Abstract
In individuals with human immunodeficiency virus type 1 (HIV-1) infection, a small reservoir of resting memory CD4+ T lymphocytes carrying latent, integrated provirus persists even in patients treated for prolonged periods with highly active antiretroviral therapy (HAART). This reservoir greatly complicates the prospects for eradicating HIV-1 infection with antiretroviral drugs. Therefore, it is critical to understand how HIV-1 latency is established and maintained. In particular, it is important to determine whether transcriptional or posttranscriptional mechanisms are involved. Therefore, HIV-1 DNA and mRNAs were measured in highly purified populations of resting CD4+ T lymphocytes from the peripheral blood of patients on long-term HAART. In such patients, the predominant form of persistent HIV-1 is latent integrated provirus. Typically, 100 HIV-1 DNA molecules were detected per 106 resting CD4+ T cells. Only very low levels of unspliced HIV-1 RNA (∼50 copies/106 resting CD4+ T cells) were detected using a reverse transcriptase PCR assay capable of detecting a single molecule of RNA standard. Levels of multiply spliced HIV-1 RNA were below the limit of detection (<50 copies/106 cells). Only 1% of the HIV-1 DNA-positive lymphocytes in this compartment could be induced to up-regulate HIV-1 mRNAs after cellular activation, indicating that most of the proviral DNA in resting CD4+ T cells either carries intrinsic defects precluding transcription or is subjected to transcriptional control mechanisms that preclude high-level production of multiply spliced mRNAs. Nevertheless, by inducing T-cell activation, it is possible to isolate replication-competent virus from resting CD4+ T lymphocytes of all infected individuals, including those on prolonged HAART. Thus, a subset of integrated proviruses (1%) remains competent for high-level mRNA production after cellular activation, and a subset of these can produce infectious virus. Measurements of steady-state levels of multiply spliced and unspliced HIV-1 RNA prior to cellular activation suggest that infected resting CD4+ T lymphocytes in blood synthesize very little viral RNA and are unlikely to be capable of producing virus. In these cells, latency appears to reflect regulation at the level of mRNA production rather than at the level of splicing or nuclear export of viral mRNAs.
In human immunodeficiency virus type 1 (HIV-1) infection, resting CD4+ T lymphocytes harboring replication-competent virus persist even in patients on highly active antiretroviral therapy (HAART) who exhibited prolonged suppression of viremia to below the limit of detection (8-10, 17, 18, 37, 43, 52, 56). Because the rescue of replication-competent virus from these resting cells has generally required the use of stimuli that induce global T-cell activation, these cells are thought to be in a state of latent infection. In most treated patients, the frequency of these latently infected cells does not decrease significantly despite several years of otherwise effective treatment (17, 43) or declines only very slowly (43, 56). This finding, together with the potential presence of other reservoirs (reviewed in reference 5), has dampened the initial hopes that the infection might be eradicated by prolonged antiretroviral therapy and has increased interest in the molecular mechanisms of HIV-1 persistence.
Two forms of HIV-1 latency can be found in resting CD4+ T cells, a labile preintegration form and a stable postintegration form (4, 7, 9, 10, 37, 47, 54). The labile form decays within the first 3 months after the initiation of therapy, leaving the stable form (4, 37). The molecular mechanisms that maintain postintegration latency are unclear. Some hypotheses implicate regulation at the level of transcription. For example, the HIV-1 promoter could potentially be silenced by the absence of activation-dependent host transcription factors (6, 13, 20, 36, 48), by epigenetic mechanisms affecting the promoter itself (3) or local chromatin structure (29, 51), or by the lack of Tat and/or Tat-associated transcriptional elongation factors (1, 22, 24, 25, 28, 31). Other hypotheses focus on regulation at the posttranscriptional level. For example, the absence of virus production from latently infected cells might reflect the fact that at low levels of transcription, the full-length viral RNA that encodes structural and enzymatic functions and that serves as the viral genome may not be efficiently exported from the nucleus in the absence of sufficient amounts of the HIV-1 Rev protein (16, 33-35, 40, 41, 45). In some studies, latent infection has been characterized by a deficiency in this unspliced viral RNA and a preponderance of multiply spliced HIV-1 mRNAs encoding regulatory proteins (41). Some studies even suggest that lymphocytes with a resting phenotype, particularly those within lymphoid tissues, may be capable of producing regulatory proteins, structural proteins, or even virus particles (14, 53, 57).
Determining the degree of viral mRNA production in latently infected resting CD4+ T cells has important implications for therapy. The lower the level of HIV-1 gene expression, the more difficult it will be to selectively target the infected cells. Numerous studies have measured HIV-1 gene expression in cells from patients on HAART (2, 21, 23, 26, 32, 55, 56, 58), but in most of these studies, resting cells were not separated from activated cells, making it difficult to assess the level of HIV-1 gene expression in the infected resting cells that comprise the latent reservoir for HIV-1. Therefore, in an effort to determine the degree of viral transcription that occurs in latently infected cells, we examined HIV-1 gene expression in highly purified populations of resting peripheral blood CD4+ T cells, the cell population in which long-term viral persistence has been most clearly demonstrated.
MATERIALS AND METHODS
Purification of resting CD4+ T cells from patients on HAART.
Peripheral blood samples were obtained from patients who achieved and maintained suppression of viral replication to <50 copies/ml with antiretroviral drugs (all patients gave informed consent before phlebotomy). Peripheral blood mononuclear cells (PBMCs) were negatively selected to remove CD8+ T cells, B cells, monocytes, NK cells, and activated CD4+ T cells as previously described (9, 18). Resting CD4+ T cells were further purified by sorting for small lymphocytes with high CD4 and low HLA-DR surface expression (9, 18).
HIV-1 DNA measurements in resting CD4+ T lymphocytes.
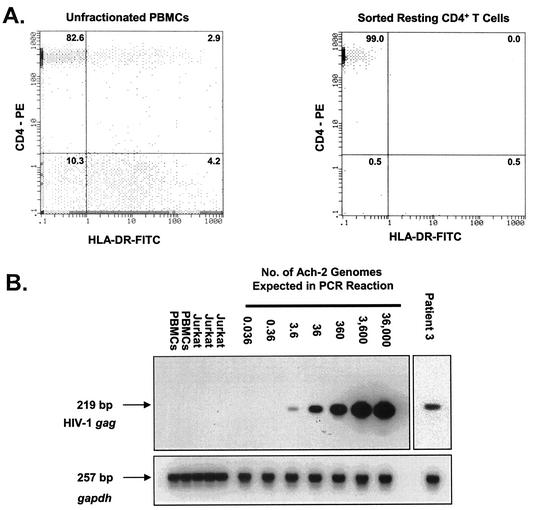
DNA was isolated from 106 purified resting CD4+ T cells using the QIAamp DNA Blood Midi kit (Qiagen) according to the manufacturer's protocol. For quantitative analysis of HIV-1 DNA, a standard curve was constructed using ACH-2 cells, which contain a single integrated copy of the HIV-1 genome (20). Known numbers of ACH-2 cells were diluted with 106 PBMCs from an uninfected donor. For both control and patient samples, 106 uninfected Jurkat cells were added to facilitate DNA isolation. In parallel to the DNA isolation procedure for patient samples and the standard curve, DNA was also isolated from multiple samples of 2 × 106 PBMCs and multiple samples of 2 × 106 Jurkat cells. DNA concentrations from these samples were used to calculate the amount of input DNA from these cellular populations in the preparation of the standard curve. In addition, the DNA concentration from Jurkat cells was used to assess what fraction of DNA in the patient sample was from Jurkat cells. In order to precisely determine the contribution of DNA from PBMCs and Jurkat cells, we performed these measurements every time that we isolated and measured HIV-1 DNA levels. DNA was quantified by absorbance at 260 nm. Viral DNA was detected by PCR using primers INT-gag-5 (5′-GGTCAGCCAAAATTACCCTATAGTGC-3′) and INT-gag-3 (5′-CTTCCTCATTGATGGTITCTTTTA-3′) (9). The cycling parameters were an initial denaturation step at 94°C for 4 min, followed by 4 cycles of PCR, with 1 cycle consisting of denaturation at 94°C for 10 s, annealing at 52°C for 10 s, and extension at 72°C for 10 s. This was followed by 26 cycles of PCR, with 1 cycle consisting of denaturation at 90°C for 10 s, annealing at 55°C for 10 s, and extension at 72°C for 10 min, and by a final extension step at 72°C for 1 min. To document the quality of the genomic DNA, the glyceraldehyde-3-phosphate dehydrogenase gene (gapdh) was also amplified in reaction with 400 ng of DNA with primers 5′GAPDH.DNA (5′-GGGAAGCTCAAGGGAGATAAAATTC-3′) and 3′GAPDH.DNA (5′-GTAGTTGAGGTCAATGAAGGGGTC-3′). PCR was performed with the high-fidelity PCR system (Roche), and hot start was employed during the PCR setup. PCR signals were confirmed by Southern hybridization. The oligonucleotide probe for HIV-1 gag was 5′-CACCTAGAACTTTAAATGCATGGG-3′, and the probe for gapdh was 5′-GGTAAGGAGATGCTGCATTCG-3′.
Isolation of mRNA from resting and activated CD4+ T lymphocytes.
A total of 106 sorted resting CD4+ T cells were lysed with 1 ml of lysis-binding buffer provided by Dynal (Dynabeads mRNA DIRECT kit). Cell lysates were stored at −70°C until analysis. mRNA was isolated according to the manufacturer's protocol (Dynabeads mRNA DIRECT kit; Dynal).
Synthesis of cDNA from RNA isolated from resting and activated CD4+ T lymphocytes.
mRNA eluted from oligo(dT)25 beads was reverse transcribed into cDNA immediately after isolation. First, mRNA was treated with DNase (amplification-grade DNase I; Gibco BRL). After heat inactivation of DNase I at 65°C for 10 min, RNA samples were placed on ice. For cDNA synthesis, one-fourth of the mRNA isolated was incubated at room temperature for 5 min with a 2 mM concentration of the relevant, spliced-form-specific primer, 6.5 mM random hexamers (Life Technology, Gibco BRL), and RNase inhibitor (28 to 56 U depending on stock titration) (RNAguard RNA inhibitor porcine; Amersham Pharmacia Biotech). Primer US.12 (5′-TGATGTCCCCCCACTGTGTTT-3′) was used for unspliced (US) RNAs, and primer MS.e7 (5′-CTGTCCCCTCAGCTACTGCTA-3′) was used for multiply spliced (MS) RNAs. Then, 100 mM concentrations of the four deoxynucleoside triphosphates (Perkin-Elmer Applied Biosystems), 10 mM dithiothreitol, and supplied reaction buffer were added. After the solution was incubated at 45°C for 55 min, reverse transcriptase (RT) was heat inactivated at 70°C for 15 min. Each reaction was done in parallel with a reaction to which all components but RT were added.
PCR amplification of cDNA from resting CD4+ T lymphocytes.
All first PCR amplifications were performed immediately after cDNA synthesis. Diethyl pyrocarbonate-treated H2O (80 μl) was added to each cDNA reaction mixture. One-fifth of diluted cDNA was used for each amplification. Two types of amplifications were performed.
For semiquantitative amplification of MS and US cDNAs, reaction mixtures were subjected to an initial denaturation step at 94°C for 4 min, followed by 32 cycles of PCR, with 1 cycle consisting of denaturation at 94°C for 30 s and annealing and extension at 65°C for 30 s, and a final extension step at 68°C for 1 min. Primers used to amplify US cDNA were primer US.1a (5′-GCTTGCTGAAGCGCGCACGG-3′) and primer US.2a (5′-CGTTCTAGCTCCCTGCTTGC-3′). Primers used to amplify MS cDNA were the previously described primer M667 (54) and primer Ms.e7 (see above).
To detect extremely low levels of HIV-1 RNA, cDNAs were also subjected to separate nested amplifications. In this case, the cycling conditions were as follows. For the first amplification, the conditions were denaturation at 94°C for 4 min, followed by 24 PCR cycles, with 1 cycle consisting of denaturation at 94°C for 30 s and annealing and extension at 63°C for 30 s, and a final extension step at 68°C for 1 min. The primer sets M667-MS.e7 and US.1a-US.12 were used for the first-stage amplification of MS and US cDNAs, respectively. Ten microliters of a 1:40 dilution of the first PCR product was subjected to a nested PCR. The cycling conditions for nested PCR follow: (i) denaturation at 94°C for 4 min; (ii) 29 PCR cycles, with 1 cycle consisting of denaturation at 94°C for 30 s and annealing and extension at 65°C for 30 s; and (iii) a final extension at 68°C for 1 min. Primer sets M667-MS.e5 (5′-TCGCTGTCTCCGCTTCTTC-3′) and US.1a-US.2a were used for this nested amplification of MS and US cDNA species, respectively.
To control for the integrity of the RNA preparations and the efficiency of the mRNA isolation and cDNA synthesis, mRNAs for cellular genes CD4 and gapdh were also analyzed by RT-PCR. Diluted cDNA (10 to 15 μl) was amplified for CD4 (5′CD4.RNA primer [5′-AAGGCGGTGTGGGTGCTG-′] and 3′CD4.RNA primer [5′-AGAAGAAGATGCCTAGCCCAATG-3′]) or gapdh (5′GAPDH.RNA primer [5′-GGAAGGTGAAGGTCGGAGTCAACGT-3′] and 3′GAPDH.RNA primer [5′-CTGTTGTCATACTTCTCATGGTTCAC-3′]).
The specificity of all PCR signals was confirmed by Southern hybridization. The oligonucleotide probe for detection of all HIV-1 RT-PCR products was 5′-GCAAGAGGCGAGGGGIGG 3′. The probe for gapdh was 5′-GGAGCCAAAAGGGTCATCATCTC-3′, and the probe for CD4 was 5′-CTGATTGTGCTGGGGGGC-3′.
All first PCRs were performed with the Expand long template PCR system (Roche). The Expand high-fidelity PCR system (Roche) was used for all nested PCRs. For DNA PCR, hot start was employed during PCR setup.
Production and quantification of synthetic mRNA standards.
Two recombinant plasmids were constructed for use in in vitro transcription, pSP64poly(A)HIVmspl and pSP64poly(A)HIVunspl. For pSP64poly(A)HIVmspl, primers M667 (50) and MS.e7 were used to amplify a 642-bp fragment from a cDNA library generated from the ACH-2 cell line. The amplified fragment contains exons 1, 5, and 7. It was subcloned into the SmaI site of the multiple cloning site of the pSP64poly(A)vector (Promega). For pSP64poly(A)HIVunspl, a plasmid carrying the reference LAI clone of HIV-1 was amplified with primers US.1a and US.12. The resulting 672-bp fragment, containing sequences around the major splice donor site, was subcloned into the SmaI site in the pSP64poly(A)vector (Promega).
Synthetic RNAs representing MS and US HIV-1 RNAs were generated from these plasmids by in vitro transcription in the presence of [3H]UTP as previously described (27). To quantify in vitro-transcribed RNA, three independent aliquots, including negative controls to which SP64 phage RNA polymerase was not added, were counted for each sample. Each aliquot was counted three times. The average counts per minute value, corrected for counting efficiency, was used to calculate the RNA copy number.
RESULTS
To study the mechanism of HIV-1 latency in the resting G0 CD4+ lymphocytes that comprise a stable reservoir for HIV-1 in vivo, we purified resting CD4+ T lymphocytes from peripheral blood samples of infected individuals who exhibited long-term suppression of viral replication on HAART (Fig. 1A). A two-stage purification procedure gave preparations of resting CD4+ T lymphocytes that contained <0.1% contamination with activated cells. In previous studies, we and others have shown that replication-competent virus persists in this population of resting CD4+ T lymphocytes despite prolonged treatment with HAART (8, 17, 18, 37, 43, 52). After the initiation of HAART, the frequency of resting CD4+ T lymphocytes harboring latent HIV-1 decreases in a biphasic fashion (4). The rapid initial decay within the first 3 months of treatment reflects the loss of virus in the labile preintegration state of latency (39, 54). The subsequent slower phase represents the persistence of lymphocytes with stably integrated provirus (9, 10, 18). Therefore, to analyze HIV-1 transcription in resting CD4+ T lymphocytes in postintegration latency, we studied patients on HAART who exhibited suppression of plasma HIV-1 RNA levels to below the limit of detection (<50 copies of viral RNA/ml) for prolonged periods of time (23 to 54 months [Table 1]).
FIG. 1.
Quantitative measurements of HIV-1 proviral DNA present in highly purified resting CD4+ T lymphocytes. (A) Isolation of resting CD4+ T lymphocytes from patients on HAART. The results of flow cytometric analysis of CD4 and HLA-DR expression on a gated lymphocyte population in unfractionated PBMCs (left panel) and on an ungated sorted population of highly purified resting CD4+ T lymphocytes (right panel) are shown. The numbers are the percentages of cells expressing CD4 and/or HLA-DR. (B) Quantification of HIV-1 DNA in highly purified resting CD4+ T lymphocytes from patients on HAART. To generate a standard curve, ACH-2 cells carrying a single integrated copy of the HIV-1 genome were diluted with HIV-1-negative PBMCs. Isolated DNA was amplified for HIV-1 gag as described in Materials and Methods. To control for DNA quality, the cellular gapdh gene was amplified in parallel. PCR products were confirmed by Southern hybridization using gene-specific probes.
TABLE 1.
Characteristics of patients and HIV-1 DNA levels in purified resting CD4+ T lymphocytes
| Patient no. | Drug regimena | Plasma HIV-1 RNA level at time of analysis (no. of copies/ml) | Duration of virologic suppression (mo) | HIV-1 DNA level (no. of copies/ 106 resting CD4+ T cells) |
|---|---|---|---|---|
| 1 | AZT/3TC/NFV | <50 | 30 | 20 |
| 2 | AZT/ddC/NVP | <50 | 58 | 100 |
| 3 | 3TC/d4T/RTV | <50 | 54 | 60 |
| 4b | AZT/3TC/EFV/ABC | <50 | 48 | 100 |
| <50 | 50 | 120 | ||
| 5 | 3TC/EFV/ABC | <50 | 23 | 120 |
| 6 | AZT/3TC/IDV/RTV | <50 | 45 | 50 |
| 7 | AZT/3TC/EFV | <50 | 45 | 50 |
| 8 | 3TC/d4T/NFV | 125 | 32 | 1,000 |
| 9 | 3TC/d4T/NFV | <50 | 51 | 120 |
| Median | <50 | 47 | 100 |
Abbreviations for antiretroviral drugs: AZT, zidovudine; 3TC, lamivudine; ddC, zalcitabine; d4T, stavudine; ABC, abacavir; NVP, nevirapine; EFV, efavirenz; IDV, indinavir; RTV, ritonavir; NFV, nelfinavir.
Analysis of HIV-1 DNA from T lymphocytes from patient 4 was performed on two separate occasions.
To determine the frequency of infected cells, we first measured proviral HIV-1 DNA in resting CD4+ T lymphocytes (Table 1 and Fig. 1B). In the patients on long-term HAART, the median number of copies of HIV-1 DNA per 106 resting CD4+ T lymphocytes was 100. For one subject (patient 8), the higher levels of HIV-1 DNA measured may have been the result of a blip in plasma HIV-1 RNA that may have allowed some new infection of resting CD4+ T cells. Using a previously described inverse PCR assay (9, 10), integration of HIV-1 DNA into the host cell genome was demonstrated in these samples (J. D. Siliciano et al., unpublished data). Replication-competent virus was rescued from resting CD4+ T lymphocytes at the same frequencies reported previously (17, 18, 43, 56) for patients on suppressive HAART regimens (0.1 to 1 infectious unit per 106 resting CD4+ T lymphocytes).
Having established the proviral burden in resting CD4+ T lymphocytes, we analyzed HIV-1 transcription. HIV-1 mRNAs are transcribed by RNA polymerase II. Alternative utilization of any one of five alternate splice donors and more than 10 alternate splice acceptors gives rise to 1.7- to 2.0-kb MS mRNAs, 4.3- to 5.5-kb singly spliced mRNAs, and 9.2-kb US mRNAs (Fig. 2A) (42, 44). We measured MS and US mRNA species (Fig. 2A). Detection of only MS mRNAs would suggest posttranscriptional mechanisms for latency (40, 41, 45), while the absence of HIV-1 mRNAs would suggest that latency operates at the level of transcription.
FIG. 2.
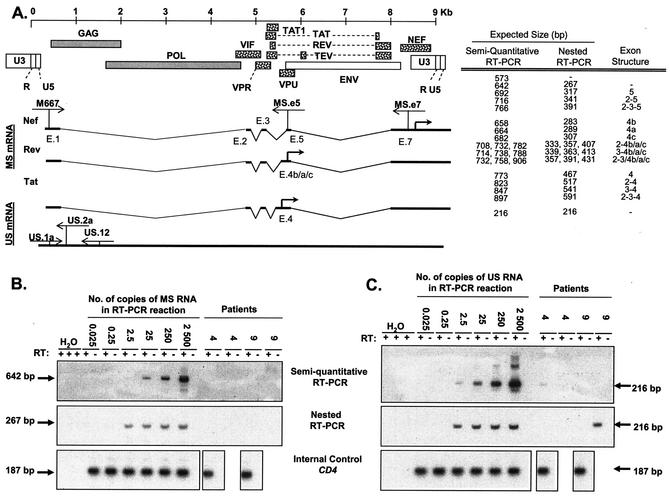
Detection of MS and US HIV-1 mRNAs associated with resting CD4+ T lymphocytes. (A) Schematic structures of MS and US HIV-1 RNAs. The HIV-1 genome and viral proteins encoded by it are shown at the top. The proteins that are translated from MS mRNA species (dotted) and proteins translated from US mRNA species (shaded) are indicated. The exon structures of MS mRNAs for nef, rev, and tat (only two-exon tat mRNA) are shown. Exon numbers (e.g., E.1) are given below each exon (42, 44). The positions of the primers are indicated by thin arrows. The thick arrow indicates the translation initiation site. The expected sizes of PCR products with their corresponding exon structures are shown to the right of the schematic structures. Exons 1 and 7 that are present in each mRNA are not included. One spliced form that has only these two exons is represented by a hyphen. (B) Single-round (top panel) and nested (middle panel) RT-PCR assays for MS HIV-1 RNAs. Assays were calibrated by diluting radiochemically quantitated, in vitro-transcribed MS RNA into lysates of resting CD4+ T lymphocytes from healthy donors and then carrying the mixture throughout RNA isolation, DNase treatment, reverse transcription, and single-round or nested PCR. Each PCR mixture contained the indicated number of copies of standard RNA in 50,000 cell equivalents of lysate. Therefore, detection of a signal at 2.5 copies/reaction mixture indicated a sensitivity of 50 copies/106 resting CD4+ T cells. Patient samples were tested in duplicate with (+) and without (−) RT. In the experiment shown, samples from patients 4 and 9 were tested, and no MS RNA species were detected. To control for RNA isolation and cDNA synthesis, CD4 RNAs were amplified over the splice junction in each sample. (C) Single-round (top panel) and nested (middle panel) RT-PCR assays for US HIV-1 RNAs. The reactions were standardized with in vitro-transcribed US RNA standard diluted into lysates of resting CD4+ T lymphocytes from healthy donors as described above for panel B. The same lysates contained serially diluted HIV-1 MS RNA standards. US RNA species were detected only sporadically in patients' samples. To control for RNA isolation and cDNA synthesis, CD4 RNAs were amplified over the splice junction in each sample, as was done for MS RT-PCR (bottom panel).
For detection of HIV-1 mRNAs, we used sensitive, semiquantitative, single-round amplifications to approximate the number of RNA molecules in each sample. To confirm the presence or absence of very low levels of HIV-1 mRNA, nested versions of these assays that can detect a single molecule were used in parallel. To validate these assays, in vitro-transcribed RNA standards were generated from plasmids carrying a spliced sequence representing exons 1, 5, and 7 for MS RNA and a sequence spanning the major splice donor site for US RNA (Fig. 2A). RNA synthesis was performed in the presence of [3H]UTP, allowing precise radiochemical quantification of synthetic RNA standards. Both MS and US synthetic mRNAs were simultaneously serially diluted in known copy numbers in lysates of 106 resting CD4+ T lymphocytes from healthy donors. After serial dilutions, these mRNA standards were carried through the entire procedure, including mRNA isolation, DNase treatment, cDNA synthesis, and single-round or nested PCR performed in parallel with patients' samples. Representative experiments are shown in Fig. 2B and C. The single-round assays were sensitive to 2.5 copies of RNA standard per 5 × 104 cell equivalents of mRNA, resulting in a dynamic range of 50 to 5,000 copies of RNA per 106 resting CD4+ T lymphocytes (Fig. 2B and C, top panels). The nested RT-PCR assays readily detected as few as 2.5 molecules of MS and US HIV-1 RNA standards in a background of mRNA isolated from 5 × 104 resting CD4+ T lymphocytes (Fig. 2B and C). Consistent with radiochemical quantification, the nested assay signal was lost as standards were diluted from 2.5 to 0.25 copy per tube.
Using these sensitive and carefully validated assays, we consistently failed to detect MS mRNA species in resting CD4+ T lymphocytes from patients on long-term HAART (Fig. 2B and Table 2). The results of these assays indicated that the number of MS mRNA molecules associated with 106 purified resting CD4+ T lymphocytes was less than 50. In contrast, we detected MS RNA in activated CD4+ T lymphocytes infected in vitro with R5 HIV-1. We also detected MS RNA in unfractionated PBMCs isolated from viremic patients because of the presence of productively infected, activated CD4+ T lymphocytes (not shown). These results suggest that the PCR primers and conditions used can readily detect HIV-1 mRNAs in infected cells and that sequence variation in the relatively conserved regions chosen as primer binding sites in HIV-1 isolates does not prevent amplification. In addition, MS RNAs were readily detected in as few as five uninduced cells from the chronically infected ACH-2 cell line serially diluted in 106 resting CD4+ T cells from healthy individuals. The final reaction mixture contained RNA from as few as one ACH-2 cell in a background of RNA from 5 × 104 uninfected resting cells. The ACH-2 cell line has been used as a model for latent virus (13, 20, 41). Although HIV-1 gene expression can be up-regulated in ACH-2 cells by activating stimuli, the basal level of transcription is much higher than the level we observed in latently infected resting lymphocytes in vivo. Thus, unlike some models of latency based on continuously proliferating cell lines, the analysis of highly purified resting G0 CD4+ T lymphocytes from patients on HAART suggests that MS HIV-1 RNA species are not produced at high levels in latently infected cells.
TABLE 2.
Levels of MS and US HIV-1 mRNAs associated with highly purified resting CD4+ T lymphocytes and ratios of HIV-1 RNA copies per HIV-1 DNA-positive resting CD4+ T cella
| Patient no. | HIV-1 mRNA molecules associated with 106 resting CD4+ T cells
|
No. of HIV-1 RNA molecules/HIV-1 DNA-positive celld
|
||||
|---|---|---|---|---|---|---|
| MS
|
US
|
MS | US | |||
| RT-PCR (no. of copies/106 cells)b | Nested RT-PCRc | RT-PCR (no. of copies/106 cells) | Nested RT-PCR | |||
| 1 | <50, <50 | −, − | <50, <50 | −, − | <2.5 | <2.5 |
| <50, <50 | −, − | 50, 50 | −, − | <2.5 | <2.5 | |
| 2 | <50, <50 | −, − | <50, <50 | −, − | <0.5 | <0.5 |
| 3 | <50, <50 | −, − | <50, <50 | −, − | <0.8 | <0.8 |
| 4e | <50, <50 | −, − | <50, <50 | −, − | <0.5 | <0.5 |
| <50, <50 | −, − | 50, <50 | −, − | <0.5 | <0.5 | |
| 4e | <50, <50 | −, − | <50, <50 | −, − | <0.4 | <0.4 |
| 5 | <50, <50 | −, − | <50, <50 | −, − | <0.4 | <0.4 |
| 6 | <50, <50 | −, − | <50, <50 | −, + | <1 | <1 |
| 7 | <50, <50 | −, − | <50, <50 | −, + | <1 | <1 |
| 8 | <50, <50 | −, − | 50, 50 | +, + | <0.05 | <0.05 |
| 9 | <50, <50 | −, − | <50, <50 | −, + | <0.4 | <0.4 |
All RT-PCR analyses were performed in duplicate. The sensitivity for both assays was 50 copies per 106 resting CD4+ T cells.
Negative signal in a semiquantitative RT-PCR is shown by <50.
Positive (+) and negative (−) signals in a final nested RT-PCR.
Number of HIV-1 mRNA molecules per one HIV-1 DNA-positive cell was calculated as follows: HIV-1 RNA level per 106 cells/HIV-1 DNA level per 106 cells. This assumes equal distribution (see text).
Analysis of HIV-1 RNA for cells from patient 4 was performed on two separate occasions.
The level of US HIV-1 RNA was also low. In four of nine patients, no US mRNAs were detected using either semiquantitative or nested RT-PCR assays (Table 2). In the remaining patients, US RNAs were detected right at the limit of detection of the assays used (50 copies/106 cells). For example, US RNA was detected in one of two semiquantitative PCRs at a level below 2.5 copies/tube for patient 4 and in one of two nested reactions for patient 9 (Fig. 2C). Therefore, the levels of both MS and US RNA species are low in purified resting CD4+ T lymphocytes from patients on long-term HAART.
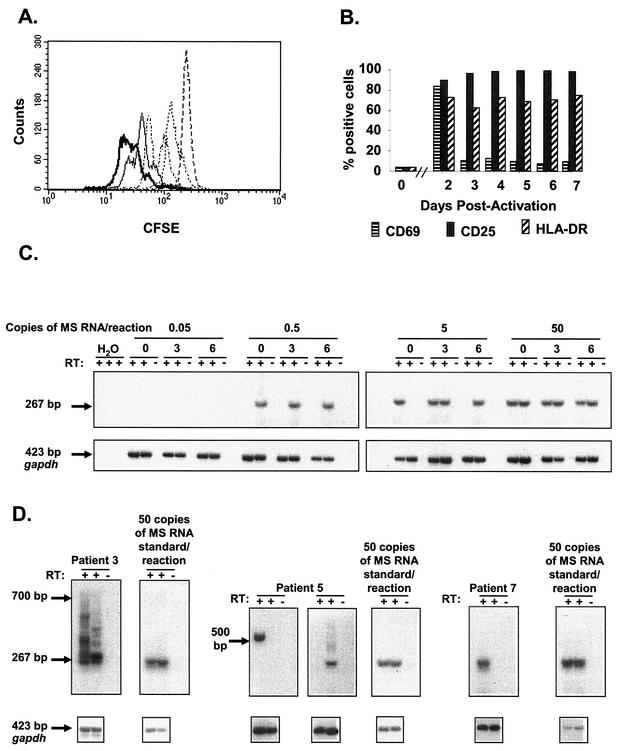
Next, we determined whether this apparent lack of transcription of HIV-1 DNA in resting CD4+ T lymphocytes could be reversed by cellular activation. Since the antigen specificity of latently infected resting CD4+ T lymphocytes is heterogeneous (12), we used the lectin phytohemagglutinin (PHA) to mimic reactivation of HIV-1 latency in vivo. Purified resting CD4+ T lymphocytes from an intensively studied subset of patients (patients 4, 5, 6, 7 and 9) were activated with PHA in the presence of irradiated allogeneic PBMCs in culture medium supplemented with interleukin-2, a procedure previously shown to induce uniform proliferation of resting CD4+ T lymphocytes (18). To demonstrate uniform cellular activation, we examined carboxy fluorescein diacetate succinimidyl ester (CFSE) dilution upon activation of CFSE-labeled resting CD4+ T lymphocytes. In all experiments, more than 95% of CFSE-labeled lymphocytes underwent at least one cell division by day 3 postactivation (Fig. 3A). To demonstrate further the uniform activation of resting CD4+ T lymphocytes in this system, we studied the surface expression of early, intermediate, and late markers of T-cell activation: CD69, CD25, and HLA-DR, respectively. As expected, these cellular markers were up-regulated with different kinetics on large proportions of the lymphocytes, with the entire population eventually giving evidence of being activated (Fig. 3B).
FIG. 3.
Reversal of transcriptional silencing by T-cell activation. (A) Activation of purified resting CD4+ T lymphocytes. Prior to activation, resting CD4+ T lymphocytes were labeled with CFSE. Proliferation was measured by a twofold dilution of CFSE. After activation, lymphocytes were harvested at 16 h (- - - -), 24 h (-----), 40 h (-○-○-○), 52 h (-·-·-·), 64 h (——), and 72 h (), and CFSE fluorescence was measured. When resting CD4+ T lymphocytes were incubated in culture medium alone, no dilution of CFSE label was observed (not shown). In all experiments, >95% of CFSE-labeled lymphocytes underwent at least one cell division by day 3 postactivation. (B) Surface expression of CD69, CD25, and HLA-DR in HIV-1 DNA-positive cells measured immediately prior to and at the indicated times after activation. (C) Assay for MS HIV-1 mRNAs associated with activated CD4+ T lymphocytes. The sensitivity of the nested RT-PCR assay was assessed using in vitro-transcribed MS RNA standards. These standards were serially diluted into the lysates from activated CD4+ T lymphocytes harvested at various time points (0, 3, and 6 days) after activation. At 0.5 copy of MS RNA standard in the final nested reaction, the nested assay is positive in one of two duplicated reactions. At 0.05 copy per reaction, the PCR signals were consistently negative. (D) Reversal of HIV-1 latency in a fraction of HIV-1 DNA-positive resting CD4+ T lymphocytes after activation. Resting CD4+ T lymphocytes were seeded at approximately 10 HIV-1 DNA-positive lymphocytes per well and activated. Lymphocytes were harvested 2 to 5 days postactivation and analyzed for MS mRNA. Induced transcription of MS HIV-1 mRNA was detected in resting CD4+ T lymphocytes isolated from all patients. Representative positive wells are shown. The amplified spliced mRNA species from patients' samples had sizes ranging from 267 to 700 bp, as expected on the basis of the MS variants produced during infection (Fig. 2A).
To analyze HIV-1 transcription after activation, we measured MS mRNA species. We chose MS mRNAs because these RNAs encode the Tat and Rev proteins that are indispensable for HIV-1 replication (11, 19, 46). Moreover, MS RNAs were not detected in latently infected resting CD4+ T lymphocytes in patients on HAART, and MS RNA production thus would represent a molecular exit from HIV-1 latency. Finally, unlike US RNA, MS RNA signals cannot be due to bound extracellular virions.
RT-PCR assays were developed to detect rare mRNA species in the larger pool of cellular RNA present in activated CD4+ T lymphocytes. MS HIV-1 RNA standards were serially diluted in lysates obtained at 0, 3, and 6 days after activation of 100,000 resting CD4+ T lymphocytes. As seen in Fig. 3C, MS HIV-1 RNA standards were readily detected at the single-molecule level in the final nested PCR.
Resting CD4+ T lymphocytes from patients on HAART were plated at cell concentrations giving approximately 10 HIV-1 DNA-positive lymphocytes per well, based on the measurements described above (Table 3). Resting CD4+ T lymphocytes were then subjected to activation under the same conditions, and individual wells were analyzed for HIV-1 transcription on days 2 to 5 postactivation. MS mRNA was detected, but only in a fraction of the wells (Table 3). Representative experiments are shown in Fig. 3D. In some HIV-1 DNA-positive wells from later time points, a wide range of spliced HIV-1 mRNAs were seen, consistent with the large number of known splice variants (Fig. 2A). In a typical experiment, only 1% of HIV-1 DNA-positive lymphocytes transcribed HIV-1 RNA after in vitro activation, even though all lymphocytes in the culture became activated. This finding, when considered together with the measured frequency of resting CD4+ T lymphocytes harboring HIV-1 DNA (Fig. 1), suggests that in patients on long-term HAART, only about 1 in 106 resting CD4+ T lymphocytes contains a provirus capable of high-level HIV-1 gene expression after cellular activation (Table 3). As expected, this frequency is consistent with and somewhat higher than the frequency of resting CD4+ T lymphocytes from which replication-competent virus can be cultured (typically 0.1 to 1 per 106 resting CD4+ T lymphocytes) (17, 18, 43, 56).
TABLE 3.
Frequency of HIV-1 DNA-positive resting CD4+ T lymphocytes that can be induced to transcribe HIV-1 mRNA after activation
| Patient no. | No. of HIV-1 DNA-positive resting CD4+ T lymphocytes/wella | Frequency of HIV-1 transcription-positive wells | % of HIV-1 DNA-positive resting CD4+ T cells competent for HIV-1 transcription after cellular activationb | Frequency of transcription- competent cells/106 resting CD4+ T cellsc |
|---|---|---|---|---|
| 4d | 10 | 1 in 5 | 2 | 2 |
| 4d | 10 | 0 in 12 | 0 | 0 |
| 6d | 5 | 2 in 21 | 2 | 1 |
| 6d | 10 | 1 in 5 | 2 | 1 |
| 5 | 36 | 1 in 8 | 0.3 | 1 |
| 7 | 10 | 1 in 5 | 2 | 1 |
| 9 | 24 | 1 in 18 | 0.2 | 0.2 |
| Mean | 1 | 1 |
A total of 100,000 or 200,000 purified resting CD4+ T lymphocytes were plated in individual wells, giving the indicated number of HIV-1 DNA-positive cells per well based on PCR measurements of HIV-1 DNA (Table 1).
The percentage of HIV-1 DNA-positive resting CD4+ T cells that are competent for HIV-1 transcription after cellular activation was calculated as follows: [(number of wells with detectable HIV-1 transcription)/(number of screened wells × number of HIV-1 DNA-positive lymphocytes per well)] × 100.
The frequency of HIV-1 transcription-competent resting CD4+ T lymphocytes in 106 resting CD4+ T lymphocytes was calculated as follows: [(percentage of HIV-1 DNA-positive resting CD4+ T cells that are competent for HIV-1 transcription/100)] × number of HIV-1 DNA-positive lymphocytes per 106 resting CD4+ T lymphocytes. HIV-1 DNA measurements were the same as in Table 1, except for patient 5 who had 360 HIV-1 DNA copies per 10b resting CD4+ T cells at the time of analysis.
Analysis of T lymphocytes from patients 4 and 6 was performed on two separate occasions.
DISCUSSION
We have analyzed HIV-1 gene expression in latently infected resting CD4+ T lymphocytes from the peripheral blood of HIV-1-infected individuals on HAART. While most evidence suggests that these cells do not release virus without cellular activation (9, 10), the mechanism underlying the lack of virus production has been unclear. Some models of virus latency suggest that the absence of activation-dependent host transcription factors (6, 13, 20, 36, 48) or chromatin structural changes (29, 51) or failure of transcriptional elongation (1, 22, 24, 25, 28, 31) precludes the expression of viral genes. However, it remained possible that posttranscriptional mechanisms were responsible for the absence of virus production. Our results suggest that few translatable HIV-1 mRNAs are made in latently infected resting CD4+ T lymphocytes from blood. In 106 resting CD4+ T cells, we found an average of 100 cells carrying HIV-1 DNA. In the same number of resting cells, the amount of MS HIV-1 mRNA was <50 molecules. Interpretation of this result is complicated by the fact that we do not know how the RNA molecules are distributed in the cells carrying HIV-1 DNA. If we assume that 50 molecules of MS RNA are present in 106 cells and that they are distributed evenly, then the steady-state level of MS RNA would be <1 molecule/cell, which is consistent with complete or near-complete transcriptional silencing. However, only 1% of the cells with HIV-1 DNA appear to be capable of producing high levels of HIV-1 RNA after cellular activation. If the basal RNA level is produced only from these cells, then the steady-state level of HIV-1 RNA in an infected cell could be as high as 50 copies/cell. This would place viral messages in the large class of low-abundance mRNAs found in the cells. Whether this level of gene expression would be sufficient to allow selective targeting of latently infected cells remains to be determined. By way of contrast, it is useful to consider that in productively infected cells, HIV-1 mRNAs are present at levels of 4,000 copies/cell (26).
While MS HIV-1 RNAs were consistently below the limit of detection (50 copies/106 cells), we were able to detect US HIV-1 RNAs in some patients at a level of approximately 50 copies/106 cells. These may represent genomic RNA molecules in virions bound to the surfaces of resting CD4+ T cells or US RNAs within the cells. The finding of a preponderance of US HIV-1 RNAs over MS HIV-1 RNAs is consistent with the results of Lewin et al. (32) who studied HIV-1 RNA levels in unfractionated PBMCs from patients on HAART. The greater abundance of US RNA relative to MS RNA demonstrated here and by Lewin et al. (32) is inconsistent with some posttranscriptional models of virus latency based on cell lines that express MS but little US RNA (41).
Several caveats to our findings should be mentioned. First, although the assays used were rigorously validated with radiochemically quantitated, in vitro-transcribed RNA standards taken through the entire procedure, it remains possible that the sensitivities of the assays for patient-derived sequences were lower than those for the RNA standards. Although primers were chosen from relatively conserved regions of the genome, sequence variation in individual patients could have reduced the sensitivity of detection in some patients. The fact that similar results were obtained in all patients studied argues against this possibility. It is also important to point out that these results do not exclude the possibility that latency is due to a block at the level of transcriptional elongation (1, 22, 24, 25, 28, 31). According to this model, transcription of viral genes initiates in latently infected cells but is prematurely terminated due to the absence in these cells of Tat and/or of the Tat-associated host factors that act on the C-terminal domain of RNA polymerase II to promote elongation. In fact, consistent with this model, we recently showed that short (∼60 nucleotides), nonpolyadenylated HIV-1 mRNAs are present in highly purified resting CD4+ T cells from patients on HAART (K. Lassen and R. F. Siliciano, unpublished results). Thus, although we could detect little MS or US HIV-1 RNA, it is possible that transcription starts at the HIV-1 long terminal repeat (LTR) in latently infected cells with subsequent premature termination. A final caveat is related to the measurement of the number of cells harboring HIV-1 DNA. Our calculation assumes one HIV-1 integration per infected cell. A recent report (30) suggests that HIV-1-infected lymphocytes may carry as many as three to four proviruses. Thus, the frequency of HIV-1 infected cells reported here may be overestimated, and the frequency of transcription-competent cells may be underestimated. Our results are consistent with the results of several other recent studies of HIV-1 gene expression in vivo. Bagnarelli et al. first noted a dramatic decline in the levels of MS HIV-1 RNA in PBMCs from patients on antiretroviral therapy (2). Zhu et al. measured HIV-1 DNA and MS and US HIV-1 RNAs in monocytes and in highly purified resting and activated CD4+ T cells from patients on HAART (58). They found low levels of MS and US HIV-1 RNAs in purified resting CD4+ T cells (10 to 50 copies/μg of total RNA). Assuming our typical yield of 1 μg of RNA/106 resting CD4+ T cells, these results are consistent with the results presented here. As mentioned above, Lewin et al. detected US but little MS HIV RNA in PBMCs from patients on HAART (32). Taken together, these studies support the idea that latently infected cells produce relatively little HIV-1 RNA.
Some groups have recently provided evidence for active HIV-1 gene expression in cells from patients on HAART. A number of studies have examined unfractionated mononuclear cells from peripheral blood or lymph nodes (21, 23, 26, 55, 57). It is likely that the HIV-1 RNA detected is from activated T cells, which express on the order of 4,000 HIV-1 RNA molecules/cell (26). Some studies have detected HIV-1 or simian immunodeficiency virus gene expression in cells with the phenotype of naive or resting cells (14, 53, 57). Zhang et al. detected viral gene expression in cells that were phenotypically similar to resting CD4+ T cells (57). The level of gene expression was lower than that observed in activated CD4+ T cells. These cells may represent infected lymphoblasts that are in the process of transitioning back to a fully quiescent state. In addition, exposure to cytokines in the context of the lymphoid tissues may render resting CD4+ T cells more permissive to viral gene expression (49). In in vitro studies of tonsil preparations, Eckstein et al. have demonstrated active infection of large fractions of resting, naive cells by X4 virus (14). This result may also reflect the use in these studies of mixed populations of cells from lymphoid tissues in which exposure to cytokines alters the state of activation of the cells. In addition, in vitro infection with X4 viruses may not provide the best model for the latent infection that occurs predominantly with R5 viruses in vivo (38). In any event, our results suggest that in rigorously purified resting CD4+ T lymphocytes from the peripheral blood of patients on long-term HAART, there is little production of translatable mRNAs by the latently infected lymphocytes present.
When we activated latently infected resting CD4+ T lymphocytes in vitro, only 1% of the HIV-1 DNA-containing resting CD4+ T cells could be induced to transcribe HIV-1 genes. Thus, the majority of HIV-1 proviral DNA present in this reservoir cannot be readily activated for the continuous transcription of HIV-1 mRNA. Some of the integrated HIV-1 DNA present in resting CD4+ T lymphocytes may be under strong epigenetic regulation that cannot be readily reversed by a short-term cellular activation (29, 51). In addition, some of these proviruses may be intrinsically defective for HIV-1 transcription due to mutations in the LTR or in Tat (15). Importantly, the frequency of transcription-competent lymphocytes (1/106 resting CD4+ T cells) is still higher than the frequency of lymphocytes from which replication-competent virus can be rescued by cellular activation (0.1 to 1 per 106 resting CD4+ T cells) (17, 18). This slightly higher frequency suggests that some transcriptionally active lymphocytes are producing defective virus.
Our study provides direct in vivo evidence that HIV-1 latency in resting CD4+ T lymphocytes operates at the level of mRNA production and not solely at the level of splicing or nuclear export of mRNAs. Further studies will be necessary to exclude the possibility that latently infected cells produce very low levels of HIV-1 mRNA. This can be done only by in vivo studies since, as demonstrated here, in vitro models, particularly those involving transformed cell lines, may not accurately mimic the transcriptional state of the resting G0 T cells that comprise the stable reservoir for HIV-1. If viral genes are transcribed only at very low levels in latently infected cells, as our study suggests, then it may be difficult to target this reservoir with HIV-1-specific RNA- and protein-directed approaches. Furthermore, the stability of the reservoir comprised of resting memory CD4+ T cells will be dictated solely by the long life span of memory CD4+ T cells. Nevertheless, even though this reservoir pool may prove very difficult to target selectively, our study indicates that the majority of resting lymphocytes carrying HIV-1 DNA cannot be induced to transcribe HIV-1 genes even after T-cell activation and may therefore be irrelevant to disease progression. Understanding the mechanisms that render most of the HIV-1 DNA in resting CD4+ T lymphocytes incompetent for transcription may facilitate efforts to completely eliminate this reservoir.
Acknowledgments
We thank the present and previous members of the laboratory for helpful suggestions and Scott Barnet for help in scheduling patients.
This work was supported in part by NIH grants AI43222 and AI51178 and by a grant from the Doris Duke Charitable Foundation.
REFERENCES
- 1.Adams, M., L. Sharmeen, J. Kimpton, J. M. Romeo, J. V. Garcia, B. M. Peterlin, M. Groudine, and M. Emerman. 1994. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc. Natl. Acad. Sci. USA 91:3862-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagnarelli, P., A. Valenza, S. Menzo, R. Sampaolesi, P. E. Varaldo, L. Butini, M. Montroni, C. F. Perno, S. Aquaro, D. Mathez, J. Leibowitch, C. Balotta, and M. Clementi. 1996. Dynamics and modulation of human immunodeficiency virus type 1 transcripts in vitro and in vivo. J. Virol. 70:7603-7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bednarik, D. P., J. A. Cook, and P. M. Pitha. 1990. Inactivation of the HIV LTR by DNA CpG methylation: evidence for a role in latency. EMBO J. 9:1157-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blankson, J. N., D. Finzi, T. C. Pierson, B. P. Sabundayo, K. Chadwich, J. B. Margolick, T. C. Quinn, and R. F. Siliciano. 2000. Biphasic decay of latently infected CD4+ T cells in acute HIV-1 infection. J. Infect. Dis. 182:1636-1642. [DOI] [PubMed] [Google Scholar]
- 5.Blankson, J. N., D. Persaud, and R. F. Siliciano. 2002. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 53:557-593. [DOI] [PubMed] [Google Scholar]
- 6.Bohnlein, E., J. W. Lowenthal, M. Siekevitz, D. W. Ballard, B. R. Franza, and W. C. Greene. 1988. The same inducible nuclear proteins regulates mitogen activation of both the interleukin-2 receptor-alpha gene and type 1 HIV. Cell 53:827-836. [DOI] [PubMed] [Google Scholar]
- 7.Bukrinsky, M. I., T. L. Stanwick, M. P. Dempsey, and M. Stevenson. 1991. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science 254:423-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. M. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chun, T.-W., L. Carruth, D. Finzi, X. Shen, J. A. Digiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y.-H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantitation of latent tissue reservoirs and total body load in HIV-1 infection. Nature 387:183-188. [DOI] [PubMed] [Google Scholar]
- 10.Chun, T.-W., D. Finzi, J. Margolick, K. Chadwich, D. Schwartz, and R. F. Siliciano. 1995. Fate of HIV-1-infected T cells in vivo: rates of transition to stable latency. Nat. Med. 1:1284-1290. [DOI] [PubMed] [Google Scholar]
- 11.Dayton, A. I., J. G. Sodroski, C. A. Rosen, W. C. Goh, and W. A. Haseltine. 1986. The trans-activator gene of the human T cell lymphotropic virus type III is required for replication. Cell 44:941-947. [DOI] [PubMed] [Google Scholar]
- 12.Douek, D. C., J. M. Brenchley, M. R. Betts, D. R. Ambrozak, B. J. Hill, Y. Okamoto, J. P. Casazza, J. Kuruppu, K. Kunstman, S. Wolinsky, Z. Grossman, M. Dybul, A. Oxenius, D. A. Price, M. Connors, and R. A. Koup. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 417:95-98. [DOI] [PubMed] [Google Scholar]
- 13.Duh, E. J., W. J. Maury, T. M. Folks, A. S. Fauci, and A. B. Rabson. 1989. Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-κB sites in the long terminal repeat. Proc. Natl. Acad. Sci. USA 86:5974-5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckstein, D. A., M. L. Penn, Y. D. Korin, D. D. Scripture-Adams, J. A. Zack, J. F. Kreisberg, M. Roederer, M. P. Sherman, P. S. Chin, and M. A. Goldsmith. 2001. HIV-1 actively replicates in naive CD4+ T cells residing within human lymphoid tissues. Immunity 15:671-682. [DOI] [PubMed] [Google Scholar]
- 15.Emiliani, S., W. Fischle, M. Ott, C. Van Lint, C. A. Amella, and E. Verdin. 1998. Mutations in the tat gene are responsible for human immunodeficiency virus type 1 postintegration latency in the U1 cell line. J. Virol. 72:1666-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felber, B. K., C. M. Drysdale, and G. N. Pavlakis. 1990. Feedback regulation of human immunodeficiency virus type 1 expression by the Rev protein. J. Virol. 64:3734-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 18.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 19.Fisher, A. G., M. B. Feinberg, S. F. Josephs, M. E. Harper, L. M. Marselle, G. Reyes, M. A. Gonda, A. Aldovini, C. Debouk, and R. C. Gallo. 1986. The trans-activator gene of HTLV-III is essential for virus replication. Nature 320:367-371. [DOI] [PubMed] [Google Scholar]
- 20.Folks, T. M., K. A. Clouse, J. Justement, A. Rabson, E. Duh, J. H. Kehrl, and A. S. Fauci. 1989. Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T-cell clone. Proc. Natl. Acad. Sci. USA 86:2365-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furtaldo, M. R., D. S. Callaway, J. P. Phair, B. S. Kunstman, J. L. Stanton, C. A. Macken, A. S. Perelson, and S. M. Wolinsky. 1999. Persistence of HIV-1 transcription in peripheral blood mononuclear cells in patients receiving potent antiretroviral therapy. N. Engl. J. Med. 340:1614-1622. [DOI] [PubMed] [Google Scholar]
- 22.Ghose, R., L. Y. Liou, C. H. Herrmann, and A. P. Rice. 2001. Induction of TAK (cyclin T1/P-TEFb) in purified resting CD4+ T lymphocytes by combination of cytokines. J. Virol. 75:11336-11343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunthard, H. F., D. V. Havlir, S. Fiscus, Z. Q. Zhang, J. Eron, J. Mellors, R. Gulick, S. D. Frost, A. J. Brown, W. Schleif, F. Valentine, L. Jonas, A. Meibohm, C. C. Ignacio, R. Isaacs, R. Gamagami, E. Emini, A. Haase, D. D. Richman, and J. K. Wong. 2001. Residual human immunodeficiency virus (HIV) type 1 RNA and DNA in lymph nodes and HIV RNA in genital secretions and in cerebrospinal fluid after suppression of viremia for 2 years. J. Infect. Dis. 183:1318-1327. [DOI] [PubMed] [Google Scholar]
- 24.Herrmann, C. H., R. G. Carroll, P. Wei, K. A. Jones, and A. P. Rice. 1998. Tat-associated kinase, TAK, activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J. Virol. 72:9881-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herrmann, C. H., and A. P. Rice. 1995. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J. Virol. 69:1612-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hockett, R. D., K. J. Michael, C. A. Derdeyn, M. S. Saag, M. Sillers, K. Squires, S. Chiz, M. A. Nowak, G. M. Shaw, and R. P. Bucy. 1999. Constant mean viral copy number per infected cell in tissues regardless of high, low, or undetectable plasma HIV RNA. J. Exp. Med. 189:1545-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hockett, R. D., Jr., K. M. Janowski, and R. P. Bucy. 1995. Simultaneous quantitation of multiple cytokine mRNAs by RT-PCR utilizing plate based EIA methodology. J. Immunol. Methods 187:273-285. [DOI] [PubMed] [Google Scholar]
- 28.Jones, K. A., and B. M. Peterlin. 1994. Control of RNA initiation and elongation at the HIV-1 promoter. Annu. Rev. Biochem. 63:713-743. [DOI] [PubMed] [Google Scholar]
- 29.Jordan, A., P. Defechereux, and E. Verdin. 2001. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 20:1726-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jung, A., R. Maier, J. P. Vartanian, G. Bocharov, V. Jung, U. Fischer, E. Meese, S. Wain-Hobson, and A. Meyerhans. 2002. Multiply infected spleen cells in HIV patients. Nature 418:144. [DOI] [PubMed] [Google Scholar]
- 31.Kao, S. Y., A. F. Calman, P. A. Luciw, and B. M. Peterlin. 1987. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330:489-493. [DOI] [PubMed] [Google Scholar]
- 32.Lewin, S. R., M. Vesanen, L. Kostrikis, A. Hurley, A. Duran, L. Zhang, D. D. Ho, and M. Markowitz. 1999. Use of real-time PCR and molecular beacons to detect virus replication in human immunodeficiency virus type 1-infected individuals on prolonged effective antiviral therapy. J. Virol. 73:6099-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malim, M. H., and B. R. Cullen. 1991. HIV-1 structural gene expression requires the binding of multiple rev monomers to the viral RRE: implications for HIV-1 latency. Cell 65:241-248. [DOI] [PubMed] [Google Scholar]
- 34.Malim, M. H., J. Hauber, R. Fenrick, and B. R. Cullen. 1988. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature 335:181-183. [DOI] [PubMed] [Google Scholar]
- 35.Malim, M. H., L. S. Tiley, D. F. McCarn, J. R. Rusche, J. Hauber, and B. R. Cullen. 1990. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell 60:675-683. [DOI] [PubMed] [Google Scholar]
- 36.Nabel, G., and D. Baltimore. 1987. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326:711-713. [DOI] [PubMed] [Google Scholar]
- 37.Persaud, D., T. Pierson, C. Ruff, D. Finzi, K. R. Chadwick, J. B. Margolick, A. Ruff, N. Hutton, S. Ray, and R. F. Siliciano. 2000. A stable latent reservoir for HIV-1 in resting CD4+ T lymphocytes in infected children. J. Clin. Investig. 105:995-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pierson, T., T. L. Hoffman, J. Blankson, D. Finzi, K. Chadwich, J. B. Margolick, C. Buck, J. D. Siliciano, R. W. Doms, and R. F. Siliciano. 2000. Characterization of chemokine receptor utilization of viruses in the latent reservoir for human immunodeficiency virus type 1. J. Virol. 74:7824-7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pierson, T. C., Y. Zhou, T. Kieffer, C. T. Ruff, C. Buck, and R. F. Siliciano. 2002. Molecular characterization of preintegration latency in infection. J. Virol. 76:8518-8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pomerantz, R. J., T. Seshamma, and D. Trono. 1992. Efficient replication of human immunodeficiency virus type 1 requires a threshold level of Rev: potential implications for latency. J. Virol. 66:1809-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pomerantz, R. J., D. Trono, M. B. Feinberg, and D. Baltimore. 1990. Cells nonproductively infected with HIV-1 exhibit an aberrant pattern of viral RNA expression: a molecular model for latency. Cell 61:1271-1276. [DOI] [PubMed] [Google Scholar]
- 42.Purcell, D. F., and M. A. Martin. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 67:6365-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramratnam, B., J. E. Mittler, L. Zhang, D. Boden, A. Hurley, F. Fang, C. A. Macken, A. S. Perelson, M. Markowitz, and D. D. Ho. 2000. The decay of the latent reservoir of replication competent HIV-1 is inversely correlated with the extent of residual viral replication during prolonged antiretroviral therapy. Nat. Med. 6:82-85. [DOI] [PubMed] [Google Scholar]
- 44.Schwartz, S., B. K. Felber, D. M. Benko, E. M. Fenyo, and G. N. Pavlakis. 1990. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol. 64:2519-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seshamma, T., O. Bagasra, D. Trono, D. Baltimore, and R. J. Pomerantz. 1992. Blocked early-stage latency in the peripheral blood cells of certain individuals infected with human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 89:10663-10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sodroski, J., W. C. Goh, C. Rosen, A. I. Dayton, E. Terwilliger, and W. Haseltine. 1986. A second post-transcriptional trans-activator gene required for HTLV-III replication. Nature 321:412-417. [DOI] [PubMed] [Google Scholar]
- 47.Spina, C. A., J. C. Guatelli, and D. D. Richman. 1995. Establishment of a stable, inducible form of human immunodeficiency virus type 1 DNA in quiescent CD4 lymphocytes in vitro. J. Virol. 69:2977-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tong-Starksen, S. E., P. A. Luciw, and B. M. Peterlin. 1987. Human immunodeficiency virus long terminal repeat responds to T-cell activation signals. Proc. Natl. Acad. Sci. USA 84:6845-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Unutmaz, D., V. N. KewalRamani, S. Marmon, and D. R. Littman. 1999. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J. Exp. Med. 189:1735-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vesanen, M., M. Markowitz, Y. Cao, D. D. Ho, and K. Saksela. 1997. Human immunodeficiency virus type-1 mRNA splicing pattern in infected persons is determined by the proportion of newly infected cells. Virology 236:104-109. [DOI] [PubMed] [Google Scholar]
- 51.Winslow, B. J., R. J. Pomerantz, O. Bagasra, and D. Trono. 1993. HIV-1 latency due to the site of proviral integration. Virology 196:849-854. [DOI] [PubMed] [Google Scholar]
- 52.Wong, J. K., M. Hezareh, H. F. Gunthard, D. V. Havlir, C. C. Ignacio, C. A. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291-1295. [DOI] [PubMed] [Google Scholar]
- 53.Wu, Y., and J. W. Marsh. 2001. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science 293:1503-1506. [DOI] [PubMed] [Google Scholar]
- 54.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Y. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213-222. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, J., and C. S. Crumpacker. 2001. Human immunodeficiency virus type 1 RNA in peripheral blood mononuclear cells of patients receiving prolonged highly active antiretroviral therapy. J. Infect. Dis. 184:1341-1344. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, L., B. Ramratnam, K. Tenner-Racz, Y. He, M. Vesanen, S. Lewin, A. Talal, P. Racz, A. S. Perelson, B. Korber, M. Markowitz, and D. D. Ho. 1999. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N. Engl. J. Med. 340:1605-1613. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, Z., T. Schuler, M. Zupancic, S. Wietgrefe, K. A. Staskus, K. A. Reimann, T. A. Reinhart, M. Rogan, W. Cavert, C. J. Miller, R. S. Veazey, D. Notermans, S. Little, S. A. Danner, D. D. Richman, D. Havlir, J. Wong, H. L. Jordan, T. W. Schacker, P. Racz, K. Tenner-Racz, N. L. Letvin, S. Wolinsky, and A. T. Haase. 1999. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science 286:1353-1357. (Erratum, 286:2273.) [DOI] [PubMed]
- 58.Zhu, T., D. Muthui, S. Holte, D. Nickle, F. Feng, S. Brodie, Y. Hwangbo, J. I. Mullins, and L. Corey. 2002. Evidence for human immunodeficiency virus type 1 replication in vivo in CD14+ monocytes and its potential role as a source of virus in patients on highly active antiretroviral therapy. J. Virol. 76:707-716. [DOI] [PMC free article] [PubMed] [Google Scholar]