Abstract
Linker histones are nonessential for the life of single-celled eukaryotes. Linker histones, however, can be important components of specific developmental programs in multicellular animals and plants. For Caenorhabditis elegans a single linker histone variant (H1.1) is essential in a chromatin silencing process which is crucial for the proliferation and differentiation of the hermaphrodite germ line. In this study we analyzed the whole linker histone complement of C. elegans by telomeric position effect variegation in budding yeast. In this assay an indicator gene (URA3) placed close to the repressive telomeric chromatin structure is subject to epigenetically inherited gene inactivation. Just one out of seven C. elegans linker histones (H1.1) was able to enhance the telomeric position effect in budding yeast. Since these results reflect the biological function of H1.1 in C. elegans, we suggest that chromatin silencing in C. elegans is governed by molecular mechanisms related to the telomere-dependent silencing in budding yeast. We confirmed this hypothesis by testing C. elegans homologs of three yeast genes which are established modifiers of the yeast telomeric chromatin structure (SIR2, SET1, and RAD17) for their influence on repeat-dependent transgene silencing for C. elegans.
Linker histones are highly abundant eukaryotic chromatin proteins. They bind to the nucleosomes, forming the 30-nm chromatin fiber. The biological function of linker histones is not sufficiently clear. In contrast to the core histones which are essential for eukaryotic life (17, 25), linker histones are dispensable in single-celled organisms, as shown by knockouts performed with Tetrahymena (40), yeast (13, 35), and in simple multicellular fungi such as Aspergillus (36) and Ascobolus (8). For multicellular eukaryotes, linker histones usually exist as a set of relatively divergent protein variants. The occurrence of a typical linker histone gene family is correlated with multicellularity in plants as well as in animals. For a long time it has been considered that different linker histone variants could fulfill different biochemical functions, but no canonical test has been established to investigate this. Only recently it has become apparent that individual linker histone variants can specifically contribute to essential aspects of multicellular life, like cell differentiation and development (for a review, see reference 23). We use Caenorhabditis elegans as a model system to investigate this question. C. elegans possesses eight different linker histone variants, the same number as for humans (3, 4, 47) or mice (12), and it allows dissection of the functions of individual linker histone genes with RNA interference (RNIi) and cytological techniques. In previous work we characterized the function of the major histone H1 variant in C. elegans, H1.1 (19). This protein is essential for the germ line-specific chromatin silencing and consequently also for hermaphrodite fertility. The phenotype of H1.1 depletion is very similar to the phenotype of the maternal-effect sterile (mes) mutants mes-2, mes-3, mes-4, and mes-6. mes-2 and mes-6 are the homologs of Drosophila Polycomb group (Pc-G) genes Enhancer of zeste and Extra sex combs. It is assumed that the mes genes function in the inheritable control of the activity state of chromatin domains, as the Pc-G does for Drosophila (18, 26).
Since the exact mechanisms of chromatin silencing in the germ line of C. elegans are not known, we tested the C. elegans linker histone gene family for their influence on telomeric position effect variegation (TPEV) in budding yeast (16). This assay quantitatively reproduces the silencing effect exerted from the telomeric heterochromatin onto an URA3 indicator gene inserted nearby. Although the endogenous budding yeast linker histone does not influence the telomeric chromatin structure (13, 35), a single C. elegans H1 protein (H1.1) enhanced the repressive effect of the telomeric chromatin. Since H1.1 is the linker histone variant involved in germ line chromatin silencing for C. elegans, our result suggests that the telomeric position effect variegation caused by this protein does indeed assay the specific biological function of this protein in the nematode. This and further observations indicate that an unexpected molecular mechanistic relation between telomeric silencing for budding yeast and germ line chromatin silencing for C. elegans may exist.
MATERIALS AND METHODS
Strains and culturing methods.
C. elegans wild-type strain is N2 (Bristol) (9). EC107 {pha-1(e2123) [pha-1(+) let-858::gfp]} and EC108 {him-8 pha-(e2123) [pha-1(+) let-858::gfp]} were constructed according to the method of Kelly and Fire (21) as described in the work of Jedrusik and Schulze (19). The general maintenance and routine culturing of C. elegans strains were performed as described by Brenner (10). Saccharomyces cerevisiae: UCC3505 (MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 ppr1::HIS3 adh4::URA3-TEL DIA5-1) (42) and UCC7007-1 (MATa ura3-52 lys2-801 ade2-101 trp1-Δ63 his3-Δ200 leu2-Δ1 ppr1::HIS3 adh4::URA3-TEL-VIIL VR-ADE-TEL sir3::KanMX4) were provided by Daniel E. Gottschling, Fred Hutchinson Cancer Research Center, Seattle, Wash. The control strain S35P-5A (URA3+ leu2) was obtained from Hans Dieter Schmitt, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany. General yeast culturing methods and yeast extract-peptone-dextrose (YPD) medium were taken from the work of Ausubel et al. (7). YPAD is YPD medium supplemented with adenine (0.1 g/l). The selective medium SC-leu is based on a synthetic complete medium (41), but without leucine.
Expression of C. elegans H1 proteins in budding yeast.
Seven C. elegans full-length H1 cDNAs (H1.1-H1.5, H1.Q, and H1.X) were excised with EcoRI and XhoI from the plasmids yk85b12, yk187f1, yk89f5, yk142d7, yk173g2, yk116f11, and yk480h7 (19), respectively, and transferred into the polylinker of the yeast expression vectors pYX142 and pYX242 (R&D Systems, Minneapolis, Minn.). This cloning step substitutes the start codon and the open reading frame of the vector polylinker with the C. elegans cDNA so that the respective histone H1 start codon becomes the first start codon in the mRNA. Both plasmids employ the constitutive promoter of the S. cerevisiae triose phosphate promoter (2) and LEU2 selection. pYX142 contains a centromeric origin of replication, generating single or a few plasmid copies per cell, whereas pYX242 uses a 2μm origin of replication for generating high copy numbers. The resulting 14 plasmids were used to transform the S. cerevisiae strains UCC3505 and S35P-5A. Selection on leucine-deficient media was used permanently. For the transformation of six plasmids into the S. cerevisiae strain UCC7007-1, a kit was used (Grow′n Glow High Efficiency Yeast Transformation kit; MoBiTec, Göttingen, Germany).
Expression of C. elegans H1::GFP fusion proteins in yeast.
The enhanced green fluorescent protein (GFP) coding sequence of pEGFP-N1 (Clontech) was cloned into the polylinker of pYX142 with EcoRI and SacI (pYX142-GFP). Complete C. elegans H1 coding sequences were amplified from cDNAs (see above) with a set of PCR primers that substituted the sequence upstream of the start codon with the sequence GGAATTCACCATG, which is identical to the sequence upstream of the start codon in pYX142. The PCR products were cloned with EcoRI (underlined above) and a different, second restriction enzyme in pYX142-GFP. This resulted in full-length C-terminal fusions of the linker histones with the N-terminal end of enhanced GFP. All expression constructs contained identical 3′ and 5′ untranslated sequences to ensure comparable levels of transcription, mRNA processing, and translation in yeast.
Test for URA3 expression.
Yeast cells were precultured in liquid SC-leu medium for 1 or 2 days. Aliquots of a decadic dilution series were plated in parallel on SC-leu medium and an SC-leu-FOA medium (1 g of 5-fluoroorotic acid/liter), on which URA3-expressing cells die (9). The plates were incubated for 2 or 3 days at 30°. Plates containing 50 to 100 colonies were used for determination of the living cell number on both media. The percentage of URA3-expressing cells was determined as 100 − ([colonies on SC-leu-FOA medium]/[colonies on SC-leu medium] × 100). Multiple independent determinations were done, and average values and standard deviations were calculated.
Indirect immunofluorescence of S. cerevisiae.
A modified version (personal communication of Werner Albig, Institute of Biochemistry, University of Göttingen) of the protocol of Kilmartin and Adams (24) was used. The cells were grown in 5-ml SC-leu medium to an optical density of 0.5 to 1.0 at 600 nm. Two hundred fifty microliters of 0.5 M potassium phosphate buffer (PPB), pH 7.4, and 3 ml of 10% paraformaldehyde in 25 mM PPB, pH 7.4, were added, and the suspension was incubated for 30 min at 25°C on a shaker. The cells were collected by centrifugation and resuspended in a solution containing 3.15 ml of 0.1 M PPB, pH 6.5, and 1.85 ml of 10% paraformaldehyde in 25 mM PPB, pH 7.4. The cells were collected by centrifugation and resuspended in 3.15 ml of 0.1 M PPB, pH 6.5, and 1.85 ml of 10% paraformaldehyde in 25 mM PPB, pH 7.4, was added. The cells were collected by centrifugation and resuspended in 1.26 ml of 0.1 M PPB, pH 6.5, and 0.74 ml of 10% paraformaldehyde in 25 mM PPB, pH 7.4, was added. Afterwards the cells were incubated for 90 min at 25°C on a shaker. The cells were washed four times with 0.1 M PPB, pH 6.5, and twice with 1.2 M sorbitol in 0.1 M PPB, pH 5.9. Ten microliters of glucoronidase-arylsulfatase (Boehringer Mannheim) and 20 μl of zymolyase 20,000 (50 mg/ml) (Medac, Hamburg, Germany) were added per ml of cell suspension. The cell walls were digested for 90 min at 30°C. The cells were washed three times with 1.2 M sorbitol in 0.1 M PPB, pH 5.9. The cells were applied onto polylysine-coated slides, dried for 30 min at room temperature, fixed in −20°C cold methanol for 6 min, fixed at −20°C cold acetone for 30 min, and dried for 20 min. The affinity-purified polyclonal anti-H1 antibody (19) was applied at a concentration of 2.7 μg/ml in phosphate-buffered saline (PBS) with 4% dry milk powder and incubated overnight at 4°C. The secondary antibody [Cy2-conjugated goat anti-rabbit immunoglobulin G F(ab′)2 fragment (Jackson ImmunoResearch Laboratories)] was diluted 1:500 in PBS with 4% dry milk powder and incubated for 2 h at 37°C. The final embedding medium contained 0.5 μg of 4′,6′-diamidino-2-phenylindole (DAPI) per ml and 0.125 M n-propyl-galate in PBS.
Chromatin immunoprecipitation.
Yeast cells were grown in YPAD medium to mid-log phase. Formaldehyde was added to a final concentration of 1%. The cell suspension was incubated for 30 min at room temperature, and 3.4 ml of 2 M glycin was added to 47 ml of fixed cell suspension. After 5 min at room temperature, the cells were pelleted by centrifugation in a tabletop centrifuge for 5 min at 800 × g. The cells were washed with 10 ml of ice-cold TBS (150 mM NaCl, 20 mM Tris-HCl [pH 7.6]) and pelleted again. The cells were resuspended in 1 ml of ice-cold TBS, centrifuged at 16,000 × g for 1 min, frozen in liquid nitrogen, and stored at −80°C. The yeast cells were resuspended in 400 μl of ice-cold lysis buffer (0.1% deoxycholic acid, 1 mM EDTA, 50 mM HEPES-KOH [pH 7.5], 140 mM NaCl, 1% Triton X-100) with 1 mM phenylmethylsulfonyl fluoride and protease inhibitors (Complete Mini, Boehringer Mannheim) and broken with an equal volume of 0.5-mm cubic zirconium beads (BioSpec Products, Inc.) by using a vortex for 10 min at 4°C. The samples were incubated on ice for 15 min, and the supernatant was transferred to 1.5-ml tubes. The pellets were resuspended for 1 min on a vortex in 400 μl of ice-cold lysis buffer. The supernatants were pooled and sonicated at 4°C with nine 10-s pulses using a Sonopuls HD 70 device with a 60% duty cycle. These lysates were centrifuged twice at 16,000 × g for 15 min at 4°C. A 30-μl bed volume of protein A-agarose macrobeads (Sigma) was added to the supernatants. The extracts were incubated on a shaking table for 50 min at 4°C and centrifuged at 5,400 × g for 2 min at 4°C. Three micrograms of polyclonal anti-H1 antibody (19)/ml was added to the supernatant and incubated on ice for 3 h. A 30-μl bed volume of protein A-agarose macrobeads was added. After a 1-h incubation on a shaking table at 4°C, the samples were centrifuged for 2 min at 5,400 × g at 4°C and the supernatants were discarded. The beads were washed consecutively with 1 ml of lysis buffer, 1 ml of lysis buffer-500 (0.1% deoxycholic acid, 1 mM EDTA, 50 mM HEPES-KOH [pH 7.5], 500 mM NaCl, 1% Triton X-100), 1 ml of LiCl detergent solution (0.5% deoxycholic acid, 1 mM EDTA, 250 mM LiCL, 0.5% NP-50, 10 mM Tris-HCl [pH 8.0]), and 1 ml of TBS. Between washes the samples were incubated on a shaking table for 5 min at 4°C and subsequently centrifuged at 5,400 × g at 4°C. The immunoprecipitates were eluted with 100 μl of 1% sodium dodecyl sulfate in TE (1 mM EDTA, 10 mM Tris-HCl [pH 8.0]). The samples were incubated at 65°C for 10 min and centrifuged briefly. The elutes were transferred to 1.5-ml tubes, and the beads were washed with 150 μl of 0.7% sodium dodecyl sulfate in TE. All elutes were pooled and incubated for 6 h at 65°C. Two hundred fifty microliters of Proteinase K solution (1 μl of 20-μg/μl glycogen, 5 μl of 20-μg/μl Proteinase K, 245 μl of TE [pH 7.6]) was added, and the extracts were incubated for 1 h at 45°C. The samples were extracted with 55 μl of 4 M LiCl and 500 μl of 25:24:1 phenol-chloroform-isoamyl alcohol, ethanol precipitated, and resuspended in 10 μl of TE. PCR analysis was performed with 1 μl as a template using the Expand Template PCR system (Boehringer Mannheim) and the conditions suggested by the manufacturer. Primers MJ21IPY (5′-GCTACATATAAGGAACGTGCTGC-3′) and MJ22IPY (5′-CTGGCCGCATCTTCTCAAATA-3′) were used to amplify a 786-nucleotide-long specific portion of the subtelomeric URA3 gene. Additionally, the primers MJ25IPY (5′-ACAGAAGGAGCGAAGTCCTTA-3′) and MJ26IPY (5′-TGTCCTTCATGTGATCGAACA-3′) were used to amplify a 1,179-nucleotide-long portion of the GEA2 gene, which resides about 40 kb from the centromere on the left arm of chromosome V.
RNAi in C. elegans.
The following cDNA clones were used: yk352g2 encoding SIR-2.1, yk472g8 encoding MRT-2, and yk25g10 encoding C26E6.9, which is SET-2. The phenotypes of protein depletion were generated with RNA-mediated interference (14). Double-stranded RNA (dsRNA) synthesis from yk352g2 was done as described by Jedrusik and Schulze (19). T7 promoter-terminated DNA templates for MRT-2 (SET-2) were created by PCR amplification of the cDNAs with the primers ESMG67 (5′-CGCGCGTAATACGACTCACTATAGGGCGAATTGCCCTCACTAAAGGGA) and T7 (5′-GTAATACGACTCACTATAGGGC) (ESMG81 [5′-TAATACGACTCACTATAGGGTCGGTGGTTCTCCGTCCGTC]).
The DNA templates were mixed together and transcribed in a single reaction using T7 and T3 RNA polymerases (Megascript T7 kit; Ambion). RNA integrity was determined by gel electrophoresis; concentrations were determined photospectrometrically. Uncapped dsRNA with a concentration of 5 mg/ml was injected into hermaphrodite gonads of C. elegans strains EC107 and EC108. Phosphate buffer M9 (22 mM KH2PO4, 42 mM Na2HPO4, 86 mM NaCl, 1 mM MgS04) or an irrelevant dsRNA was injected in control experiments. The F1 progeny derived from 0 to 48 h after injection were raised at 25°C and scored for desilencing of let-858::gfp.
Trichostatin exposure of C. elegans.
L3 or L4 larva of strain EC107 were injected with 500 nM trichostatin in M9 phosphate buffer. The injected animals were incubated at 25°C and scored as adults. Control animals received an M9 phosphate buffer injection without trichostatin.
Microscopy.
Conventional and confocal light microscopy were performed with a Zeiss Axioplan 2 microscope equipped with a Zeiss confocal laser-scanning module LSM 510, Zeiss laser scanning software LSM 510 release 2.01, a Spot RT charge-coupled-device camera (Diagnostic Instruments, Sterling Heights, Mich.), Nomarski differential interference contrast, and epifluorescence optics. Green fluorescent images were acquired at an excitation wavelength of 488 nm with an emission filter bandpass of 505 to 550 nm; Hoechst and DAPI DNA staining were carried out with an excitation wavelength of 365 nm and an emission filter bandpass of 395 nm. Figures 1, 2, 5, and 7 are laser-scanning micrographs, whereas Fig. 6 was recorded in a conventional microscopic setup. DNA staining of living yeast cells was done by adding 10 μm Hoechst 33258 to a yeast cell suspension.
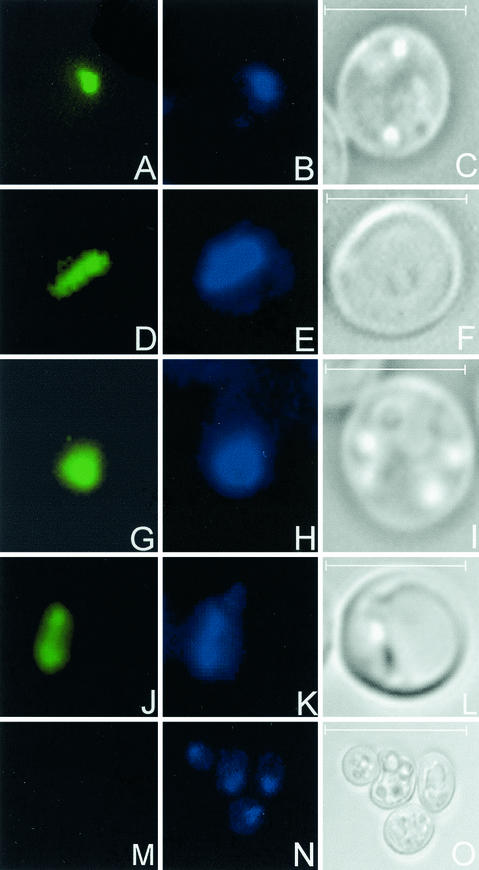
FIG. 1.
Indirect immunofluorescence detection of C. elegans histone H1 variants expressed in the yeast strain UCC3505. pYX142-H1.1 (A), pYX242-H1.1 (D), pYX142-H1.4 (G), pYX242-H1.4 (J), and pYX142 (M), the unmodified vector, used as a negative control, are shown. Panels B, E, H, K, and N show the corresponding DAPI DNA staining, and panels C, F, I, L, and O represent the corresponding Nomarski micrographs. Bar, 3 μm in panels A to L and 10 μm in panels M to O.
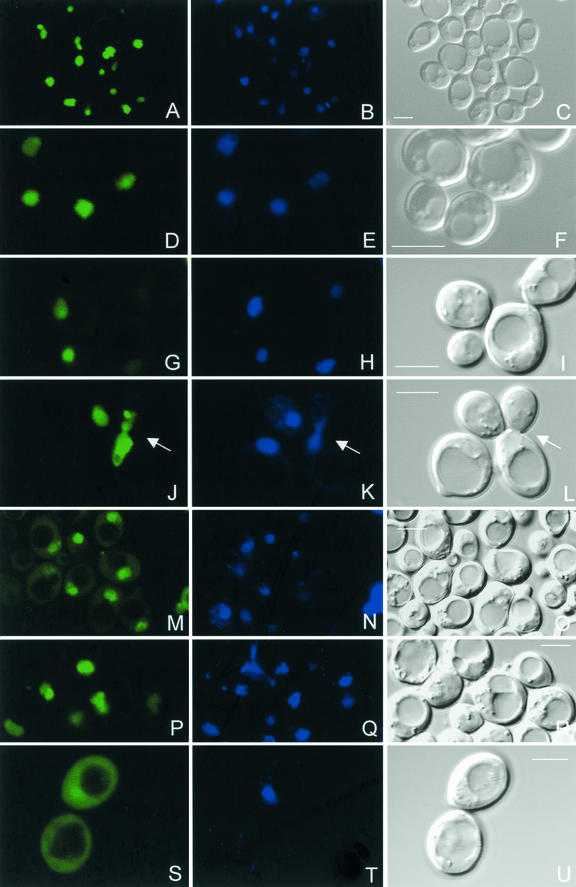
FIG. 2.
Detection of C. elegans H1 variant-GFP fusion proteins in living cells of the yeast strain UCC3505. pYX142-H1.1::GFP (A), pYX142-H1.2::GFP (D), pYX142-H1.3::GFP (G), pYX142-H1.4::GFP (J), pYX142-H1.5::GFP (M), pYX142-H1.Q::GFP (P), and pYX142-H1.X::GFP (S) are shown. The middle column shows the corresponding Hoechst vital DNA staining, and the third column represents the corresponding Nomarski micrographs. A small subfraction of H1.5::GFP and a larger fraction of H1.X::GFP localize to the cytoplasm. Panels J to L also show H1.4::GFP in a mitotic cell (arrows). The GFP fluorescence is localized to the condensed chromosomes. Bar, 5 μm.
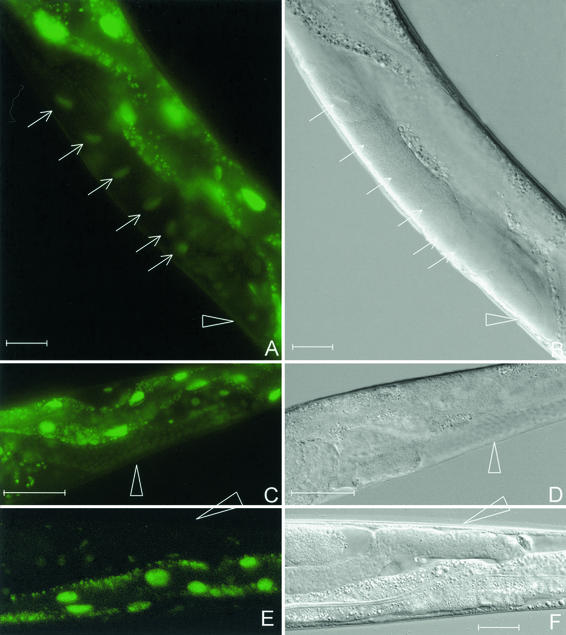
FIG. 5.
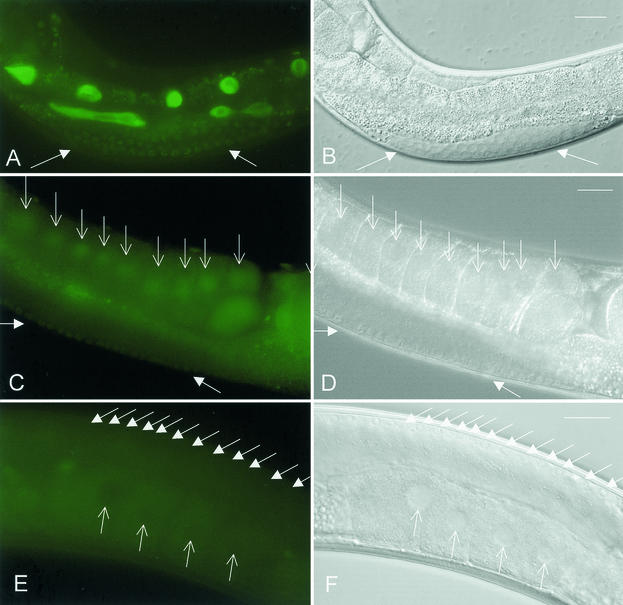
Desilencing of the repetitive transgene let-858::gfp by sir-2.1 RNAi in C. elegans. Depletion of SIR-2.1 induces fluorescence in oocyte nuclei (arrows) (A) and in undifferentiated germ nuclei (triangles in panels A to F). (E) LET-858::GFP fluorescence in a control animal. Here the silenced germ nuclei remain nonfluorescent. An irrelevant dsRNA was used in the control experiment. Panels B, D, and F represent corresponding Nomarski micrographs. Panels A, C, and E show gut autofluorescence in addition to the nuclear LET-858::GFP signal. Bar, 20 μm.
FIG. 7.
Injection of the histone deacetylase inhibitor trichostatin leads to an enhanced somatic expression of the repetitive let-858::gfp transgene but does not desilence the germ line. (A) LET-858::GFP fluorescence of a trichostatin-injected hermaphrodite (arrows). The left arrow points to an extruded gonad arm, which does not show any LET-858::GFP fluorescence. The other hermaphrodite (triangle) is a control animal injected with M9 phosphate buffer. The LET-858::GFP fluorescence of the somatic nuclei is much weaker than of the trichostatin-injected animal. (B) The corresponding Nomarski micrograph. Bar, 20 μm.
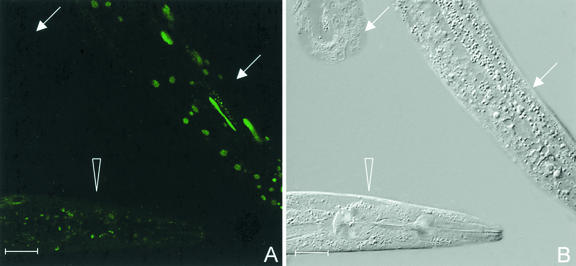
FIG. 6.
Desilencing of the repetitive let-858::gfp transgene by set-2 (A) and mrt-2 (C) RNAi in C. elegans. The experiment induces fluorescence in oocyte nuclei (open arrows in panel C) and in undifferentiated germ nuclei (solid arrows in panels A and C). (E) The silenced germ line of a control animal. At high exposure the autofluorescence of the cytoplasm is brighter than the nuclear fluorescence, so that syncytial germ nuclei (solid arrows) and oocyte nuclei (open arrows) appear as black circles on a green background. Panels B, D, and F represent the corresponding Nomarski micrographs. Panels A and C show gut autofluorescence in addition to the nuclear LET-858::GFP signal. Bar, 20 μm.
Flow cytometry.
Yeast strains were grown overnight in selective medium until they reached an optical density at 600 nm of 0.3. The cells were pelleted by centrifugation in a tabletop centrifuge for 1 min and resuspended in 1 ml of PBS. A total of 100,000 events were collected on a Cytomation MoFlo cell sorter. Parameters were set as follows: forward scatter, side scatter, 730 (LIN mode, amplification factor 6); FL1 (fluorescein isothiocyanate), 600 (LOG mode). The sample flow rate was adjusted to an event rate of approximately 30,000 s−1. Bipartite cell populations were separated at the numerical minimum between them in the histogram.
Computer software.
Database searches in GenBank were done with the BLAST program suite version 2.2.1, and database searches in the C. elegans databases were done with BLAST version 2.0a13MP (5). CLUSTALX (44) was used for the generation of a multiple alignment, and MEGA2 (27) was used to calculate and print the unweighted pair group method with arithmetic mean similarity tree. Flow-cytometric data were analyzed with Microsoft Excel. Final procession of micrographs was done with the spot camera software 3.1 and Adobe PhotoShop 5.5.
RESULTS
A single nematode H1 protein modifies the position effect variegation in yeast.
In the yeast strain UCC3505 (42), the URA3 gene is placed close to the left telomere of chromosome VII, which results in an epigenetically inheritable inactivation of the URA3 gene in about 50% of the cells by the repressively condensed chromatin of the telomere. The ratio of URA3 expression can be determined experimentally by comparing the plating efficiency on a standard medium with the plating efficiency on a medium containing the suicide substrate 5-fluoroorotic acid.
The ratio of URA3 expression was determined for 39 different yeast lines which either expressed one of the C. elegans linker histone proteins from the constitutive triose phosphate isomerase gene promoter or were transformed with the unmodified expression vectors and served as controls.
The results are given in Table 1. In the series using the single (or low-copy-number) vector pYX142, only two constructs considerably reduced the ratio of URA3-expressing cells. These are H1.1 (reduction from 48 to 16%) and H1.1::GFP (reduction from 48 to 19%). H1.4 expression reduced URA3 expression slightly (from 48 to 42%), as the C-terminal GFP fusion proteins of H1.2::GFP-H1.5::GFP and H1.Q::GFP did. No construct increased the ratio of URA3-expressing cells. In the series using the high-copy-number vector pYX242, only two constructs reduced the ratio of URA3-expressing cells. These are H1.1 (reduction from 31 to 11%) and H1.1::GFP (reduction from 31 to 19%). The expression of all other H1 proteins with the high-copy-number vector, including H1.4 and H1.4::GFP, slightly increased the ratio of URA3-expressing cells. All H1 plasmids expressing unfused linker histones were additionally tested in a control strain (S35P-5A), which contains URA3 at the wild-type position distant from telomeres. In this strain the plasmids did not significantly influence the expression of the URA3 gene. In S35P-5A derivatives expressing H1.1, 100% of the cells did express the URA3 gene. In a further control experiment, H1.1 and H1.4 were expressed in S. cerevisiae strain UCC7007-1, which is a derivative of UCC3505 with a disrupted sir3 gene. In this genetic background, which has no telomeric silencing, all cells expressed URA3. This indicates that the changes of the ratio of URA3-expressing cells caused by H1.1 in the TPEV strain UCC3505 are due to a specific influence of this linker histone protein to the telomeric chromatin structure and not due to a direct interaction of the linker histone protein with the URA3 promoter.
TABLE 1.
URA3 expression for S. cerevisiae cell linesa
| Protein expressed | % of URA3-expressing cells for strain:
|
|||||
|---|---|---|---|---|---|---|
| UCC3505(pYX142) (n) | UCC3505(pYX242) (n) | S35P-5A(pYX142) | S35P-5A(pYX242) | UCC7007-1(pYX142) | UCC7007-1(pYX242) | |
| Unmod. vector | 48.4 ± 1.1 (5) | 31.4 ± 3.4 (3) | 100 | 100 | 100 | 100 |
| H1.1 | 16.2 ± 2.0 (3) | 11.2 ± 4.1 (3) | 100 | 100 | 100 | 100 |
| H1.2 | 46.9 ± 4.3 (2) | 32.8 ± 3.0 (2) | 100 | 100 | ND | ND |
| H1.3 | 50.0 ± 0.0 (2) | 37.2 ± 0.4 (2) | 99 | 100 | ND | ND |
| H1.4 | 41.9 ± 1.5 (3) | 32.9 ± 2.0 (3) | 100 | 100 | 100 | 100 |
| H1.5 | 49.8 ± 1.2 (2) | 39.1 ± 0.0 (2) | 99.5 | 99 | ND | ND |
| H1.Q | 48.8 ± 0.1 (2) | 35.2 ± 2.0 (2) | 100 | 100 | ND | ND |
| H1.X | 51.0 ± 0.6 (2) | ND | 99.5 | ND | ND | ND |
| H1.1::GFP | 18.5 ± 1.9 (4) | 18.9 (1) | ND | ND | ND | ND |
| H1.2::GFP | 43.3 ± 1.7 (3) | ND | ND | ND | ND | ND |
| H1.3::GFP | 43.8 ± 1.1 (3) | ND | ND | ND | ND | ND |
| H1.4::GFP | 43.1 ± 0.9 (4) | 35.2 (1) | ND | ND | ND | ND |
| H1.5::GFP | 47.8 ± 1.3 (3) | ND | ND | ND | ND | ND |
| H1.Q::GFP | 40.2 ± 0.7 (3) | ND | ND | ND | ND | ND |
| H1.X::GFP | 52.4 ± 0.9 (3) | ND | ND | ND | ND | ND |
The ratio of URA3-expressing cells was determined on leucine-deficient media with or without the suicide substrate 5′-fluoroorotic acid. The numbers represent the percentages of URA3-expressing cells in yeast transformants of the TPEV strain UCC3505 (n, number of independent determinations), of the URA3 (wild-type) strain S35P-5A (single determination), and of the strain UCC7007-1 (single determination), which is a sir3 mutant in the genetic background of UCC3505. The transformants express either one of the seven C. elegans linker histones, the C-terminal GFP fusion H1.1::GFP (H1.4::GFP), or the unrelated polypeptide encoded by the polylinker of the vector (unmodified [unmod.] vector). A significant change in the ratio of URA3-expressing cells is caused only by H1.1 and H1.1::GFP. This change is specific for the strain UCC3505, in which URA3 expression is partially suppressed by the left telomere of chromosome VII. pYX142 is a low-copy-number vector, whereas pYX242 is a high-copy-number vector. Numbers indicating the maximal URA3 repression are set in boldface. ND, not determined.
Nematode linker histones are expressed and translocated into the nucleus and participate in interphase and mitotic chromatin in S. cerevisiae.
The expression of recombinant linker histones was characterized by indirect immunofluorescence of H1.1 and H1.4 expressed in the yeast strain UCC3505 by the single (or low)-copy-number vector as well as by the high-copy-number vector (Fig. 1). The immunofluorescence signal is strictly nuclear and colocalizes to the DAPI-stained DNA in all cells. The control staining done with the line transformed with the unmodified vector produced no immunofluorescence signal (Fig. 1M). Correspondingly, the fluorescence of H1.1::GFP to H1.5::GFP and H1.Q::GFP colocalized in living yeast cells with the DNA fluorescence obtained by vital Hoechst staining (Fig. 2). Mitotic cells displayed H1::GFP fluorescent mitotic chromosomes as shown in Fig. 2J. A small fraction of H1.5::GFP and a major fraction of H1.X::GFP localizes to the cytoplasm. In all cases the expression of nematode H1 variants or of H1::GFP fusion proteins did not detectably reduce the growth rate of S. cerevisiae.
The expression levels of nematode linker histone-GFP fusion proteins in S. cerevisiae do not correlate with telomeric position effect variegation.
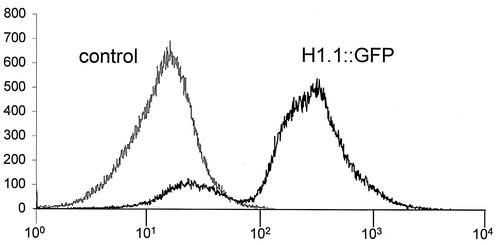
The polyclonal anti-H1 antibody reacts preferentially with H1.4 and cannot be used for quantitative comparisons. Therefore, the expression of H1::GFP fusion proteins was quantified with flow cytometry (Table 2). Histogram analysis revealed a bipartite cell population for all fusion proteins (Fig. 3 and Table 2). Although the cells were grown in selective medium, a fraction of cells, typically ranging from 14 to 17.7%, does not show green fluorescence. Only for H1.2::GFP, the ratio of nonfluorescing cells is considerably larger (53%). These observations are characteristic for each plasmid and were reproduced in multiple independent experiments. When diluted cell suspensions were plated on YPD medium, the nonfluorescent cells grew to nonfluorescent colonies, which had lost the plasmid. No nonfluorescent colonies were observed on selective SC-leu medium. These results indicate that the nonfluorescent cells originated from unusually high rates of plasmid losses. As the exact fraction of nonfluorescent cells was construct specific and reproduced in many independent transformation experiments, the plasmid loss is most likely a consequence of linker histone expression. The mean relative fluorescence of all cells is highest for H1.4::GFP (20.6) and lowest for H1.2::GFP (11.4). Three constructs produce very similar mean fluorescence intensities: H1.1, H1.4, and H1.Q. The mean fluorescence intensities of the fluorescing population (instead of the total population) are similar for H1.1, H1.2, H1.3, H1.4, and H1.Q. We conclude that the level of H1::GFP fusion protein expression is very similar for all proteins besides H1.5 and H1.X, which show a reduced level of fluorescence.
TABLE 2.
Fluorescence intensities of H1:GFP-expressing S. cerevisiae cellsa
| Protein expressed | % of fluorescing cells | Relative avg fluorescence (A) | Relative avg fluorescence (B) |
|---|---|---|---|
| Unmod. vector | 0.0 | 1.0 | 1.0 |
| H1.1::GFP | 83.2 | 19.8 | 23.5 |
| H1.2::GFP | 47.2 | 11.4 | 22.8 |
| H1.3::GFP | 85.2 | 16.8 | 18.8 |
| H1.4::GFP | 82.6 | 20.6 | 24.5 |
| H1.5::GFP | 82.3 | 12.6 | 14.1 |
| H1.Q::GFP | 85.9 | 18.9 | 21.0 |
| H1.X::GFP | 86.0 | 12.4 | 14.2 |
The fluorescence intensity of H1::GFP-expressing yeast cells was determined with flow cytometry. The corresponding histogram of control cells and the bipartite histogram of H1.1::GFP-expressing cells are shown in Fig. 3. All yeast lines producing H1::GFP fusion proteins contained a fraction of nonfluorescing cells. The first column of the table gives the percentage of cells in the second peak, which produce GFP fluorescence. The second column (A) presents the average fluorescence of the total number of cells relative to the nonfluorescing control sample. The third column (B) gives the average fluorescence intensity of the cells effectively expressing GFP fluorescence, which are found in the right peak of the histogram. Unmod., unmodified.
FIG. 3.
Flow-cytometric analysis of H1.1::GFP fluorescence in S. cerevisiae UCC3505 cells (black line). The vector is pYX142. Nonfluorescing control cells (UCC3505 transfected with the unmodified vector) are represented by the gray line. Each curve is a histogram of 105 cells, and cell numbers (y axis) are plotted with respect to green fluorescence intensity (x axis). A small fraction of the H1.1::GFP-transfected cell population does not fluoresce. The corresponding histograms of the remaining six H1::GFP-expressing lines in vector pYX142 are comparable. A quantitative analysis of all flow-cytometric measurements is presented in Table 2.
Nematode linker histones bind to subtelomeric DNA in S. cerevisiae.
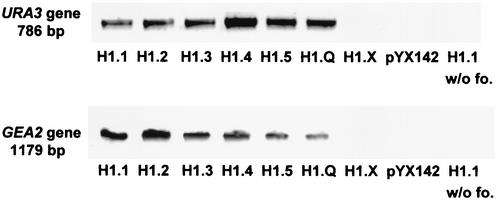
The linker histone-expressing derivatives of yeast strain UCC3505 were analyzed with chromatin immunoprecipitation after formaldehyde cross-linking. We used the polyclonal anti-C. elegans H1 antibody to investigate if and which C. elegans linker histones are associated with subtelomeric DNA. A PCR analysis of the precipitated chromatin (Fig. 4) shows that all C. elegans linker histones (beside H1.X) are associated with the subtelomeric URA3 gene of UCC3505, as well as with the GEA2 gene, which is not subtelomeric. The control experiments (no H1 expressed, no formaldehyde added) and H1.X produced no PCR band, indicating the specificity of the analysis. The negative result for H1.X is not informative, since the polyclonal anti-H1 antibody does not bind to it.
FIG. 4.
Chromatin immunoprecipitation after formaldehyde cross-linking. A polyclonal anti-C. elegans H1 antibody was used to precipitate chromatin from yeast strains (UCC3505) transfected with expression constructs of all C. elegans linker histones. The precipitate was analyzed by PCR amplification of the subtelomeric URA3 gene and of the GEA2 gene, which resides about 40 kb distant from the centromere on the left arm of chromosome V. In control cells (UCC3505 transfected with pYX142), in experiments without formaldehyde cross-linking (lanes indicated as w/o fo.), and in H1.X-expressing cells, no URA3 and GEA2 bands are produced. The latter is an expected result, because the anti-C. elegans H1 antibody does not react with H1.X.
The C. elegans proteins SIR-2.1, SET-2, and MRT-2 are involved in the silencing of repetitive transgenes in C. elegans.
Since only the C. elegans linker histone variant H1.1, but no other linker histone variant of C. elegans, enhanced the telomeric position effect in S. cerevisiae, it seems possible that mechanistically related molecular functions govern the chromatin silencing in both organisms. To elucidate this aspect, we searched for S. cerevisiae proteins which are essential for telomeric chromatin silencing in yeast and which have identifiable homologs in C. elegans. The yeast proteins Sir2p, Set1p (33), and Rad17p (30) fulfill these criteria. The corresponding C. elegans genes are sir-2.1 (45), set-2 (46), and mrt-2 (1). As a reporter for germ line silencing, we used the approach introduced by Kelly and Fire (21). A GFP-tagged version of a ubiquitously expressed C. elegans gene (let-858) is established as a repetitive extrachromosomal transgenic array in a genetic background [pha-1(e2123)], which allows positive selection for the transgene at 25°C. The endogenous let-858 promoter is active in the germ line of C. elegans. However, when the let-858::GFP reporter is established as a replicating transgene after a small number of animal generations, the expression in the germ line is lost due to chromatin silencing acting on the repetitive structure, and as a consequence the green fluorescence is lost in germ cells. This is used to monitor the status of the silencing system. dsRNA was injected into the gonads of young hermaphrodite animals carrying the array, and LET-858::GFP fluorescence was scored for adult F1 progeny. Seven hermaphrodites of the let-858::gfp reporter strain EC107 and eight hermaphrodites of the let-858::gfp reporter strain EC108 were injected with sir-2.1 dsRNA. Forty-one out of a total of 104 F1 hermaphrodites (39%) and all of a total of 13 males (100%) showed LET-858::GFP fluorescence in oocyte and sperm nuclei as well as in undifferentiated germ nuclei (Fig. 5). Depletion of SET-2 led to desilencing in 9 out of 32 (28%) hermaphrodites and 12 out of 14 males (86%). LET-858::GFP fluorescence was observed in oocyte and sperm nuclei as well as in undifferentiated germ nuclei (Fig. 6A). Depletion of MRT-2 led to desilencing in 6 out of 17 (35%) observed hermaphrodites and 6 out of 8 (86%) males (Fig. 6B). LET-858::GFP fluorescence was observed in oocyte and sperm nuclei as well as in undifferentiated germ nuclei (Fig. 6C). In all three depletion experiments, fertility and germ line cytology of all hermaphrodites and males were normal. The control experiment was done with the strain EC107. In 215 F1 control hermaphrodites, no desilencing was observed (Fig. 6E) when M9 phosphate buffer was injected. In a further control experiment, an irrelevant dsRNA was injected into five F0 hermaphrodite animals. Of 247 scored F1 hermaphrodites, no animal showed desilencing of the germ line (Fig. 5E). This shows that activation of the RNAi mechanism alone does not lead to desilencing of the germ line.
Trichostatin does not influence the chromatin silencing in the germ line of C. elegans but increases the somatic expression of let-858::gfp.
The histone deacetylase inhibitor trichostatin was injected into 11 L3 or L4 larva of the indicator strain EC107. The animals were raised at 25°C, and gonad expression of the let-858::gfp transgene was scored in comparison to that for M9 phosphate buffer-injected control animals. The let-858::gfp transgene was increasingly expressed in the somatic cells of the trichostatin-injected animals but not in the controls. The oocyte and sperm nuclei, as well as the undifferentiated meiotic and mitotic germ nuclei, did not show any fluorescence in the trichostatin-treated animals or in the control animals (Fig. 7). This indicates that the germ line chromatin silencing of C. elegans is not dependent on trichostatin-sensitive histone deacetylases.
DISCUSSION
For C. elegans, the maintenance of the germ line depends on a specific epigenetic system. This is represented by the four mes genes mes-2, mes-3, mes-4, and mes-6. Two of these genes (mes-2 and mes-6) are the C. elegans homologs of Pc-G genes (18, 26) which were described for Drosophila previously. The exact molecular mechanism of mes gene-mediated gene repression is not known, but it has recently been shown that differential histone modifications of autosomes and the sex chromosome are most likely part of the underlying mechanism, which is supposed to inactivate a part or all of the X chromosome (15, 22, 38). Most phenotypic aspects of the mes genes are copied with H1.1 RNAi, suggesting that H1.1 participates in this process. Position effect variegation has served as a productive tool to identify and analyze chromatin-modifying genes in Drosophila (39) and in yeast (37). We show here that H1.1 expression, but not expression of any of the other C. elegans linker histones, enhances the telomeric position effect in yeast, whereas the endogenous histone H1 of budding yeast does not influence telomeric silencing (13, 35).
Nuclear translocation and chromatin binding of recombinant linker histones in yeast is not self-evident. Human linker histone, which was abundantly expressed with the GAL10 promoter in budding yeast (3), did not enter the nuclear compartment and formed aggregates in the cytoplasm instead (W. Albig, personal communication). Expression of sea urchin linker histone in yeast caused cell death or greatly reduced the survival rate (29, 31). In a control strain for which the URA3 gene is located on its wild-type position distant from telomeres, H1.1 or H1.1::GFP expression did not affect URA3 expression. In a sir3 mutant of the TPEV strain UCC3505 with abolished telomeric silencing, H1.1 expression did not affect URA3 expression. These observations indicate that the reduction of URA3 expression in the TPEV strain UCC3505 is dependent on the telomeric chromatin and does not result from a specific interaction with the URA3 promoter.
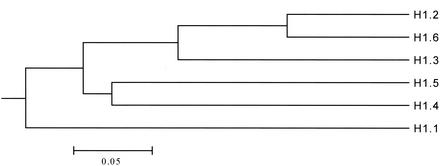
Although the canonical tripartite C. elegans linker histones (H1.1 to H1.6) are similar to each other, a systematic sequence comparison shows that H1.1 is the most divergent isoform in this gene family (Fig. 8). It contains 35% more alanine residues than the remaining canonical linker histones of C. elegans (28% Ala in H1.1 contrast to 20.8% ± 1.2% in H1.2 to H1.6). In H1.1 the highest local concentration of alanine is found in a repeated oligopeptide (AAAKK, four times) in the first half of the C-terminal domain. This oligopeptide occurs only once in H1.3, H1.4, and H1.Q. It is completely absent from H1.2, H5, H1.6, and H1.X. More details are shown in Table 3. Moreover, H1.1 is also the only linker histone without an identifiable sister gene, to which it would show a stronger relation than to the other linker histones. Our results suggest that variant specific structural features of H1.1 could dedicate this protein to an epigenetic function, which enables it to act as an element of structure-specific gene repression for C. elegans as well as for yeast. Consequently, this would imply that the molecular mechanisms of germ line silencing in C. elegans and of telomeric repression in S. cerevisiae should be functionally related. Therefore, we tested whether telomeric silencing in yeast can be used as a model for germ line silencing in C. elegans. A number of chromatin proteins have been identified as modifiers of TPEV in budding yeast, most of which interestingly are also involved in the suppression of the silent mating type loci (6). Three of these proteins (Sir2p, Rad17p, and Set1p) have homologs in higher eukaryotes. The depletion of the homologous C. elegans proteins SIR-2.1, MRT-2, and SET-2 caused desilencing of the let-858::gfp transgene in the germ line of C. elegans. The relevance of this finding is emphasized by the recent finding that SET-2 enhances the cytological phenotype of mes-3 and mes-4 in C. elegans (46). This proves that not only is SET-2 involved in the silencing of the germ line as we show here, it is additionally involved in germ line development for C. elegans. MRT-2 is required for telomerase function and consequently for long-term survival of the germ line for C. elegans (1).
FIG. 8.
Similarity tree calculated from a protein alignment of C. elegans linker histones which have the canonical three-domain structure. H1.X is not included, because it is not a chromatin protein in C. elegans (20).
TABLE 3.
Oligopeptide composition of the C. elegans linker histonesa
| Peptide | No. of occurrences in:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| H1.1 | H1.2 | H1.3 | H1.4 | H1.5 | H1.6 | H1.Q | H1.X | |
| AAAKKP | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| PAAAKK | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| PAAAKKP | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| AAAKK | 4 | 0 | 1 | 1 | 0 | 0 | 1 | 0 |
| AAKK | 6 | 1 | 1 | 1 | 0 | 1 | 1 | 0 |
H1.1 contains 35% more alanine residues than the remaining canonical linker histones of C. elegans. The additional alanine residues are located in the first half of the C-terminal domain, where they contribute to a repeated oligopeptide structure. The numbers of occurrences of different possible overlapping versions of this oligopeptide are presented. All indicated peptides are repeated in H1.1, but they are unique or absent in the remaining proteins.
We noted, however, that in contrast to our previous extensive experiments with H1.1 depletion, the penetrance of the desilenced phenotype as well as the intensity of the LET-858::GFP fluorescence was considerably lower and no cytological damage was observed. This could indicate that these proteins serve less-central functions in the chromatin silencing or are partially redundant. A redundant function for SIR-2.1 would be understandable, since three further SIR2 homologs exist in the C. elegans genome (45). For C. elegans the repression of transgenes is typically far more sensitive to the depletion of silencing proteins than germ line development (19). Interestingly, in yeast the silencing genes SIR3, SIR4, and RAD17 are also required for telomere maintenance (34). SIR2 is the only member of the yeast SIR gene class which has homologs in distantly related organisms, ranging from bacteria to humans. Our data show now for the first time the involvement of a Sir2 protein in chromatin silencing in a multicellular eukaryote.
Sir2p is a NAD-dependent histone deacetylase which cannot be inhibited by the drug trichostatin. Injections of trichostatin into larval-stage C. elegans enhanced the somatic transgene expression of the LET-858::GFP reporter but did not modify the silenced status of this transgenic array in the germ line. We conclude that if histone deacetylation is involved in germ line silencing, this task is completely accomplished by trichostatin-insensitive histone deacetylases, such as, for example, SIR-2.1. Histone deacetylation by Sir2p is required for silencing and efficient spreading of the SIR complex along the chromatin fiber adjacent to the telomeres of S. cerevisiae. Only one of the Sir proteins is a limiting component of telomeric silencing in budding yeast: overexpression of Sir2p has no effect on telomeric silencing and overexpression of Sir3p enhances the telomeric position effect (37), whereas overexpression of Sir4p or Sir1p disrupts telomeric silencing (11). Sir3p is thought to link the Sir2p- and Sir4p-containing silencing complex to the local chromatin by binding to Rap1p, which is associated with the telomeric DNA, and to the core histone proteins adjacent to it (43). Since the expression of the C. elegans linker histone H1.1 specifically enhances telomeric silencing and since H1.1 is an abundant DNA binding protein, we suggest that this protein may function by recruiting the Sir2p-containing silencing complex or other histone-modifying factors to the local DNA.
Our work identifies a new and unexpected linkage between the epigenetics of the C. elegans germ line and of telomeric gene repression in S. cerevisiae. Yeast cells have one of two possible mating types, a or α. In budding yeast most genes involved in telomeric silencing are also essential for the suppression of the silent mating type locus (6). The mating type specification is a binary process of cell differentiation. In C. elegans sex determination is chromosomal, and sexual development is controlled by a complex genetic network (28). The distinction between germ line and soma, which is also a binary distinction, is at least in part controlled by an evolutionary old system related to the chromatin silencing machinery of single-celled eukaryotic organisms. This evolutionarily older system now provides the basis for the specific analysis of linker histone variant functions of C. elegans. We propose that the complex linker histone gene families of higher eukaryotes contribute to specific aspects of multicellular life, such as cell differentiation and development. This functional specialization is demonstrated here by linker histone variant-specific enhancement of the telomeric position effect variegation in budding yeast. This will help to extend the current progress in the understanding of the generality of epigenetic mechanisms (32) to the linker histones.
Acknowledgments
We thank Ulrich Grossbach for his support of the C. elegans group, Sabine Steinhoff and Bettina Schulze for the establishment of the basic yeast protocols, Werner Albig for the yeast immunofluorescence protocol, Hans Dieter Schmitt for the yeast strain S35P-5A, Daniel E. Gottschling for the yeast strains UCC3505 and UCC7007-1, Yuji Kohara, Mishima, for the yk cDNA clones, Harald Kolmar for performing the flow cytometry, Bettina Schulze for critically reading the manuscript, and the students of our summer 1998 and 2000 advanced level practical course in developmental biology for their contributions to this project. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources. We acknowledge the excellent technical assistance of Sabine Pitzel and Barbara Lage.
This work was supported by the Deutsche Forschungsgemeinschaft grant GRK 242-2 (Graduiertenkolleg “Molekulare Genetik der Entwicklung”) to U. Grossbach, grant SCHU 1033/3-3 to E. Schulze and U. Grossbach, and grant SFB271 A14 to E. Schulze.
REFERENCES
- 1.Ahmed, S., and J. Hodgkin. 2000. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature 13:159-164. [DOI] [PubMed] [Google Scholar]
- 2.Alber, T., and G. Kawasaki. 1982. Nucleotide sequence of the triose phosphate isomerase gene of Saccharomyces cerevisiae. J. Mol. Appl. Genet. 1:419-434. [PubMed] [Google Scholar]
- 3.Albig, W., D. M. Runge, M. Kratzmeier, and D. Doenecke. 1998. Heterologous expression of human H1 histones in yeast. FEBS Lett. 435:245-250. [DOI] [PubMed] [Google Scholar]
- 4.Albig, W., T. Meergans, and D. Doenecke. 1997. Characterization of the H1.5 gene completes the set of human H1 subtype genes. Gene 184:141-148. [DOI] [PubMed] [Google Scholar]
- 5.Altschul, S. F., D. Gish, W. Mille, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 6.Aparicio, O. M., B. L. Billington, and D. E. Gottschling. 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 20:1279-1287. [DOI] [PubMed] [Google Scholar]
- 7.Ausubel, F. M., R. B. Brent, R. E. Kingston, R. E. Moore, J. G. Seidmann, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology, 2nd ed., suppl. 12. John Wiley & Sons, New York, N.Y.
- 8.Barra, J. L., L. Rhounim, J. L. Rossignol, and G. Faugeron. 2000. Histone H1 is dispensable for methylation-associated gene silencing in Ascobolus immersus and essential for long life span. Mol. Cell. Biol. 20:61-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 10.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chien, C. T., S. Buck, R. Sternglanz, and D. Shore. 1993. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell 75:531-541. [DOI] [PubMed] [Google Scholar]
- 12.Drabent, B., K. Franke, C. Bode, U. Kosciessa, U. H. Bouterfa, H. Hameister, and D. Doenecke. 1995. Isolation of two murine H1 histone genes and chromosomal mapping of the H1 gene complement. Mamm. Genome 6:505-511. [DOI] [PubMed] [Google Scholar]
- 13.Escher, D., and W. Schaffner. 1997. Gene activation at a distance and telomeric silencing are not affected by yeast histone H1. Mol. Gen. Genet. 256:456-461. [DOI] [PubMed] [Google Scholar]
- 14.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 15.Fong, Y., L. Bender, W. Wang, and S. Strome. 2002. Regulation of the different chromatin states of autosomes and X chromosomes in the germ line of C. elegans. Science 296:2235-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottschling, D. E., O. M. Aparicio, B. L. Billington, and V. A. Zakian. 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 16:751-762. [DOI] [PubMed] [Google Scholar]
- 17.Han, M., M. Chang, U. J. Kim, and M. Grunstein. 1987. Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell 48:589-597. [DOI] [PubMed] [Google Scholar]
- 18.Holdeman, R., S. Nehrt, and S. Strome. 1998. MES-2, a maternal protein essential for viability of the germline in Caenorhabditis elegans, is homologous to a Drosophila Polycomb group protein. Development 125:2457-2467. [DOI] [PubMed] [Google Scholar]
- 19.Jedrusik, M. A., and E. Schulze. 2001. A single histone H1 isoform (H1.1) is essential for chromatin silencing and germline development in Caenorhabditis elegans. Development 128:1069-1080. [DOI] [PubMed] [Google Scholar]
- 20.Jedrusik, M. A., S. Vogt, P. Claus, and E. Schulze. 2002. A novel linker histone-like protein is associated with cytoplasmic filaments in Caenorhabditis elegans. J. Cell Sci. 15:2881-2891. [DOI] [PubMed] [Google Scholar]
- 21.Kelly, W. G., and A. Fire. 1998. Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans. Development 125:2451-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly, W. G., C. E. Schaner, A. F. Dernburg, M. H. Lee, S. K. Kim, A. M. Villeneuve, and V. Reinke. 2002. X-chromosome silencing in the germline of C. elegans. Development 129:479-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khochbin, S. 2001. Histone H1 diversity: bridging regulatory signals to linker histone function. Gene 271:1-12. [DOI] [PubMed] [Google Scholar]
- 24.Kilmartin, J. V., and A. E. Adams. 1984. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J. Cell Biol. 98:922-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, U. J., M. Han, P. Kayne, and M. Grunstein. 1988. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J. 7:2211-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korf, I., Y. Fan, and S. Strome. 1998. The Polycomb group in Caenorhabditis elegans and maternal control of germline development. Development 125:2469-2478. [DOI] [PubMed] [Google Scholar]
- 27.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 28.Kuwabara, P. E., and M. D. Perry. 2001. It ain't over till it's ova: germline sex determination in C. elegans. Bioessays 23:596-604. [DOI] [PubMed] [Google Scholar]
- 29.Linder, C., and F. Thoma. 1994. Histone H1 expressed in Saccharomyces cerevisiae binds to chromatin and affects survival, growth, transcription, and plasmid stability but does not change nucleosomal spacing. Mol. Cell. Biol. 14:2822-2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Longhese, M. P., V. Paciotti, H. Neecke, and G. Lucchini. 2000. Checkpoint proteins influence telomeric silencing and length maintenance in budding yeast. Genetics 155:1577-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miloshev, G., P. Venkov, K. van Holde, and J. Zlatanova. 1994. Low levels of exogenous histone H1 in yeast cause cell death. Proc. Natl. Acad. Sci. USA 91:11567-11570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moazed, D. 2001. Common themes in mechanisms of gene silencing. Mol. Cell 8:489-498. [DOI] [PubMed] [Google Scholar]
- 33.Nislow, C., E. Ray, and L. Pillus. 1997. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol. Biol. Cell 8:2421-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palladino, F., T. Laroche, E. Gilson, A. Axelrod, L. Pillus, and S. M. Gasser. 1993. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell 75:543-555. [DOI] [PubMed] [Google Scholar]
- 35.Patterton, H. G., C. C. Landel, D. Landsman, C. L. Peterson, and R. T. Simpson. 1998. The biochemical and phenotypic characterization of Hho1p, the putative linker histone H1 of Saccharomyces cerevisiae. J. Biol. Chem. 273:7268-7276. [DOI] [PubMed] [Google Scholar]
- 36.Ramon, A., M. I. Muro-Pastor, C. Scazzocchio, and R. Gonzalez. 2000. Deletion of the unique gene encoding a typical histone H1 has no apparent phenotype in Aspergillus nidulans. Mol. Microbiol. 35:223-233. [DOI] [PubMed] [Google Scholar]
- 37.Renauld, H., O. M. Aparicio, P. D. Zierath, B. L. Billington, S. K. Chhablani, and D. E. Gottschling. 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7:1133-1145. [DOI] [PubMed] [Google Scholar]
- 38.Reuben, M., and R. Lin. 2002. Germline X chromosomes exhibit contrasting patterns of histone H3 methylation in Caenorhabditis elegans. Dev. Biol. 245:71-82. [DOI] [PubMed] [Google Scholar]
- 39.Reuter, G., and P. Spierer. 1992. Position effect variegation and chromatin proteins. Bioessays 14:605-612. [DOI] [PubMed] [Google Scholar]
- 40.Shen, X., L. Yu, J. W. Weir, and M. A. Gorovsky. 1995. Linker histones are not essential and affect chromatin condensation in vivo. Cell 82:47-56. [DOI] [PubMed] [Google Scholar]
- 41.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 42.Singer, M. S., and D. E. Gottschling. 1994. TLC1 template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404-409. [DOI] [PubMed] [Google Scholar]
- 43.Stone, E. M., and L. Pillus. 1998. Silent chromatin in yeast: an orchestrated medley featuring Sir3p. Bioessays 20:30-40. [DOI] [PubMed] [Google Scholar]
- 44.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tissenbaum, H. A., and L. Guarente. 2001. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410:227-230. [DOI] [PubMed] [Google Scholar]
- 46.Xu, L., and S. Strome. 2001. Depletion of a novel SET-domain protein enhances the sterility of mes-3 and mes-4 mutants of C. elegans. Genetics 159:1019-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamamoto, T., and M. Horikoshi. 1996. Cloning of the cDNA encoding a novel subtype of histone H1. Gene 173:281-285. [DOI] [PubMed] [Google Scholar]