Abstract
Rotavirus infection seems to be a multistep process in which the viruses are required to interact with several cell surface molecules to enter the cell. The virus spike protein VP4, which is cleaved by trypsin into two subunits, VP5 and VP8, is involved in some of these interactions. We have previously shown that the neuraminidase-sensitive rotavirus strain RRV initially attaches to a sialic acid-containing cell molecule through the VP8 subunit of VP4 and subsequently interacts with integrin α2β1 through VP5. After these initial contacts, the virus interacts with at least two additional proteins located at the cell surface, the integrin αvβ3 and the heat shock cognate protein Hsc70. In this work, we have shown that rotavirus RRV and its neuraminidase-resistant variant nar3 interact with Hsc70 through a VP5 domain located between amino acids 642 and 658 of the protein. This conclusion is based on the observation that a recombinant protein comprising the 300 carboxy-terminal amino acids of VP5 binds specifically to Hsc70 and a synthetic peptide containing amino acids 642 to 658 competes with the binding of the RRV and nar3 viruses to the heat shock protein. The VP5 peptide also competed with the binding to Hsc70 of the recombinant VP5 protein, and an antibody to Hsc70 reduced the binding of the recombinant protein to the surface of MA104 cells. The fact that the synthetic peptide blocks the infectivity of rotaviruses RRV and nar3 but not their binding to cells indicates that the interaction of VP5 with Hsc70 most probably occurs at a postattachment step during the virus entry process.
Rotaviruses, the single most important cause of severe dehydrating diarrhea in young children worldwide, are formed by a triple-layered protein capsid (17). The outermost layer, which is responsible for the initial interactions of the virus with the cell surface, consists of two proteins, VP7, a glycoprotein that forms the smooth surface of the virion, and VP4, which forms the spikes that extend from the surface of the virus particle (8). VP4 has an essential role during the early interactions of the virus with the cell surface, including receptor binding and cell penetration (1, 6, 7). To be infectious, rotaviruses depend on the proteolytic cleavage of VP4 (776 amino acids) into subunits VP8 (amino acids 1 to 247) and VP5 (amino acids 248 to 776); this cleavage does not affect cell binding (4, 9, 16) and has been associated with the entry of the virus into the cell's cytoplasm (4, 9, 16, 22, 27). The role of VP7 during the early interactions of the virus with the cell is not clear, although it has been shown that it can modulate some of the VP4-mediated virus phenotypes, including receptor binding (20).
In vivo, rotaviruses have a very specific cell tropism, infecting primarily mature enterocytes at the tip of intestinal villi, and the susceptibility of these cells seems to be limited to a narrow age window (17). In cell culture, the infectivity of some rotavirus strains has been shown to be sensitive to neuraminidase treatment of cells, indicating that the presence of sialic acid on the cell surface is relevant for virus infectivity. However, the interaction with sialic acid does not seem to be essential for rotavirus infection, since neuraminidase-resistant variants have been isolated from neuraminidase-sensitive rotavirus strains (19); in addition, some animal and most human rotavirus isolates are naturally neuraminidase resistant (3).
The establishment of multiple interactions between cell surface molecules and viruses seems to be a common theme during the cell infection process (28). Rotaviruses are not the exception, and at least four different cellular molecules have been proposed to interact sequentially with the virion during cell entry. Thus, it has been suggested that rotavirus strains that are sensitive to neuraminidase treatment of cells bind in the first place to a sialic acid-containing receptor, probably ganglioside GM3 (26). After this initial interaction, a second interaction with integrin α2β1, which is apparently shared by the neuraminidase-sensitive and neuraminidase-resistant strains, takes place (30). In addition to these two interactions, integrin αvβ3 and the heat shock protein Hsc70 have also been shown to be involved in rotavirus cell entry, most probably at a postattachment step (1, 10, 11).
We have previously characterized the binding to the surface of MA104 cells of the neuraminidase-sensitive simian rotavirus strain RRV and that of its neuraminidase-resistant variant nar3 (19), which no longer requires sialic acid to attach to and infect the cells. It was shown that RRV binds to a sialic acid-containing cell molecule through the VP8 domain of VP4 and subsequently interacts with integrin α2β1 through a DGE recognition motif located in the VP5 subunit of this protein (5, 30). On the other hand, the nar3 virus was found to initially interact with the cell surface through the VP5 DGE motif. In this work, we report that there is a second cell binding domain in VP5, different from the DGE integrin binding motif, which is used by both the RRV and nar3 viruses to interact with the heat shock protein Hsc70 on the surface of MA104 cells after their interaction with integrin α2β1.
MATERIALS AND METHODS
Cells and viruses.
MA104 cells were cultured in Eagle's minimal essential medium (MEM) supplemented with 10% fetal bovine serum. RRV was obtained from H. B. Greenberg, Stanford University, Stanford, Calif., and rotavirus variant nar3 has been described previously (19). RRV and nar3 were propagated in MA104 cells as described previously (25). Purified viral particles were prepared by CsCl gradients as described previously (31).
Antibodies.
H. B. Greenberg (Stanford University, Stanford, Calif.) generously provided the monoclonal antibodies (MAbs) 2G4, specific for VP5, and 159, directed against VP7. Nonneutralizing MAb HS2 directed to VP5 was described previously by Padilla-Noriega et al. (24). MAbs to integrins α2β1 (P1E6) and α4β1 (P1H4) were obtained from Chemicon and used at 10 μg/ml. The antibody to Hsc70 used in this work is a rabbit polyclonal serum described by Guerrero et al. (10); this serum was used at 80 μg/ml.
Proteins and peptides.
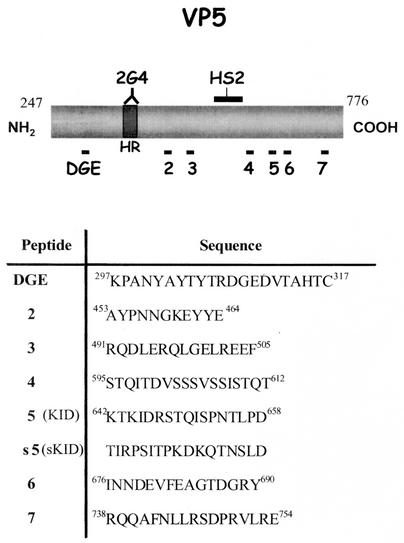
The cloning of RRV VP5 protein and the mutant VP5D308A as fusion proteins with glutathione S-transferase (GST) has been described previously (30, 31). These fusion proteins were expressed in Escherichia coli and purified by affinity chromatography as previously described by Isa et al. (14). VP5-His (amino acids 249 to 776), VP5-NH2 (amino acids 249 to 474), and VP5-COOH (amino acids 474 to 776) from rotavirus RRV VP4 and Hsc70 of human origin were cloned as fusions with a six-histidine tail at their carboxy termini in the pET 28 expression vector (Novagen) and affinity purified with the AKTA system on HiTrap chelating columns (Pharmacia). Peptides were chemically synthesized by Research Organics and used at the concentrations indicated in the figure legends. The amino acid sequences of the peptides used in this work are shown in Fig. 1.
FIG. 1.
Schematic representation of the RRV-VP5 protein. (A) The positions of the DGE tripeptide binding motif for α2β1 integrin and the hydrophobic region (HR) are shown. 2G4 and HS2 indicate the binding sites for these two MAbs. The relative positions of the synthetic peptides (numbered 2 to 7) used in this work are shown. (B) Amino acid sequence and amino acid positions of the synthetic peptides; s5 (sKID) is a scrambled version of peptide 5 (KID).
Infectivity assay.
MA104 cells grown in 96-well plates were washed twice with MEM, and 2,000 focus-forming units (FFU) of RRV or nar3 virus (previously activated with trypsin [10 mg/ml] for 30 min 37°C) were adsorbed to the cell surface for 1 h at 37°C. After the adsorption period, the virus inoculum was removed, the cells were washed twice with MEM, and cultures were maintained for 14 h at 37°C. Infected cell cultures were fixed and immunostained as previously described (2). The FFU were counted with the aid of a Visiolab 1000 station (12). This station, which was used for both image acquisition and analysis, is configured with a Matrox Meteor RGB frame grabber and a 8295 Cohu RGB charge-coupled device color TV camera. Motorized stages (Marzhauser, Berlin, Germany) were adapted to an inverted Nikon Diaphot 300 microscope. The stage control unit was a Marzhauser Multicontrol MC2000, piloted by Explo (Biocom, Paris, France). Macro command files for Explo were developed in order to perform semiautomated counting of the infected cells. In this manner, accurate positioning in the center of each well is achieved automatically for later predefined scanning and visual counting of infected cells within a selected well area. Usually, with this system, one tenth of the well is counted. When the infected cells are too scarce, the area counted is increased to one third of the well to obtain statistically significant numbers.
Binding assay.
MA104 cells grown in 48-well plates were washed twice and incubated with MEM without serum for 30 min at 37°C. After this time, the MEM was removed, and 500 μl of a solution of 1% of bovine albumin in phosphate-buffered saline (PBS) was added to the cells and incubated for 1 h at room temperature. The cells were then washed with an ice-cold solution of 0.5% bovine serum albumin in PBS and incubated with the indicated amount of virus or recombinant protein diluted in ice-cold MEM for 1 h at 4°C. After this time, the cells were washed three times with ice-cold PBS, and finally 120 μl of lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% TritonX-100) was added. The cells were frozen and thawed twice, and the amount of virus or protein present in the lysate was determined by an enzyme-linked immunosorbent assay (ELISA).
Capture ELISAs for rotavirus and rotavirus recombinant proteins.
To detect the virus, goat and rabbit antisera to rotavirus were used as capture (diluted 1:10,000) and detection (diluted 1:1,500) antibodies, respectively. The GST fusion proteins were captured with a goat antirotavirus serum and detected with rabbit polyclonal serum to GST (diluted 1:1,500). His tail proteins were captured with a goat antirotavirus serum and detected with MAb 2G4 for VP5-NH2 or MAb HS2 for VP5-His and VP5-COOH (both diluted 1:1,000). The corresponding alkaline phosphatase-conjugated anti-immunoglobulin was used (Kirkergaard and Perry, diluted 1:1,500), and finally the substrate (Sigma 104 [1 mg/ml]) diluted in diethanolamine buffer (100 mM diethanolamine [pH 9.4], 1 mM MgCl2, 5 mM sodium azide) was added. The absorbance at 405 nm was recorded in a Microplate Autoreader EL311 (Bio-Tek Instruments).
Binding to Hsc70.
To study the interaction between rotavirus particles and Hsc70, ELISAs were carried out essentially as described previously by Guerrero et al. (10). Briefly, 96-well ELISA plates (Costar) were coated with 500 ng of purified recombinant Hsc70 protein in PBS per well for 2 h at 37°C. To avoid nonspecific binding, 200 μl of a solution of 1% bovine serum albumin in PBS was added to the plate and incubated for 2 h at 37°C. The plates were washed twice and incubated with the indicated amount of peptide, protein, or virus for 1 h at 37°C, and the presence of the recombinant protein or virus was determined by using specific antibodies. To detect RRV, a rabbit polyclonal serum was used (diluted 1:1,500). The VP5-NH2 protein was detected with MAb 2G4 (diluted 1:1,000). The VP5-His and VP5-COOH proteins were detected with MAb HS2 (diluted 1:1,000). The corresponding alkaline phosphatase-conjugated antibodies and substrate were added as described above.
RESULTS
Using a recombinant VP5 protein expressed in bacteria as a fusion to GST, we previously showed that VP5 binds to integrin α2β1 on the surface of MA104 cells through a DGE motif located at amino acid residues 308 to 310 (the VP5 numbering used in this work corresponds to that of the complete RRV VP4 protein). The recombinant GST-VP5 protein was shown to block the binding of rotavirus strain nar3 when incubated with the cells before virus infection (31). In contrast, a mutant protein in which the integrin binding motif DGE was replaced by AGE (GST-VP5D308A), despite still being able to bind to the cell surface in a dose-dependent manner, was no longer able to block the binding of nar3. Furthermore, the binding of the mutant protein was not blocked by antibodies to integrin α2β1, as opposed to the binding of wild-type GST-VP5 (30). These data suggested that, in addition to the DGE motif, VP5 could also interact with the cell surface through a second binding site.
Synthetic peptides of VP5 block the infectivity of rotaviruses.
To evaluate if there was a functional domain on VP5, different from the DGE integrin binding motif, which might be involved in the initial interactions of the virus with the cell surface, a set of peptides representing discrete regions of VP5 were synthesized (Fig. 1), and the effect of these peptides on the infectivity of rotavirus RRV and its variant nar3 was tested. These peptides were chosen by considering the degree of conservation of a given region among the different VP4 rotavirus sequences that were available in GenBank in 1997 (57 sequences listed in reference 14) and on the solvent accessibility of a given region. These predictions were made by the profile-fed neural network systems from Heidelberg (PHD; http://www.embl-heidelberg.de/predictprotein/phd_pred.html). The peptides chosen were those that were the most conserved and the most accessible to the solvent according to the PHD program.
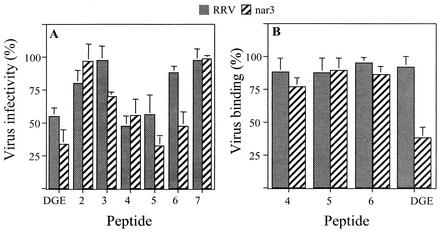
Incubation of MA104 cell monolayers with the various peptides before infection showed that peptides 4 and 5 were able to decrease the infectivity of both the RRV and nar3 viruses by about 50%, peptide 6 decreased the infectivity of nar3 but not that of RRV, and peptides 2, 3, and 7 did not have a significant effect on the infectivity of either virus (Fig. 2A). A peptide containing the α2β1 integrin binding motif DGE was used as control because we have already shown that this peptide is able to block the infectivity of the RRV and nar3 viruses by about 50 and 40%, respectively (30).
FIG. 2.
Effect of peptides on infectivity and binding of RRV and nar3 viruses. (A) MA104 cells grown in 96-well plates were incubated with 8 mg of the indicated peptides per ml for 1 h at 37°C, and then 2,000 FFU of the corresponding virus were added and adsorbed for 1 h at 37°C. The excess virus was removed, and the infection was allowed to proceed for 14 h at 37°C. Finally, the cells were fixed and immunostained as described in Materials and Methods. Data are expressed as the percentage of the virus infectivity obtained when the cells were incubated with PBS as a control. (B) MA104 cells grown in 48-well plates were incubated with the indicated peptides (8 mg/ml) for 1 h at 4°C, 500 ng of purified virus particles was added, and the cells were further incubated for 1 h at 4°C. The excess unbound virus was removed, and the amount of virus attached to the cells was determined by an ELISA. Data are expressed as the percentage of virus bound to the cells when the cells were incubated with PBS as a control. The arithmetic means and standard deviations for at least two independent experiments performed in duplicate are shown.
To determine if peptides 4, 5, and 6 decreased virus infectivity by preventing attachment of the viral particles to the cell surface, MA104 cells were incubated with peptides, 500 ng of purified virus was added, and the amount of virus attached to the cells was determined by an ELISA (see Materials and Methods). None of the peptides tested was found to affect the binding of either RRV or nar3 except peptide DGE, which, as previously reported (30), blocked the binding of nar3 but did not affect the cell attachment of RRV (Fig. 2B). The fact that peptides 4, 5, and 6 blocked rotavirus infectivity but did not affect the binding of the virus suggests that these peptides might be inhibiting an interaction between the virus and the cell surface that occurs at a postattachment step.
VP5 protein interacts with the cell surface through two different domains.
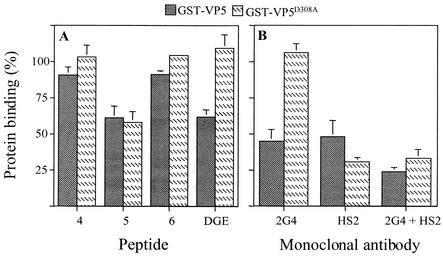
As mentioned, the characterization of the mutant recombinant protein GST-VP5D308A suggested the existence of a binding site in this protein in addition to the DGE motif (30). To determine if this additional site was represented by the synthetic peptides used in the previous experiments, a binding assay with the recombinant proteins was performed. In this assay, wild-type GST-VP5 and the mutant GST-VP5D308A proteins were added to cells that had been incubated with peptides 4, 5, 6, or DGE, and the amount of recombinant protein that remained attached to the cells was determined by an ELISA. We found that while peptide DGE blocked the binding of wild-type GST-VP5 by about 50%, peptide 5 blocked the binding of both the GST-VP5 and GST-VP5D308A proteins to a similar level (Fig. 3A). The two other peptides tested, peptides 4 and 6, did not significantly affect the binding of either recombinant protein. These results suggest that there could be a second cell binding domain on VP5, located in the region around amino acids 642 to 658, which is present in peptide 5 (hereafter called peptide KID).
FIG. 3.
Binding of GST-VP5 and GST-VP5D308A fusion proteins to cells in the presence of synthetic peptides and antibodies to VP5. (A) MA104 cell monolayers were incubated with 4 mg of the indicated peptides per ml for 1 h at 4°C, 1.5 μg of the purified recombinant proteins was added, and the cells were incubated for 1 h at 4°C. The amount of fusion protein bound to the cells was determined by an ELISA. Data are expressed as the percentage of protein bound to the cells when the cells were incubated with PBS as a control. (B) Recombinant GST-fusion proteins (1.5 μg) were incubated with the indicated MAbs for 1 h at room temperature, and this mixture was then added to monolayers of MA104 cells and incubated for 1 h at 4°C. The amount of bound protein was determined by an ELISA. Data are expressed as the percentage of protein bound to the cells when the proteins were incubated with PBS as a control. The arithmetic means and standard deviations for at least two independent experiments performed in duplicate are shown.
We also characterized the cell binding domains of VP5 with two MAbs. MAb 2G4 is a neutralizing antibody that recognizes an epitope located around amino acid residue 393 of VP4 (18), which has been shown to block the cell binding of nar3 as well as the binding of the wild-type GST-VP5 recombinant protein (31). MAb HS2 is a heterotypic, nonneutralizing antibody (24) that recognizes VP5 by both ELISA and immunoblot and which, by mutant deletion analysis, has been shown to bind VP5 in the region located between amino acids 540 and 593 (unpublished data) (Fig. 1). The effect of these two antibodies on the binding of the wild-type and mutant GST-VP5 proteins to the surface of MA104 cells was evaluated. When the recombinant proteins were incubated with MAb 2G4, the binding of wild-type GST-VP5 was found to be blocked by about 50%, while the attachment of GST-VP5D308A was unaffected. In contrast, MAb HS2 blocked the binding of both wild-type and mutant fusion proteins, although the latter protein was blocked to a larger extent (Fig. 3B). Incubation of the recombinant proteins with a mixture of both MAbs resulted in a more pronounced inhibition in the binding of wild-type GST-VP5, while the binding of GST-VP5D308A was very similar to that obtained in the presence of MAb HS2 alone. These results confirm that two distinct cell surface binding domains are present in the VP5 protein of rotaviruses.
Two domains of VP5 bind independently to the cell surface.
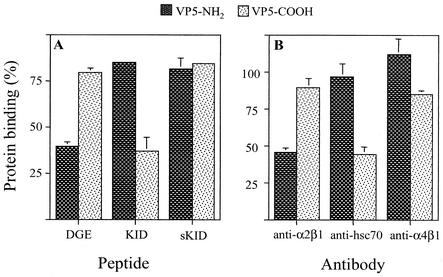
To find out if the two domains of VP5 bound independently of each other, the VP5 protein was expressed in E. coli as two separate halves. One construct, VP5-NH2, represents the amino-terminal half of VP5 (amino acids 248 to 474); this recombinant protein contains the integrin α2β1 binding motif DGE and the epitope recognized by MAb 2G4. The second construct, VP5-COOH, represents the carboxy-terminal half of VP5 (amino acids 474 to 776); it contains the region recognized by MAb HS2 and the region represented by peptide KID. Both proteins were expressed as fusions with a histidine tail and were purified by affinity chromatography. In a binding assay, both recombinant proteins were able to bind to the surface of MA104 cells to a similar extent (not shown). The binding of these proteins in the presence of peptides DGE, KID, and sKID (a scrambled version of peptide KID) was determined. We found that peptide DGE blocked the binding of protein VP5-NH2, without affecting the binding of VP5-COOH. Conversely, peptide KID blocked the binding of VP5-COOH without altering the attachment of the VP5-NH2 protein (Fig. 4A). The peptide sKID, used as a control, did not affect the binding of either recombinant protein. These results indicate that the two halves of VP5 are able to bind to the cell surface independently and specifically, probably attaching to different cell molecules.
FIG. 4.
Binding of recombinant VP5-NH2 and VP5-COOH proteins to cells in the presence of peptides and antibodies directed to cell surface molecules. MA104 cell monolayers were incubated with 4 mg of peptide per ml (A) or with antibodies against integrins α2β1 and α4β1 or to Hsc70 (B) for 1 h at 4°C. Then, 200 ng of purified recombinant VP5-NH2 and VP5-COOH proteins was added and incubated for 1 h at 4°C. The amount of protein bound to the cells was determined by an ELISA. Data are expressed as the percentage of protein bound to the cells when the cells were incubated with PBS as a control. The arithmetic means and standard deviations for at least two independent experiments performed in duplicate are shown.
Carboxy-terminal domain of VP5 interacts with the heat shock protein Hsc70.
To further characterize the cell binding of the carboxy-terminal half of VP5, we tested the capacity of antibodies to integrin α2β1 and to the heat shock protein Hsc70 to block the attachment of the recombinant proteins to the cell surface. An antibody to integrin α4β1 was also included because antibodies to this integrin have been shown to block virus infectivity, and it has been reported that VP5 contains a potential α4β1 binding domain (IDA tripeptide) at amino acids 538 to 540 (5, 13).
In this assay, MA104 cells were incubated with the indicated antibodies, and then either VP5-NH2 or VP5-COOH was added. The amount of protein that remained bound to the cell surface was determined by an ELISA. As expected, a MAb to integrin α2β1 decreased the binding of VP5-NH2 but did not affect the attachment of VP5-COOH. On the other hand, the antibody directed to Hsc70 was found to decrease the binding of the VP5-COOH protein, while it did not alter the attachment of VP5-NH2 (Fig. 4B). The MAb to integrin α4β1 did not significantly affect the binding of either protein.
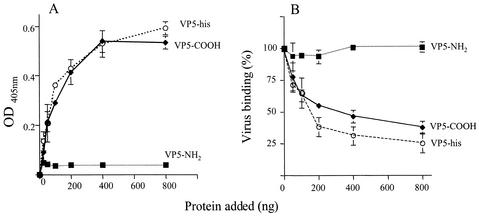
We have previously shown that triple-layered rotaviruses but not double-layered particles bind to purified Hsc70 protein immobilized on an ELISA plate (10). The same kind of assay was used to test if the VP5 protein was indeed able to bind to Hsc70. In this assay, recombinant proteins VP5-NH2 and VP5-COOH, and a full-length VP5 protein, also constructed as a fusion with a histidine tail (VP5-His), were used. While VP5-NH2 was found not to bind Hsc70, the complete VP5 protein and VP5-COOH bound in a saturable and concentration-dependent manner (Fig. 5A). We then tested whether these proteins were able to block the interaction of RRV with the heat shock protein. For this, an ELISA plate covered with Hsc70 was incubated with increasing amounts of the recombinant proteins, and purified RRV particles were added. The virus bound to the plate was detected with a MAb directed to VP7. As shown in Fig. 5B, the recombinant proteins VP5-His and VP5-COOH blocked the binding of rotavirus RRV to Hsc70 in a concentration-dependent manner, while protein VP5-NH2, as expected, had no effect on the binding of the virus.
FIG. 5.
Effect of the two domains of VP5 on binding of rotaviruses to Hsc70. (A) ELISA plates coated with Hsc70 (500 ng/well) were incubated for 1 h at 37°C with increasing amounts of recombinant VP5-His, VP5-NH2, or VP5-COOH proteins, and the amount of recombinant protein bound to Hsc70 was detected with MAbs to VP5 as described in Materials and Methods. The amount of recombinant protein added to each well is plotted against the reading for optical density at 405 nm (OD405) obtained in the ELISA plate. (B) In a parallel plate, purified RRV virus particles (300 ng/well) were added after removing the unbound recombinant proteins, and the plate was further incubated for 1 h at 37°C. The amount of virus bound to Hsc70 was determined with an antibody to VP7 rotavirus protein (MAb 159). Data are expressed as the percentage of virus bound to Hsc70 when this protein was incubated with PBS as control. The arithmetic means and standard deviations for at least two independent experiments performed in duplicate are shown.
Altogether, these data indicate that the VP5 protein of rotaviruses contains at least two domains that interact with the cell surface. One domain is represented by tripeptide DGE, at amino acids 308 to 310, which mediates the binding of the virus to integrin α2β1 (30), while the second is in a region located toward the carboxy terminus of VP5, at amino acids 642 to 658, and mediates the interaction of the virus with the heat shock protein Hsc70. This second interaction apparently takes place at a postattachment step during rotavirus infection.
DISCUSSION
Hsc70, a constitutive member of the heat shock-induced Hsp70 protein family that functions in normal cellular physiology, has been proposed to serve as a postattachment receptor for both neuraminidase-resistant and neuraminidase-sensitive rotavirus strains (10). In this work, we have shown that rotaviruses interact with Hsc70 through the VP5 subunit of VP4, and we also showed that this interaction is mediated by a VP5 domain located between amino acids 642 and 659 of the protein. This conclusion is based on the observation that a synthetic peptide which mimics this VP5 region (peptide KID) competes with the binding of the RRV and nar3 viruses to this protein. Peptide KID also competed with the binding to Hsc70 of the recombinant protein VP5-COOH, which contains the carboxy-terminal 300 amino acids of VP5. Furthermore, an antibody to Hsc70 reduced the binding of VP5-COOH to the surface of MA104 cells. The fact that peptide KID blocks the infectivity of rotaviruses RRV and nar3 but not their binding indicates that the interaction of VP5 with Hsc70 most probably occurs at a postattachment step, during the virus entry process. This finding is consistent with the previous observation that antibodies to neither Hsc70 nor the Hsc70 protein itself block the binding of rotaviruses to cells, although they efficiently inhibit virus infectivity (10).
The data described above indicate that the viruses are able to bind Hsc70 in the solid-phase ELISA despite the fact that the same viruses would appear not to bind Hsc70 on the cell surface. There are several possible explanations for these observations. (i) Hsc70 on the cell surface may not be accessible to directly bind the virus due to steric hindrance caused, for instance, by association with another cell membrane molecule. The initial binding of the virus to a different cell molecule could somehow favor making Hsc70 available to interact with the virus particle, or the initial interactions of the virus with the attachment receptor could induce a conformational change on VP4 so that it can interact more efficiently with Hsc70. (ii) The concentration of Hsc70 in the ELISA might be much higher than that on the cell surface; thus, if the binding of the virus to Hsc70 is not of high affinity, we would be able to detect virus-Hsc70 interaction in the ELISA but not on the cell binding assay. (iii) If the binding to sialic acid, in the case of rotavirus RRV, or to the primary receptor, for nar3, is more rapid because of a higher concentration of the primary receptor molecule on the cell surface or due to a higher affinity for this receptor compared to Hsc70, the virus will initially bind more efficiently to the primary receptor, and direct binding to cell surface Hsc70 would not be detected. On the other hand, the observation that the various recombinant VP5 proteins can apparently use the KID domain to attach directly to cells probably indicates either that this domain has the correct conformation on the recombinant proteins to interact efficiently with Hsc70 or that the size of the individual VP5 molecules might allow them to interact with the heat shock protein, overcoming a potential steric blockage.
Despite their typical nucleocytoplasmic residence, members of this family of chaperones have been reported to be present on the surface of several cells (21), including the MA104 cell line used in this work (10). However, the role of Hsc70 during rotavirus infection is not clear. As a postattachment cell receptor for rotaviruses, Hsc70 could serve only as an anchor on the membrane for the viruses during their transit to the cell's cytoplasm, or the chaperone function of the protein could be relevant for this event. It is tempting to speculate that the chaperone activity of Hsc70 plays a more active role during rotavirus entry, perhaps by triggering conformational changes on the particle that allow the virus to reach the cytoplasm or by promoting the uncoating of the viral particle. This idea is consistent with the known functions of the Hsp70 protein family, which include protein folding, translocation across biological membranes, and assembly and disassembly of oligomeric complexes. In particular, it is of interest that the ATP-dependent activity of Hsc70 has been described to be essential for the release of clathrin from endocytic clathrin-coated vesicles; thus, for instance, the expression of dominant negative mutants of Hsc70 blocks the recycling of the transferrin receptor because the clathrin coat cannot be removed from the vesicles (23).
Recently, using a phage display library of the VP4 protein of porcine rotavirus strain CRW8, Jolly et al. reported several regions of VP4 which were able to bind to the surface of MA104 cells. Three independent recombinant phages were found to contain VP4 sequences in the VP5 region between amino acids 650 and 657 (15), which is present in the peptide KID reported in this work. The identification of this VP5 cell binding domain by two different approaches and for two different virus strains supports its participation in the virus entry process. It is interesting, however, that the sequence of this region is somewhat variable among the different strains of rotavirus that have been sequenced so far, suggesting that the interaction between this region and Hsc70 is not strictly sequence specific.
The binding sites for MAbs 7A12 and 2G4, directed to the VP4 sialic acid binding site located in VP8 and the membrane permeabilization domain in VP5, respectively, were recently mapped by cryoelectron microscopy and difference map analysis (29). It was found that MAb 7A12 binds at the tips of the dimeric heads of VP4, while MAb 2G4 binds in the cleft between the two heads of the spike. These results, together with the secondary-structure prediction of the complete VP4 protein, suggest that the VP4 region that interacts with integrin α2β1 should be located near the head of the spike. On the other hand, based on these findings and on the fact that no neutralizing antibodies induced by the complete virus particle have been mapped to the carboxy-terminal region VP5, the domain represented by peptide KID would seem to be located at the bottom of the spike, probably near the surface VP7 protein layer. It is tempting to speculate that the initial contact of the virus with the cell surface triggers a conformational change in the viral particle that facilitates the interaction of the carboxy-terminal region of VP5 with Hsc70, an interaction that might be important for the virus to enter the cell. Further experiments to elucidate the role of Hsc70 in rotavirus infection are currently under way.
Acknowledgments
We are grateful to Paul Gaytán and Eugenio López for help in the synthesis of oligonucleotides.
This work was partially supported by grants 55003662 and 55000613 from the Howard Hughes Medical Institute, G37621N from the National Council for Science and Technology-Mexico, and IN201399 from DGAPA-UNAM.
REFERENCES
- 1.Arias, C. F., C. A. Guerrero, E. Mendez, S. Zárate, P. Isa, R. Espinosa, P. Romero, and S. Lopez. 2001. Early events of rotavirus infection: the search for the receptor(s). Novartis Found. Symp. 238:47-60. [DOI] [PubMed] [Google Scholar]
- 2.Arias, C. F., M. Lizano, and S. López. 1987. Synthesis in Escherichia coli and immunological characterization of a polypeptide containing the cleavage site associated with trypsin enhancement of rotavirus SA11 infectivity. J. Gen. Virol. 68:633-642. [DOI] [PubMed] [Google Scholar]
- 3.Ciarlet, M., and M. K. Estes. 1999. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J. Gen. Virol. 80:943-948. [DOI] [PubMed] [Google Scholar]
- 4.Clark, S. M., J. R. Roth, M. L. Clark, B. B. Barnett, and R. S. Spendlove. 1981. Trypsin enhancement of rotavirus infectivity: mechanism of enhancement. J. Virol. 39:816-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coulson, B. S., S. H. Londrigan, and D. J. Lee. 1997. Rotavirus contains integrin ligand sequences and a disintegrin-like domain implicated in virus entry into cells. Proc. Natl. Acad. Sci. USA 94:5389-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dector, M. A., P. Romero, S. Lopez, and C. F. Arias. 2002. Rotavirus gene silencing by small interfering RNAs. EMBO Rep. 3:1175-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estes, M. K. 1996. Rotaviruses and their replication, p. 1625-1655. In B. N. Fields, D. N. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Virology, vol. 2. Raven Press, New York, N.Y.
- 8.Estes, M. K., and J. Cohen. 1989. Rotavirus gene structure and function. Microbiol. Rev. 53:410-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukuhara, N., O. Yoshie, S. Kitaoka, and T. Konno. 1988. Role of VP3 in human rotavirus internalization after target cell attachment via VP7. J. Virol. 62:2209-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerrero, C. A., D. Bouyssounade, S. Zárate, P. Isa, T. López, R. Espinosa, P. Romero, E. Méndez, S. López, and C. F. Arias. 2002. The heat shock cognate protein 70 is involved in rotavirus cell entry. J. Virol. 76:4096-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerrero, C. A., E. Mendez, S. Zarate, P. Isa, S. Lopez, and C. F. Arias. 2000. Integrin αvβ3 mediates rotavirus cell entry. Proc. Natl. Acad. Sci. USA 97:14644-14649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerrero, C. A., S. Zárate, G. Corkidi, S. López, and C. F. Arias. 2000. Biochemical characterization of rotavirus receptors in MA104 cells. J. Virol. 74:9362-9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hewish, M. J., Y. Takada, and B. S. Coulson. 2000. Integrins α2β1 and α4β1 can mediate SA11 rotavirus attachment and entry into cells. J. Virol. 74:228-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isa, P., S. Lopez, L. Segovia, and C. F. Arias. 1997. Functional and structural analysis of the sialic acid-binding domain of rotaviruses. J. Virol. 71:6749-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jolly, C. L., J. A. Huang, and I. H. Holmes. 2001. Selection of rotavirus VP4 cell receptor binding domains for MA104 cells using a phage display library. J. Virol. Methods 98:41-51. [DOI] [PubMed] [Google Scholar]
- 16.Kaljot, K. T., R. D. Shaw, D. H. Rubin, and H. B. Greenberg. 1988. Infectious rotavirus enters cells by direct cell membrane penetration, not by endocytosis. J. Virol. 62:1136-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapikian, A. Z., and R. M. Chanock. 1996. Rotaviruses, p. 1657-1708. In B. N. Fields, D. N. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Virology, vol. 2. Raven Press, New York, N.Y.
- 18.Mackow, E. R., R. D. Shaw, S. M. Matsui, P. T. Vo, M. N. Dang, and H. B. Greenberg. 1988. The rhesus rotavirus gene encoding protein VP3: location of amino acids involved in homologous and heterologous rotavirus neutralization and identification of a putative fusion region. Proc. Natl. Acad. Sci. USA 85:645-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mendez, E., C. F. Arias, and S. Lopez. 1993. Binding to sialic acids is not an essential step for the entry of animal rotaviruses to epithelial cells in culture. J. Virol. 67:5253-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendez, E., C. F. Arias, and S. Lopez. 1996. Interactions between the two surface proteins of rotavirus may alter the receptor-binding specificity of the virus. J. Virol. 70:1218-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Multhoff, G., and L. E. Hightower. 1996. Cell surface expression of heat shock proteins and the immune response. Cell Stress Chaperones 1:167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nandi, P., A. Charpilliene, and J. Cohen. 1992. Interaction of rotavirus particles with liposomes. J. Virol. 66:3363-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newmyer, S. L., and S. L. Schmid. 2001. Dominant-interfering Hsc70 mutants disrupt multiple stages of the clathrin-coated vesicle cycle in vivo. J. Cell Biol. 152:607-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Padilla-Noriega, L., R. Werner-Eckert, E. M. Mackow, M. Gorziglia, G. Larralde, K. Taniguchi, and H. B. Greenberg. 1993. Serologic analysis of human rotavirus serotypes P1A and P2 by using monoclonal antibodies. J. Clin. Microbiol. 31:622-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pando, V., P. Isa, C. F. Arias, and S. Lopez. 2002. Influence of calcium on the early steps of rotavirus infection. Virology 295:190-200. [DOI] [PubMed] [Google Scholar]
- 26.Rolsma, M. D., T. B. Kuhlenschmidt, H. B. Gelberg, and M. S. Kuhlenschmidt. 1998. Structure and function of a ganglioside receptor for porcine rotavirus. J. Virol. 72:9079-9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruiz, M. C., S. R. Alonso-Torre, A. Charpilliene, M. Vasseur, F. Michelangeli, J. Cohen, and F. Alvarado. 1994. Rotavirus interaction with isolated membrane vesicles. J. Virol. 68:4009-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider-Schaulies, J. 2000. Cellular receptors for viruses: links to tropism and pathogenesis. J. Gen. Virol. 81:1413-1429. [DOI] [PubMed] [Google Scholar]
- 29.Tihova, M., K. A. Dryden, R. Bellamy, H. B. Greenberg, and M. Yeager. 2001. Localization of membrane permeabilization and receptor binding sites on the VP4 hemagglutinin of rotavirus: implications for cell entry. J. Mol. Biol. 314:985-992. [DOI] [PubMed] [Google Scholar]
- 30.Zárate, S., R. Espinosa, P. Romero, C. A. Guerrero, C. F. Arias, and S. Lopez. 2000. Integrin a2B1 mediates the cell attachment of the rotavirus neuraminidase-resistant variant nar3. Virology 278:50-54. [DOI] [PubMed] [Google Scholar]
- 31.Zárate, S., R. Espinosa, P. Romero, E. Méndez, C. F. Arias, and S. López. 2000. The VP5 domain of VP4 can mediate attachment of rotaviruses to cells. J. Virol. 74:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]