivo. First, no consensus NES has been identified in the PBX/EXD N terminus. Although NESs may be heterogeneous, the existence of NES-containing partners of PBC proteins cannot be excluded, since PBX/EXD may translocate to the cytoplasm through the binding of an intermediary protein that possesses an NES. The binding of this adapter to PBX/EXD could be regulated by MEIS/HTH. Second, inhibition of CRM1-mediated nuclear export by LMB does not induce full nuclear localization of EXD (2), indicating the possible involvement of cytoplasmic retention factors that are insensitive to LMB.
To identify additional factors controlling the subcellular distribution of PBX, we performed a yeast two-hybrid screen using the PBX1 N terminus as bait. One resulting clone encoded a fragment of nonmuscle myosin II heavy chain B (NMHCB). We verified the association between PBX1 and NMHCB in mammalian cells and revealed the colocalization of these two proteins in the cytoplasm of mouse distal limb bud cells. Overexpression of the NMHCB fragment induced cytoplasmic accumulation of PBX/EXD and MEIS in both mammalian cells and Drosophila S2 cells. Furthermore, the subcellular localization of EXD was deregulated in Drosophila zipper mutants, which have a defective nonmuscle myosin heavy chain. Together, these results suggest that NMHCB promotes the cytoplasmic localization of PBX and EXD in vivo.
MATERIALS AND METHODS
Expression vectors.
Regions of human PBX1A encoding residues 1 to 96, 137 to 232, and 1 to 232 were amplified by PCR using pBSKpbx1a as a template and cloned into NcoI and SmaI sites of the pAS2-1 vector (Clontech). The constructs were verified by sequencing. pGAL-O45 is a mammalian expression vector for the GAL4 DNA-binding domain (GAL-DBD) and was described previously (33). pPacPL is a Drosophila expression vector (described in reference 37). pcDNA3FLAG is a FLAG-tagged expression vector driven by a cytomegalovirus promoter. pVP16 is a LEU-selectable yeast expression vector in which VP16 coding sequences (fused to an NLS to direct nuclear localization) are 5′ to the cloning site. The mouse 9.5-day-postcoitum (dpc) embryo prey library (a generous gift of S. Hollenberg) was constructed by random-primed cDNA synthesis; products between 350 and 700 nucleotides were selected and cloned into the NotI site of pVP16. Expression vectors for PBX1A-HA, PBX1A(Δ1-89)-HA, PBX1A(Δ172-219)-HA, PBX1A-HA(Δ172-295), pPacPLPBX1A-HA, and MEIS1A have been described previously (37). PBX1A(Δ90-219)-HA was constructed by subsequently cloning the coding region for PBX1A residues 1 to 89 and hemagglutinin-tagged residues 219 to 430 into the pCS2+ vector.
Yeast two-hybrid screen and cloning.
pAS2-1PBX1A1-232 was transformed into the yeast strain PJ69-2A and mated to strain Y187 expressing a cDNA prey library derived from 9.5-dpc mouse embryos. Colony selection, plasmid DNA isolation from positive clones, and specificity tests were performed according to the Clontech Matchmaker manual. Positive clones were sequenced and screened by BLAST analysis of the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov:80/BLAST/). The insert encoding the NMHCB fragment (fNMHCB; see Results) was excised and subcloned into the pGAL-O45 or pcDNA3 FLAG vector as pGAL-fNM and pFLAG-fNM, respectively. GAL-DBD and GAL-DBD-tagged fNMHCB were subsequently subcloned into the pPacPL vector.
Cell culture and transfection.
HEK293T and Cos-7 cells were cultured in α minimal essential medium and Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, respectively. Drosophila S2 cells were cultured in Schneider's Drosophila medium (GIBCO) with 10% fetal bovine serum. For immunoprecipitation, HEK293T cells were seeded at 3 × 106 per 100-mm-diameter dish. The cells were allowed to attach overnight and then transfected by the calcium phosphate coprecipitation method or by Lipofectamine 2000 reagent (Invitrogen). For immunostaining, cells were plated on glass coverslips in 35-mm-diameter dishes.
Antibodies.
Rabbit polyclonal antibody against PBX1 and mouse monoclonal antibody against GAL-DBD were purchased from Santa Cruz. FLAG M2 agarose beads were purchased from Sigma. A monoclonal antibody against NMHCB was obtained from the Developmental Studies Hybridoma Bank. The anti-EXD monoclonal antibody and anti-ZIPPER antibodies were provided by R. A. White and D. Kiehart, respectively. The secondary antibodies were horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) (Santa Cruz) and goat anti-rabbit IgG (Sigma), fluorescein isothiocyanate-linked goat anti-rabbit IgG (Sigma), and rhodamine-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories Inc.). Anti-MEIS1 antibody was generated by the immunization of rabbits with a fusion of maltose binding protein to the first 34 residues of MEIS1. Antiserum was purified by immunoaffinity methods.
Immunoprecipitation assays.
Forty to 48 h posttransfection, cells were harvested and lysed in 500 μl of EBC lysis buffer (120 mM NaCl, 50 mM Tris-Cl [pH 8.0], 0.5% NP-40, and protease inhibitors) at 4°C for 30 min, followed by brief sonication. Whole-cell extracts were precleared with protein G-Sepharose for 30 min; 0.5 to 1 mg of precleared lysate was incubated with 0.5 to 1 μg of primary antibody from 2 h to overnight at 4°C, followed by the addition of 30 μl of a 50% slurry of protein G-Sepharose and further incubation for 2 h at 4°C. The precipitates were washed two times with high-salt NETN (20 mM Tris-Cl, pH 8.0, 1 mM EDTA, 0.5% NP-40, 0.5 M NaCl) and one time with NETN (without 0.5 M NaCl) and eluted with 1× sodium dodecyl sulfate (SDS) sample buffer. The eluted proteins were separated by SDS-polyacrylamide gel electrophoresis and analyzed by Western blotting. The secondary antibodies were conjugated with horseradish peroxidase and detected by enhanced chemiluminescence (MEN Life Science). To immunoprecipitate FLAG epitope-tagged proteins, lysates were incubated with Anti-FLAG M2 agarose beads (Sigma) instead of protein G-Sepharose. FLAG peptide (0.2 mg/ml; Sigma) was used to elute the precipitates for 1 h at 4°C.
Immunofluorescence staining.
Cells were cultured on 12-mm-diameter circular coverslips and fixed with 4% paraformaldehyde. After incubation with appropriate primary antibodies for 2 h at room temperature, the cells were washed with 0.1% Tween 20 in phosphate-buffered saline (PBS) and incubated with fluorescein isothiocyanate- or rhodamine-conjugated secondary antibody for 1 h at room temperature in the dark. DAPI (4′,6′-diamidino-2-phenylindole; 1 μg/ml in PBS) was added to the last wash solution. Samples were analyzed under a Nikon ECLIPSE E800 fluorescence microscope with a Nikon DXM1200 digital camera. Mouse embryos (11.5 dpc) were sectioned by cryostat and immunolabeled with anti-PBX1 and anti-NMHCB antibodies. Samples were examined with a Zeiss LSM510 laser scanning confocal microscope. The images were further processed using Adobe PhotoShop.
Drosophila stocks
The wild-type Drosophila stock used was Oregon R, and the zipper mutant stock used was zi ID16/CyO (P[w+mC= GAL4-Kr.C]DC3,P[w+mC= UAS-GFP.S65T]DC7). Homozygous zipper embryos were distinguished from their heterozygous siblings by the absence of green fluorescent protein signal.
Immunolabeling of Drosophila embryos.
Eggs were collected from Oregon R and zipper mutant flies for 13 h and then dechorionated and fixed for 15 min in 4% paraformaldehyde in 1× PBS- 0.2% Tween 20 and heptane. The embryos were rehydrated, washed with 1× PBS-0.2% Tween 20, and blocked with 1× PBS- 1% bovine serum albumin- 0.2% Triton X-100 overnight. After being blocked, the embryos were incubated with anti-EXD and anti-ZIPPER primary antibodies. The secondary antibodies were Alexa Fluor-568-conjugated anti-mouse IgG and Alexa Fluor-633-conjugated anti-rabbit IgG (Molecular Probes). Immunostainings were visualized using a Leica TCS-SP2 confocal microscope.
RESULTS
A yeast two-hybrid screen identifies a PBX N terminus-interacting clone highly related to rat NMHCB.
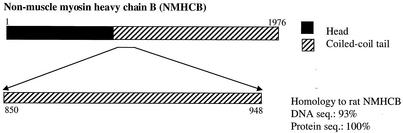
To search for proteins modulating PBX subcellular distribution, we used residues 1 to 232 of human PBX1 as bait in a yeast two-hybrid screen of a mouse 9.5-dpc embryo cDNA library. Of 3 × 108 clones screened, 21 true-positive clones were obtained. Sequence analysis showed that the majority of them were MEIS/PREP family proteins. However, one clone had 93% nucleotide identity and 100% amino acid identity with the Rattus norvegicus NMHCB. This clone included a 297-bp DNA fragment encoding 99 amino acids in the NMHCB tail region (hereafter referred to as fNMHCB), as shown in Fig. 1.
FIG. 1.
Fragment of murine NMHCB identified in a yeast two-hybrid screen as an interaction partner of the PBX N terminus. The head and coiled-coil tail domains are indicated. The murine prey clone contained a 297-bp insert encoding a peptide 100% identical to rat NMHCB residues 850 to 948, as indicated. The DNA sequences (seq.) are 93% identical.
The NMHCB interaction domain in PBX1 includes the PBC-B region.
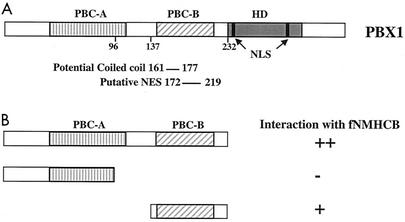
PBC-A and PBC-B are two conserved domains in the N-terminal half of PBC family members (Fig. 2A). PBC-A is important for interaction with MEIS/PREP1 family proteins. PBC-B includes a region corresponding to the predicted NES of EXD and other functional motifs (2, 10). To further map the NMHCB interaction region in the PBX1A N terminus, we performed a yeast two-hybrid assay using the PBX deletion mutants PBX1 1-232 (NT), PBX1 1-96 (PBC-A) and PBX1 137-232 (PBC-B) as bait. Protein expression was verified by Western blotting (data not shown). After mating was performed with yeast expressing fNMHCB prey peptides, >30 colonies bearing the bait plasmids encoding the entire PBX N terminus or the PBC-B region appeared on selective medium after 3 and 5 days, respectively. By contrast, no colonies appeared after 10 days of incubation when the PBC-A region was used as bait. β-Galactosidase filter lift assays verified activation of an independent reporter carrying the lacZ gene (Fig. 2B). A primary-structure prediction program, COILS, identified a potential coiled-coil motif at residues 161 to 177 in the PBC-B domain of PBX1A. The myosin II heavy chain dimerizes via its coiled-coil α-helical tail, suggesting a means of interaction with a structurally similar domain in PBC-B.
FIG. 2.
The PBC-B region interacts with fNMHCB. (A) Diagram of PBX1A. PBC-A and PBC-B are two domains conserved among PBC family proteins. The HD, NLS, putative NES, and a potential coiled-coil structure are indicated. (B) The interaction domain was further mapped to PBC-B using the yeast two-hybrid assay. The interaction strength was scored as time to colony formation on selective medium. The expression of GAL-PBX fusion proteins in yeast was confirmed by Western blot analysis. ++, +, and −, high, intermediate, and undetectable interactions, respectively.
NMHCB associates with PBX1 in vivo.
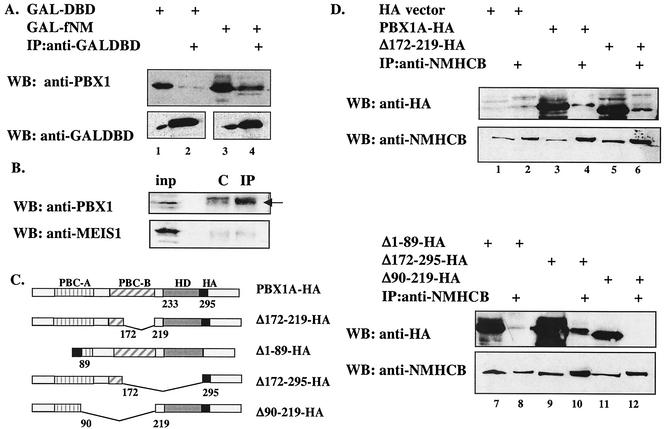
NMHCB-PBX1 interaction was tested by coimmunoprecipitation in mammalian HEK293T cells. A fusion of the GAL-DBD to fNMHCB (GAL-fNM) coprecipitated with transfected PBX1A. As shown in Fig. 3A, PBX1A associated with the GAL-fNM fusion protein but not GAL-DBD alone (compare lanes 2 and 4). Similar results were obtained using a FLAG-tagged NMHCB fragment, FLAG-fNM (data not shown). To confirm this interaction in untransfected cells, we precipitated endogenous NMHCB with a monoclonal antibody. Results showed weak but detectable association with endogenous PBX1A as revealed with an anti-PBX1 antibody (Fig. 3B, top). By contrast, we detected no association of MEIS1 with NMHCB (Fig. 3B, bottom). To further define the NMHCB interaction domain in PBX, hemagglutinin-tagged PBX1A and a series of PBX1A deletion mutants (Fig. 3C) were expressed in HEK293T cells, and their interactions with NMHCB were examined. From the results shown in Fig. 3D, we conclude that residues 90 to 172 in the PBC-B region (Fig. 3D, lane 12), with contributions from the PBC-A region (Fig. 3D, lane 8), are required for the interaction between PBX1A and NMHCB.
FIG. 3.
PBX1A associates with fNMHCB, as well as endogenous NMHCB, in vivo. (A) The NMHCB fragment interacts with PBX1A in HEK293T cells. Cells were transfected with mammalian expression vectors for GAL-DBD (lanes 1 and 2) or GAL-fNM (lanes 3 and 4) with PBX1A. The cell lysates were immunoprecipitated (IP) with anti-GAL-DBD antibody and resolved by SDS-10% polyacrylamide gel electrophoresis. PBX1A was detected with an anti-PBX1 antibody. Below is the Western blot (WB) of the same membrane demonstrating comparable levels of GAL species in input (lanes 1 and 3) and IP (lanes 2 and 4) samples. +, present. (B) Endogenous NMHCB associates with PBX1A but not MEIS1 in HEK293T cells. Cell lysate (input [inp]) was immunoprecipitated with nonspecific antibody (lane C) or anti-NMHCB antibody (lane IP). Anti-PBX1 (top) and anti-MEIS1 (bottom) antibodies were used for Western blot analysis. The arrow points to PBX1. (C) Schematic illustration of wild-type and mutant PBX1A proteins. PBC-A and PBC-B are two regions conserved among PBC family proteins. HA, hemagglutinin tag. (D) Finer mapping of the NMHCB interaction domain in PBX1A. Deletion of the putative NES (Δ172-219) or HD (Δ172-295) did not affect PBX1A and NMHCB interaction, while deletion of PBC-A (Δ1-89) and PBC-B (Δ90-219) reduced or abolished the interaction. Expression vectors for 3XHA empty vector (lanes 1 and 2), PBX1A-HA (lanes 3 and 4), PBX1AΔ172-219-HA (lanes 5 and 6), PBX1AΔ1-89-HA (lanes 7 and 8), PBX1AΔ172-295-HA (lanes 9 and 10), and PBX1AΔ90-219-HA (lanes 11 and 12) were transfected into HEK293T cells, coimmunoprecipitation was performed with anti-NMHCB, and Western blotting was carried out with anti-HA antibody (top) and anti-NMHCB antibody (bottom).
fNMHCB promotes cytoplasmic retention of PBX.
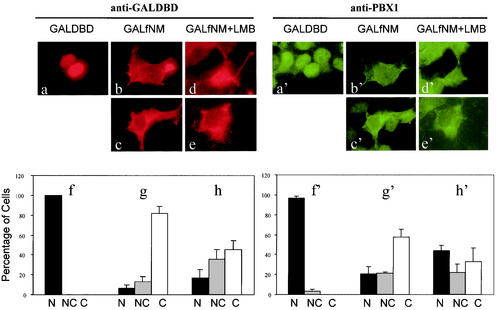
NMHCB is a cytoplasmic protein and a major component of the cytoskeleton. To investigate whether NMHCB directs the cytoplasmic localization of PBX, we first investigated the cytoplasmic retention properties of the NMHCB fragment obtained in the yeast two-hybrid screen. In contrast to the exclusively nuclear localization of GAL-DBD (Fig. 4a and f), we observed that the GAL-fNM fusion protein is mainly cytoplasmically localized in HEK293T (Fig. 4b, c, and g) and Cos-7 cells (data not shown). Both cell lines express MEIS proteins (data not shown), and thus, endogenous PBX1 protein is normally localized in the nucleus (Fig. 4a′ and f′). However, GAL-fNM induced a marked nuclear-to-cytoplasmic shift of endogenous PBX1 (Fig. 4b′, c′, and g′). Moreover, these two proteins appeared to colocalize in the cytoplasm (compare b to b′ and c to c′).
FIG. 4.
fNMHCB has cytoplasmic retention properties. HEK293T cells were transfected with GAL-DBD or GAL-fNM and double labeled with anti-GAL-DBD (a to h) and anti-PBX1 (a′ to h′) antibodies. The two sets of photomicrographs show the same cell(s) visualized for GAL and PBX1 immunoreactivities (e.g., a and a′). The bar graphs display the percentages of cells showing nuclear (N), nuclear and cytoplasmic (NC), and cytoplasmic (C) localization of GAL species (f, g, h) and PBX1 (f′, g′, h′). The values are the averages of three independent experiments; 150 to 200 cells from each transfection were scored for the localization of the protein of interest. The error bars show standard deviations.
In contrast, GAL-fNM did not affect the nuclear localization of TBP (data not shown), indicating that this cytoplasmic retention effect is specific to PBX1.
The results described above suggest that the NMHCB fragment has cytoplasmic retention properties, since its overexpression causes PBX to accumulate in the cytoplasm. However, an alternative explanation could invoke nuclear export. For example, fNMHCB could harbor an NES or expose the NES in PBX. To test whether CRM1-mediated nuclear export plays a role in this process, we expressed GAL-fNM in HEK293T cells and treated them with a high concentration (200 nM) of LMB, an inhibitor of CRM1-dependent export. While LMB did cause a moderate shift toward nuclear accumulation of both GAL-fNM and PBX, a substantial fraction of cells showed a strict cytoplasmic accumulation of both proteins (Fig. 4d, e, and h and d′, e′, and h′). These results support a role for NMHCB that is independent of export functions.
fNMHCB competes with MEIS for translocation of PBX1A or EXD in Drosophila S2 cells.
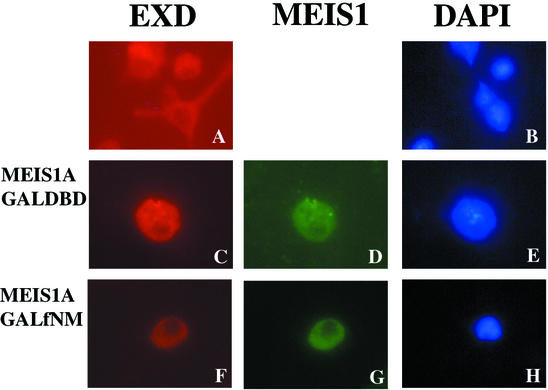
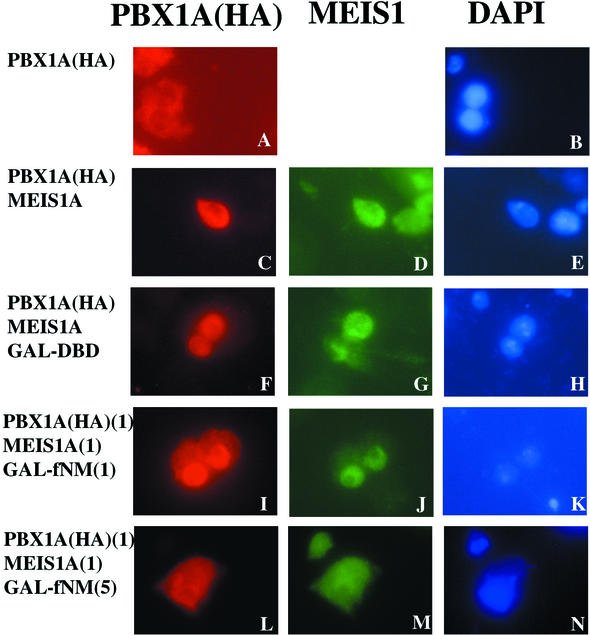
To determine whether the interaction between MEIS and PBX can be affected by fNMHCB, we used Drosophila S2 cells, in which endogenous EXD is localized in the cytoplasm due to the absence of HTH (Fig. 5A and B). After expression of MEIS1A in S2 cells, both EXD and MEIS1A localized to the nucleus (Fig. 5C to E). Similar to EXD, PBX1A localized to the cytoplasm of S2 cells in the absence of MEIS (Fig. 6A and B). Coexpression of MEIS1A led to nuclear accumulation of both PBX1A and MEIS1A (Fig. 6C to E). However, overexpression of GAL-fNM, but not GAL-DBD, induced cytoplasmic retention of EXD, PBX1A, and MEIS1A (Fig. 5C to H and Fig. 6F to N). A higher ratio of GAL-fNM to MEIS1A expression vector was required for cytoplasmic retention of PBX1A than for that of EXD and was likely due to the greater abundance of transfected PBX1A over endogenous EXD. Thus, fNMHCB can compete against the action of MEIS to direct the localization of PBX/EXD to the cytoplasm.
FIG. 5.
fNMHCB retains EXD in the cytoplasm of Drosophila S2 cells. Drosophila S2 cells were transfected with expression vectors for MEIS1A, GAL-DBD, and GAL-fNM individually or in combination. Immunostaining was performed with anti-EXD and anti-MEIS1 antibodies. DAPI staining reveals nuclei. (A and B) Endogenous EXD is located in the cytoplasm of S2 cells (which lack HTH, a Drosophila MEIS homologue). (C to E) Cotransfection of MEIS1A induces nuclear translocation of EXD. (F to H) GAL-fNM opposes the action of MEIS1A, inducing retention of EXD and MEIS1A in the cytoplasm.
FIG. 6.
fNMHCB retains PBX1A in the cytoplasm of Drosophila S2 cells. Drosophila S2 cells were transfected with insect expression vectors for HA-PBX1A, MEIS1A, GAL-DBD, and GAL-fNM individually or in combination. Immunostaining was performed with anti-HA and anti-MEIS1 antibodies. DAPI staining reveals nuclei. (A and B) PBX1A is located in the cytoplasm of S2 cells. (C to E) Expression of MEIS1A induces nuclear translocation of PBX1A. (F to H) GAL-DBD has no effect on the subcellular localization of either PBX1A or MEIS1A, whereas GAL-fNM opposes the action of MEIS1A, inducing cytoplasmic retention of PBX1A, in a dose-dependent manner. (I to K) Equal amounts of expression vectors for GAL-fNM and MEIS1A were transfected. (L to N) The ratio of transfected GAL-fNM to MEIS1A expression vectors was 5:1.
Colocalization of NMHCB and PBX1 in the mouse distal limb bud.
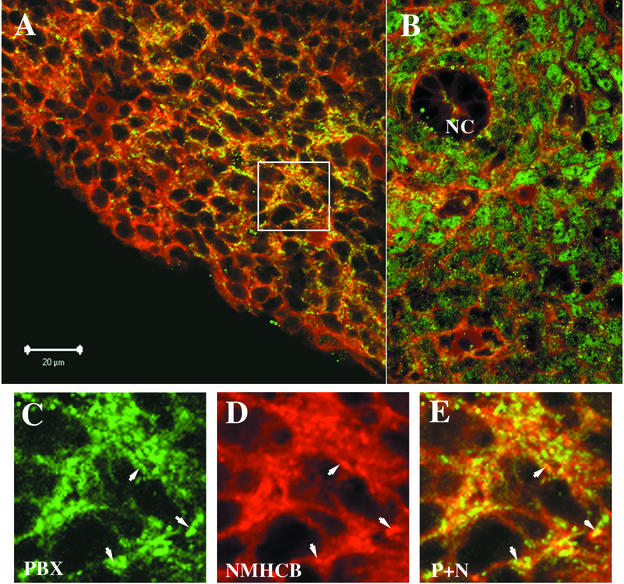
PBX1 is expressed in many tissues in the mouse embryo. However, the subcellular distribution of PBX1 is not uniform. For example, PBX is localized to the cytoplasm in distal limb bud mesenchymal cells but is localized to the nucleus in the proximal limb bud, where MEIS proteins are also expressed (19). By immunofluorescence staining, we confirmed the subcellular distribution of PBX1 in the mouse limb bud and elsewhere. As shown in Fig. 7B, PBX1 is concentrated in the nucleus of mesenchymal cells surrounding the notochord. By contrast, PBX1 is cytoplasmically localized in the gut epithelium and neural tube (data not shown). To study whether there is colocalization of cytoplasmic PBX1 and NMHCB, 11.5-dpc mouse embryonic sections were double stained with anti-PBX1 and anti-NMHCB, and the samples were examined by confocal microscopy. In the distal limb bud region, the majority of PBX1 is colocalized with NMHCB in the cytoplasm (Fig. 7 A and C to E). By contrast, mesenchymal cells surrounding the notochord show nuclear PBX1 staining and minimal overlap with NMHCB (Fig. 7B).
FIG. 7.
Colocalization of PBX1 and NMHCB in mouse distal limb bud. PBX1 (green) and NMHCB (red) expression were detected by confocal microscopy of immunostained 11.5-dpc mouse embryo sections. (A) Lower magnification of the distal limb bud. PBX1 is in the cytoplasm and largely colocalized with NMHCB, as demonstrated by yellow signals in regions of green-red overlap. (B) In mesenchymal cells surrounding the notochord (NC), PBX1 shows nuclear staining and minimal overlap with NMHCB. (C and D) Higher magnification of PBX1 and NMHCB distribution in the distal limb bud (boxed in panel A). (E) Superposition of images in panels C and D. Colocalization is revealed by the yellow signal. The arrows in panels C to E point to the overlapping signals of PBX1 and NMHCB.
The subcellular localization of EXD in Drosophila zipper mutant embryos is partially deregulated.
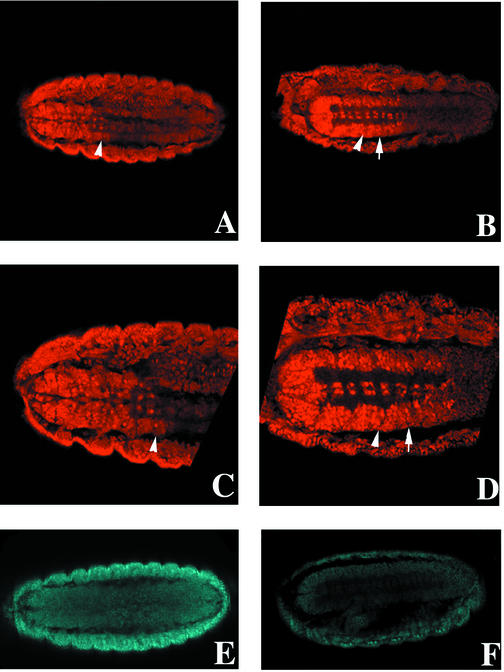
In Drosophila, EXD is broadly expressed but localized to the cytoplasm at early embryonic stages. Nuclear accumulation first becomes apparent in the anterior ventral epidermis from stages 8 to 10 and more significantly in the thoracic epidermis, thoracic CNS, and midgut constrictions after germ band retraction (stage 15) (4). zipper encodes the Drosophila nonmuscle myosin heavy chain. Zygotic zipper transcripts and protein peak 4 to 12 h after egg laying (41). To study whether the zipper mutation shifts EXD to the nucleus, we analyzed zipper mutants at stage 15, when the contribution of maternal zipper begins to diminish (41). As previously shown (4), nuclear accumulation of EXD in the CNS of stage 15 wild-type embryos was predominantly in the thoracic segments T1, T2, and T3. In the abdominal CNS, most cells expressed a low level of EXD that was restricted to the cytoplasm (Fig. 8A and C), consistent with the repression of exd and hth in abdominal segments (5, 25, 34). However, in homozygous zipper mutant embryos, the boundary of nuclear versus cytoplasmic EXD, and high versus low levels of EXD expression, was shifted at least one segment posterior to encompass the first abdominal segment, A1 (Fig. 8B and D). Reduced accumulation of ZIPPER protein in stage 15 mutant embryos was confirmed (compare Fig. 8E and F). These results demonstrate that the nonmuscle myosin promotes the cytoplasmic retention of EXD and that it is one of several mechanisms that constrain EXD availability in the nucleus.
FIG. 8.
EXD expression in the CNSs of Drosophila wild-type and zipper mutant embryos. EXD was visualized in wild-type (A and C) and zipper mutant (B and D) embryos with an anti-EXD antibody. The arrowheads indicate the boundary between the third thoracic (T3) and first abdominal (A1) segments. Anterior is to the left. (A) Ventral view of EXD staining in the CNS of a stage 15 wild-type embryo. The boundary between cells expressing high nuclear versus low cytoplasmic EXD is located at the T3-A1 border. (C) Closer examination of the thoracic CNS shows that most cells express EXD at intermediate to high levels in both the cytoplasm and nucleus, or mainly in the nucleus. In the abdominal CNS, most cells contain low levels of strictly cytoplasmic EXD. (B) EXD staining in the CNS of a stage 15 zipper mutant embryo. The boundary between nuclear versus cytoplasmic EXD (arrow) is shifted posteriorly to encompass A1. (D) Closer view of CNS cells in a zipper mutant. Cells in A1 (between the arrowhead and the arrow) express high levels of EXD in both the cytoplasm and nucleus. Several “z” planes were examined for each embryo to ensure that differences in EXD expression were not artifacts of the plane of section (data not shown). Four out of four zipper mutant embryos for which ventral views of the CNS were possible displayed the same posteriorized EXD nuclear localization. (E and F) The expression level of ZIPPER in a zipper mutant embryo (F) is greatly reduced compared to that in a wild-type embryo (E).
DISCUSSION
A novel function of NMHCB.
In this study, we demonstrate that nonmuscle myosin regulates the subcellular localization of PBX and EXD in cultured cells and the fly embryo. It seems that nonmuscle myosin plays important roles in development and oncogenesis by controlling not only such processes as cell shape and motility but also the availability of at least one transcription factor family, PBX/EXD. These apparently disparate functions could provide a means to coordinate morphogenetic programs at the cellular and transcriptional levels. There is a precedent for a role for cytoskeletal elements in this regard, since signaling through Rho GTPases induces the activity of the transcriptional regulator serum response factor via actin polymerization and the depletion of G-actin pools (18, 38). In addition, microtubule association controls the nucleocytoplasmic compartmentalization of the Smad and MIZ-1 transcription factors (15, 42). We have shown colocalization of PBX1 and NMHCB in the mouse distal limb bud. It is conceivable that signaling pathways controlling other aspects of MEIS and PBX functions in the limb (11, 29) also regulate this interaction.
NMHCB functions through cytoplasmic retention, not as an NES-containing adapter.
The putative NES in PBX may function alternatively as a docking site for NES-containing partners. We first mapped NMHCB interaction to the PBC-B domain of PBX, which includes the putative NES, suggesting that NMHCB could be such a partner. However, a region in PBC-B (residues 172 to 219) largely coextensive with the putative NES of EXD (residues 178 to 220) is not required for interaction with NMHCB. In addition, NMHCB is unlikely to ever act in the nuclear compartment, since its large molecular mass (∼220 kDa) would prohibit diffusion through the nuclear pore complex, and its distribution is not affected by LMB treatment (our unpublished observation). This is in contrast to the CRM1-sensitive compartmentalization of actin (40). Additional findings also argue against export-based mechanisms in the action of NMHCB, since the cytoplasmic localization of GAL-fNM and the cytoplasmic targeting of PBX by GAL-fNM are both largely refractory to LMB treatment. Despite the predominantly cytoplasmic localization of GAL-fNM in mammalian cells, the original VP16-fNM prey construct isolated in a yeast two-hybrid screen must have some access to the yeast nucleus. This may be due to the NLS supplied by the VP16 vector or because yeast cells do not express endogenous PBX, which may be required for the mutual retention of PBX and fNMHCB in the cytoplasm (see below).
NMHCB and PBX interact through coiled-coil motifs.
The tail region of NMHC is characterized by a coiled-coil motif involved in self-dimerization. However, this coiled-coil motif may also bridge the interaction between NMHCB and other partners. In this study, we defined the NMHCB interaction domain as residues 90 to 171 in PBX1 and showed that it overlapped with a potential coiled-coil motif (residues 161 to 177). Finer mutation of this region will help to elucidate its role in NMHCB-PBX interaction.
Multiple mechanisms control EXD subcellular localization in fly embryos.
The BX-C group of homeotic genes (Ubx, abd-A, and Abd-B) has been shown to downregulate the nuclear accumulation of EXD through repression of exd and hth (5, 25, 34). The effects of downregulation would be exacerbated by the requirement for EXD in stabilization of HTH (1). Nonetheless, the normal functions of Ubx and abd-A still require EXD activity (5, 13, 19, 32, 35). Thus, the amount of available nuclear EXD in abdominal segments is tightly regulated within narrow limits.
In addition to the influences of gene expression, protein stability, nuclear import, and nuclear export, we show here that cytoplasmic retention by nonmuscle myosin provides yet another level of control of EXD subcellular localization. The loss of ZIPPER results in nuclear accumulation of EXD in the first abdominal segment. The same phenotype (increased nuclear localization of EXD in A1) is seen in Ubx mutants (5), suggesting that ZIPPER and UBX make equally important contributions to the control of EXD compartmentalization. ZIPPER's role in this process may extend well beyond A1 but could be obscured by perduring maternal product. The lethality of zipper mutants precludes examination at later stages, when maternal stores might be further depleted. Together, these findings emphasize that multiple nonredundant controls govern EXD/PBX subcellular localization. They also demonstrate that interaction with nonmuscle myosin is an evolutionarily conserved strategy for controlling the intracellular distribution of EXD/PBX.
The role of NMHCB in MEIS and PBX interaction.
While a number of investigations have focused on elucidating the mechanism of PBX/EXD localization, the regulation of MEIS subcellular distribution has not been well studied. However, one recent report suggests that zebrafish MEIS3 must interact with PBX in order to translocate to the nucleus (39). Two leucine-rich regions, LFPLLALIF (residues 83 to 91) and LDNLMIQVL (residues 143 to 151), in the conserved PBX interaction domains of MEIS1 (HM1 and HM2) may function as NESs. The heterodimerization of PBX and MEIS may therefore mask NESs in both partners. PBX-MEIS interaction could also expose one or more NLSs within the MEIS HD, as has been demonstrated for PBX (37).
In this study, we showed that overexpression of GAL-fNM leads to the cytoplasmic localization of both PBX and MEIS. Thus, GAL-fNM may disrupt PBX-MEIS interaction in the cytoplasm, resulting in the intramolecular masking of NLSs in both partners. Our data argue against a cytoplasmic NMHCB-PBX complex retaining MEIS in this compartment. No potential coiled-coil structure has been identified in MEIS, suggesting that it is unlikely to interact with NMHCB in the same manner as PBX. Moreover, yeast two-hybrid assays indicate that the MEIS1 N terminus (residues 1 to 231) does not interact with fNMHCB (data not shown), and MEIS is not coimmunoprecipitated with NMHCB-PBX complexes (Fig. 3B).
In the normal course of development, NMHCB may retain PBX in the cytoplasm, or stabilize the conformation through which the PBX N terminus masks the NLSs in the HD (37). Entry to the nucleus would therefore depend on sufficient levels of MEIS (or PREP1) proteins to effectively compete with NMHCB for interaction with PBX. PBX-MEIS interaction would then lead to unmasking of their NLSs, occlusion of their NESs, and stable nuclear accumulation of both partners. This competition between nonmuscle myosin and MEIS family members could be influenced by patterning signals, such as DPP (3).
Acknowledgments
We thank M. Kamps for the human PBX1A cDNA, A. Buchberg for the murine Meis1a cDNA, R. White and D. Kiehart for the anti-EXD and anti-ZIPPER antibodies, K. Yoshida for LMB, and U. Stochaj and A. Percival-Smith for helpful discussions. The CMII23 anti-NMHCB antibody generated by G. W. Conrad and A. H. Conrad was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the Department of Biological Sciences, University of Iowa, Iowa City.
H.H. is the recipient of an Internal Scholarship of the Faculty of Medicine, McGill University. M.P. holds a Doctoral Research Award from the Canadian Institutes of Health Research (CIHR). P.L. is a CIHR Investigator. M.F. is a Chercheur-National of the Fonds de la Recherche en Santé du Québec. This work was funded by grants to P.L. from the National Cancer Institute of Canada and to M.F. from the CIHR.
REFERENCES
- 1.Abu-Shaar, M., and R. S. Mann. 1998. Generation of multiple antagonistic domains along the proximodistal axis during Drosophila leg development. Development 125:3821-3830. [DOI] [PubMed] [Google Scholar]
- 2.Abu-Shaar, M., H. D. Ryoo, and R. S. Mann. 1999. Control of the nuclear localization of Extradenticle by competing nuclear import and export signals. Genes Dev. 13:935-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arquier, N., L. Perrin, P. Manfruelli, and M. Semeriva. 2001. The Drosophila tumor suppressor gene lethal(2)giant larvae is required for the emission of the Decapentaplegic signal. Development 128:2209-2220. [DOI] [PubMed] [Google Scholar]
- 4.Aspland, S. E., and R. A. White. 1997. Nucleocytoplasmic localisation of extradenticle protein is spatially regulated throughout development in Drosophila. Development 124:741-747. [DOI] [PubMed] [Google Scholar]
- 5.Azpiazu, N., and G. Morata. 1998. Functional and regulatory interactions between Hox and extradenticle genes. Genes Dev. 12:261-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berthelsen, J., C. Kilstrup-Nielsen, F. Blasi, F. Mavilio, and V. Zappavigna. 1999. The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev. 13:946-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bürglin, T. 1997. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res 25:4173-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bürglin, T. R. 1998. The PBC domain contains a MEINOX domain: coevolution of Hox and TALE homeobox genes? Dev. Genes Evol. 208:113-116. [DOI] [PubMed] [Google Scholar]
- 9.Bürglin, T. R., and G. Ruvkin. 1992. A new motif in PBX genes. Nat. Genet. 1:319-320. [DOI] [PubMed] [Google Scholar]
- 10.Calvo, K. R., P. Knoepfler, S. McGrath, and M. P. Kamps. 1999. An inhibitory switch derepressed by pbx, hox, and Meis/Prep1 partners regulates DNA-binding by pbx1 and E2a-pbx1 and is dispensable for myeloid immortalization by E2a-pbx1. Oncogene 18:8033-8043. [DOI] [PubMed] [Google Scholar]
- 11.Capdevila, J., T. Tsukui, C. Rodriquez Esteban, V. Zappavigna, and J. C. Izpisua Belmonte. 1999. Control of vertebrate limb outgrowth by the proximal factor Meis2 and distal antagonism of BMPs by Gremlin. Mol. Cell 4:839-849. [DOI] [PubMed] [Google Scholar]
- 12.Cecconi, F., G. Proetzel, G. Alvarez-Bolado, D. Jay, and P. Gruss. 1997. Expression of Meis2, a Knotted-related murine homeobox gene, indicates a role in the differentiation of the forebrain and the somitic mesoderm. Dev. Dynam. 210:184-190. [DOI] [PubMed] [Google Scholar]
- 13.Chan, S. K., L. Jaffe, M. Capovilla, J. Botas, and R. S. Mann. 1994. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell 78:603-615. [DOI] [PubMed] [Google Scholar]
- 14.Chang, C. P., Y. Jacobs, T. Nakamura, N. A. Jenkins, N. G. Copeland, and M. L. Cleary. 1997. Meis proteins are major in vivo DNA binding partners for wild-type but not chimeric Pbx proteins. Mol. Cell. Biol. 17:5679-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong, C., Z. Li, R. Alvarez, Jr., X. H. Feng, and P. J. Goldschmidt-Clermont. 2000. Microtubule binding to Smads may regulate TGF beta activity. Mol. Cell 5:27-34. [DOI] [PubMed] [Google Scholar]
- 16.Featherstone, M. HOX proteins and their cofactors in transcriptional regulation. In T. Lufkin (ed.), Murine homeobox gene control of embryonic patterning and organogenesis, in press. Elsevier, Amsterdam, The Netherlands.
- 17.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 18.Gineitis, D., and R. Treisman. 2001. Differential usage of signal transduction pathways defines two types of serum response factor target gene. J. Biol. Chem. 276:24531-24539. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez-Crespo, S., M. Abu-Shaar, M. Torres, A. C. Martinez, R. S. Mann, and G. Morata. 1998. Antagonism between extradenticle function and Hedgehog signalling in the developing limb. Nature (London) 394:196-200. [DOI] [PubMed] [Google Scholar]
- 20.Graba, Y., D. Aragnol, and J. Pradel. 1997. Drosophila Hox complex downstream targets and the function of homeotic genes. Bioessays 19:379-388. [DOI] [PubMed] [Google Scholar]
- 21.Jaw, T. J., L. R. You, P. S. Knoepfler, L. C. Yao, C. Y. Pai, C. Y. Tang, L. P. Chang, J. Berthelsen, F. Blasi, M. P. Kamps, and Y. H. Sun. 2000. Direct interaction of two homeoproteins, homothorax and extradenticle, is essential for EXD nuclear localization and function. Mech. Dev. 91:279-291. [DOI] [PubMed] [Google Scholar]
- 22.Knoepfler, P. S., K. R. Calvo, H. Chen, S. E. Antonarakis, and M. P. Kamps. 1997. Meis1 and pKnox1 bind DNA cooperatively with Pbx1 utilizing an interaction surface disrupted in oncoprotein E2a-Pbx1. Proc. Natl. Acad. Sci. USA 94:14553-14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krumlauf, R. 1994. Hox genes in vertebrate development. Cell 78:191-201. [DOI] [PubMed] [Google Scholar]
- 24.Kudo, N., N. Matsumori, H. Taoka, D. Fujiwara, E. P. Schreiner, B. Wolff, M. Yoshida, and S. Horinouchi. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96:9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurant, E., C. Y. Pai, R. Sharf, N. Halachmi, Y. H. Sun, and A. Salzberg. 1998. Dorsotonals/homothorax, the Drosophila homologue of meis1, interacts with extradenticle in patterning of the embryonic PNS. Development 125:1037-1048. [DOI] [PubMed] [Google Scholar]
- 26.Mann, R. S., and M. Abu-Shaar. 1996. Nuclear import of the homeodomain protein extradenticle in response to Wg and Dpp signalling. Nature 383:630-633. [DOI] [PubMed] [Google Scholar]
- 27.Mann, R. S., and M. Affolter. 1998. Hox proteins meet more partners. Curr. Opin. Genet. Dev. 8:423-429. [DOI] [PubMed] [Google Scholar]
- 28.Mercader, N., E. Leonardo, N. Azpiazu, A. Serrano, G. Morata, C. Martinez, and M. Torres. 1999. Conserved regulation of proximodistal limb axis development by Meis1/Hth. Nature 402:425-429. [DOI] [PubMed] [Google Scholar]
- 29.Mercader, N., E. Leonardo, M. E. Piedra, A. C. Martinez, M. A. Ros, and M. Torres. 2000. Opposing RA and FGF signals control proximodistal vertebrate limb development through regulation of Meis genes. Development 127:3961-3970. [DOI] [PubMed] [Google Scholar]
- 30.Oulad-Abdelghani, M., C. Chazaud, P. Bouillet, V. Sapin, P. Chambon, and P. Dollé. 1997. Meis2, a novel mouse Pbx-related homeobox gene induced by retinoic acid during differentiation of P19 embryonal carcinoma cells. Dev. Dyn. 210:173-183. [DOI] [PubMed] [Google Scholar]
- 31.Pai, C. Y., T. S. Kuo, T. J. Jaw, E. Kurant, C. T. Chen, D. A. Bessarab, A. Salzberg, and Y. H. Sun. 1998. The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, extradenticle, and suppresses eye development in Drosophila. Genes Dev. 12:435-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peifer, M., and E. Wieschaus. 1990. Mutations in the Drosophila gene extradenticle affect the way specific homeo domain proteins regulate segmental identity. Genes Dev. 4:1209-1223. [DOI] [PubMed] [Google Scholar]
- 33.Rambaldi, I., E. Nagy Kovàcs, and M. S. Featherstone. 1994. A proline-rich transcriptional activation domain in murine HOXD-4 (HOX-4.2). Nucleic Acids Res. 22:376-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rauskolb, C., M. Peifer, and E. Wieschaus. 1993. extradenticle, a regulator of homeotic gene activity, is a homolog of the homeobox-containing human proto-oncogene pbx1. Cell 74:1101-1112. [DOI] [PubMed] [Google Scholar]
- 35.Rauskolb, C., and E. Wieschaus. 1994. Coordinate regulation of downstream genes by extradenticle and the homeotic selector proteins. EMBO J. 13:3561-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rieckhof, G. E., F. Casares, H. D. Ryoo, M. Abu-Shaar, and R. S. Mann. 1997. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell 91:171-183. [DOI] [PubMed] [Google Scholar]
- 37.Saleh, M., H. Huang, N. C. Green, and M. S. Featherstone. 2000. A conformational change in PBX1A is necessary for its nuclear localization. Exp. Cell Res. 260:105-115. [DOI] [PubMed] [Google Scholar]
- 38.Sotiropoulos, A., D. Gineitis, J. Copeland, and R. Treisman. 1999. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell 98:159-169. [DOI] [PubMed] [Google Scholar]
- 39.Vlachakis, N., D. R. Ellstrom, and C. G. Sagerstrom. 2000. A novel pbx family member expressed during early zebrafish embryogenesis forms trimeric complexes with Meis3 and Hoxb1b. Dev. Dyn. 217:109-119. [DOI] [PubMed] [Google Scholar]
- 40.Wada, A., M. Fukuda, M. Mishima, and E. Nishida. 1998. Nuclear export of actin: a novel mechanism regulating the subcellular localization of a major cytoskeletal protein. EMBO J. 17:1635-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young, P. E., A. M. Richman, A. S. Ketchum, and D. P. Kiehart. 1993. Morphogenesis in Drosophila requires nonmuscle myosin heavy chain function. Genes Dev. 7:29-41. [DOI] [PubMed] [Google Scholar]
- 42.Ziegelbauer, J., B. Shan, D. Yager, C. Larabell, B. Hoffmann, and R. Tjian. 2001. Transcription factor MIZ-1 is regulated via microtubule association. Mol. Cell 8:339-349. [DOI] [PubMed] [Google Scholar]