Abstract
The virus-encoded nonstructural protein 5B (NS5B) of hepatitis C virus (HCV) is an RNA-dependent RNA polymerase and is absolutely required for replication of the virus. NS5B exhibits significant differences from cellular polymerases and therefore has become an attractive target for anti-HCV therapy. Using a high-throughput screen, we discovered a novel NS5B inhibitor that binds to the enzyme noncompetitively with respect to nucleotide substrates. Here we report the crystal structure of NS5B complexed with this small molecule inhibitor. Unexpectedly, the inhibitor is bound within a narrow cleft on the protein's surface in the “thumb” domain, about 30 Å from the enzyme's catalytic center. The interaction between this inhibitor and NS5B occurs without dramatic changes to the structure of the protein, and sequence analysis suggests that the binding site is conserved across known HCV genotypes. Possible mechanisms of inhibition include perturbation of protein dynamics, interference with RNA binding, and disruption of enzyme oligomerization.
Hepatitis C virus (HCV), a member of the family Flaviviridae, is a positive single-stranded RNA virus that is the primary causative agent of parenterally transmitted non-A/non-B hepatitis (9). It is estimated that there are 170 million chronically infected HCV carriers worldwide (2 to 3% of the global population), and many of these individuals are expected to develop serious HCV-related liver diseases, including hepatocellular carcinoma. Unfortunately, clinically proven prophylactic and/ortherapeutic anti-HCV vaccines are not presently known, and currently utilized HCV treatments (combinations of various interferons and the nucleoside analog ribavirin) are associated with suboptimal response rates and high incidences of side effects. Thus, there is an urgent need to identify and develop additional antiviral agents to improve the effectiveness and tolerability of HCV therapy. Recent research has focused on the structural and functional aspects of essential HCV replication components in order to define useful targets.
The HCV genome comprises approximately 9,600 nucleotides, which encode a polyprotein precursor of ∼3,000 amino acid residues. Co- and posttranslational proteolytic cleavage of this polypeptide by cellular and viral enzymes yields the specific HCV proteins required for virus replication and assembly (recently reviewed in reference 32). The nonstructural protein NS5B has been characterized as an RNA-dependent RNA polymerase (RdRp) based on in vitro experiments using recombinant material (13, 21), and its activity was shown to be required for HCV infectivity in chimpanzees (16). NS5B is a 66-kDa membrane-associated protein containing motifs shared by all RdRps, including the active site Gly-Asp-Asp sequence, which is believed to bind magnesium ions and is essential for enzymatic activity. It has a highly hydrophobic C-terminal region that is responsible for membrane anchoring (14, 33, 35). Crystal structures of unligated NS5B that incorporate a 21- or 55-residue C-terminal deletion to improve solubility have been independently reported by three groups (2, 6, 20). These studies show that the protein contains canonical features found in most polynucleotide polymerases, such as three structural domains denoted “fingers,” “palm,” and “thumb.” However, the NS5B structures, along with those of polymerases derived from bacteriophage φ6 (8) and rabbit hemorrhagic disease virus (27), demonstrate the unique architecture of the viral RdRp class, which includes connected finger and thumb domains. Because NS5B is structurally distinct from related mammalian DNA and RNA polymerase enzymes, it represents an attractive target for the development of novel anti-HCV agents (3).
Reports of ligand-bound forms of NS5B include the observation of a nucleoside triphosphate (NTP) moiety and two metals within the active site (2, 5), as well as the binding of rGTP to a site on the surface of the enzyme at the interface between the finger and thumb domains (5). The interaction of these ligands with NS5B was not found to inhibit the enzyme's catalytic activity, although rGTP could potentially play a regulatory role. A recent description of noncompetitive, nonnucleoside inhibitors of NS5B suggests that the enzyme possesses binding sites outside the catalytic region, which could be exploited for purposes of drug design (11). However, no reports have provided structural details of interactions between NS5B inhibitors and the polymerase. Here we describe efforts that led to the crystal structure of a novel NS5B-inhibitor complex, present details of the observed protein-ligand interactions, and propose possible mechanisms for the inhibition of the NS5B enzyme.
MATERIALS AND METHODS
Mutagenesis, expression, and purification.
The HCV NS5B construct employed for this study was the BK isolate (genotype 1b), which had a 21-amino-acid C-terminal deletion and a six-histidine C-terminal tag (12, 35). The truncated gene construct was cloned into a proprietary Escherichia coli expression vector, and protein expression was controlled by a Lac promoter/suppressor system. Site-directed mutagenesis was performed with this plasmid as a template. The presence of the desired single-site mutation K114R or K106Q single mutant (SM) or the L47Q/F101Y/K114R triple mutant (TM) was confirmed by complete cDNA sequencing. Protein expression was carried out in the DH5a strain of E. coli and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 30°C. The E. coli cell pellets were lysed by microfluidization (Microfluidizer; Microfluidics, Newton, Mass.) in lysis buffer (20 mM Tris [pH 8.0], 5 mM MgCl2, 300 mM NaCl, 10% glycerol, 12 mM 2-mercaptoethanol, 1× complete EDTA-free protease inhibitor tablets [Roche Molecular Biochemicals]) followed by ultracentrifugation. The supernatant was loaded on a SuperFlow Ni-nitrilotriacetic acid affinity column (Qiagen), and the HCV polymerase was eluted with an imidazole gradient (60 to 250 mM). The protein was further purified by SP Fast Flow ion-exchange chromatography and Sephacryl S-100 gel filtration chromatography (Amersham Pharmacia Biotechnology). The purified proteins were concentrated to 15 to 30 mg/ml in a buffer containing 10 mM HEPES (pH 7.5), 400 mM NaCl, and 2 mM TCEP [Tris(2-carboxyethyl) phosphine hydrochloride]; quick-frozen in liquid N2; and stored at −80°C prior to use.
Inhibitor synthesis.
Compound 1 was prepared by coupling 6-cyclopentyl-4-hydroxy-6-[2-(4-hydroxyphenyl)ethyl]-5,6-dihydro-pyran-2-one with toluene-4-thiosulfonic acid S-(4-amino-2-tert-butyl-5-methylphenyl) ester or the corresponding hydrochloride salt by methods analogous to those described for synthesizing related 5,6-dihydropyran-2-ones (4, 30): 1H NMR (CDCl3, 300 MHz) δ 1.28 to 1.85 (m, 8H), 1.54 (s, 9H), 1.83 (s, 3H), 2.02 to 2.09 (m, 2H), 2.40 to 2.46 (m, 1H), 2.61 to 2.67 (m, 2H), 2.73 (d, 1H, J = 18.0 Hz), 2.93 (d, 1H, J = 18.0 Hz), 6.68 to 6.73 (m, 4H), 6.95 to 6.98 (m, 2H).
Assay of polymerase and inhibitor.
Recombinant HCV NS5B polymerase was tested for its ability to perform primer/template-directed transcription in assays that contained 30 mM Tris-HCl (pH 7.2), 10 mM MgCl2, 20 mM NaCl, 1 mM dithiothreitol, 0.05% Tween 20, 1% glycerol, 5 pmol of biotin-dG12 (primer), 0.5 pmol of poly(rC)300 (template), 1 μM GTP, 0.1 to 0.3 μCi of [α-32P]GTP, and 2.5 pmol (0.15 μg) of HCV polymerase protein in a final volume of 75 μl. Reactions were initiated by addition of enzyme and incubated for 30 min at 30°C. Reactions were stopped by addition of 33 mM EDTA, and polynucleotide products were collected by filtration through DEAE Filtermat papers (Wallac); unincorporated triphosphate was removed by washing the filters with 5% dibasic sodium phoshate. The filters were counted in a Packard Tri-Lux Microbeta scintillation counter. Compounds to be tested were added at various concentrations from stocks in 10% dimethyl sulfoxide (DMSO)-water (final DMSO concentration = 1% of the reaction mixture). Fifty percent inhibitory concentration (IC50) values were estimated from the primary cpm data (collected in triplicate) by using the formula cpm(I) = cpm (no inhibitor) · {1 − [I]/([I] + IC50)}. Km and Vmax values at inhibitor concentrations of 0, 1, 2, and 4 μM were calculated from the data shown in Fig. 1D by using the formula velocity (S) = Vmax {[S]/([S] + Km)}.
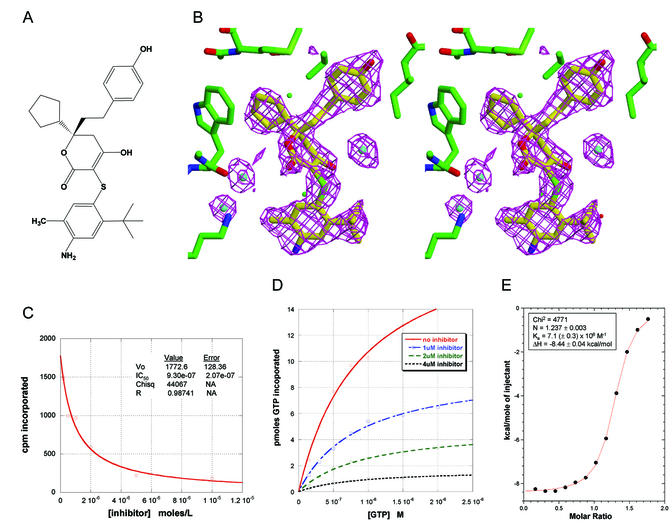
FIG. 1.
Characterization of the inhibitor used to cocrystallize with HCV NS5B polymerase. (A) Structure of racemic inhibitor 1. (B) Calculated 2.2-Å omit-refine (Fobs − Fcalc) electron density map generated with inhibitor and nearby water molecules removed, contoured at a 3σ level. (C) NS5B polymerase activity as a function of inhibitor concentration. IC50 was calculated as described in Materials and Methods. (D) NS5B polymerase activity as a function of GTP concentration and inhibitor concentration. Km and Vmax were calculated for each curve as described in Materials and Methods. For no inhibitor, Km = 0.89 μM GTP and Vmax = 20.6; for 1 μM inhibitor, Km = 0.88 μM GTP and Vmax = 9.5; for 2 μM inhibitor, Km = 0.95 μM GTP and Vmax = 5.0; for 4 μM inhibitor, Km = 0.82 μM GTP and Vmax = 1.7. (E) Isothermal titration calorimetry data for the binding of inhibitor 1, as described in Materials and Methods.
Biophysical analysis.
Binding of inhibitor 1 to HCV NS5B polymerase was measured by isothermal titration calorimetry with a MicroCal VP-ITC. The polymerase was prepared by dialysis against a mixture containing 20 mM HEPES (pH 7.0), 20 mM NaCl, 5 mM MgCl2, 1.0 mM dithiothreitol, and 2.0% glycerol. The ligand solution was prepared by diluting stock inhibitor 1 (in DMSO) with dialysis buffer (final concentration, 2.0% DMSO [vol/vol]). Inhibitor 1 at a concentration of 200 μM was titrated into 10 μM NS5B at 15°C. To minimize the heat of dilution, an appropriate volume of DMSO was first added to the dialyzed polymerase. The calorimetric data were analyzed with the ORIGIN program (MicroCal). Prior to analysis, a correction factor was subtracted from the data to correct for the heat of dilution.
Crystallization.
Initial crystallization studies of uncomplexed enzyme by using a 21-amino-acid C-terminal deletion of wild-type NS5B (BK isolate) resulted in thin crystals with a high propensity for twinning. Single-site mutations of K106Q and K114R improved crystal quality, and the X-ray structures for these mutants were solved by isomorphous replacement and multiwavelength anomalous diffraction methods (22), since NS5B coordinates were unavailable at the time. Additional mutations were then chosen based on surface exposure of hydrophobic amino acids in the SM structures. A TM construct, K114R/L47Q/F101Y, was incubated with inhibitor 1 for 1 h at 4°C and then mixed with precipitating reagents at equal volumes and equilibrated with well solutions by the hanging drop vapor diffusion method. The conditions under which crystals grew are as follows: (i) 0.2 M ammonium sulfate, 0.1 M ammonium acetate, and 30% monomethyl polyethylene glycol 2000 (pH 4.6); (ii) 2.0 M ammonium sulfate only (pH 5.0); or (iii) 1.2 to 1.8 M potassium phosphate and 100 mM sodium citrate (pH 5.5). Tetragonal crystals appeared at 20°C after about 1 week. The crystals belong to space group P41212, with unit cell dimensions of a = b = 83 Å and c = 180 Å, and contain 1 molecule per asymmetric unit.
Diffraction analysis.
The tetragonal crystals of the TM NS5B-inhibitor complex described above were used to collect X-ray diffraction data at 100 K on a Mar345 image plate with a Rigaku rotating anode X-ray generator. Data were processed with Denzo/Scalepack (29). Statistics for data over 20 to 2.2 Å (2.3 to 2.2 Å) were as follows: completeness, 97% (72); Rmerge = 3.4% (10.8), <I/σI> = 26 (15), and redundancy of measurements, 6.7 (3). Our previously determined structures of K106Q and K114R NS5B (see above) served as molecular replacement models with AmoRe (26). The atomic coordinates of the TM enzyme were refined by using XPLOR (7), and electron density for the bound inhibitor was identified. Additional refinement was done with ARP/wARP (25), which improved the definition of several surface protein loops and identified water molecules. Model refitting was done with XFIT (23). Refinement of the enzyme-inhibitor complex (4,338 nonhydrogen atoms for protein amino acids 1 to 563 and ligand; 560 water molecules) by using XPLOR over 10 to 2.2 Å gave a crystallographic Rwork of 20.7% (29,165 reflections with F > 2σF) and Rfree of 25.1% (1,535 reflections, or 5%), with root mean square deviation (r.m.s.d.) in bond length and bond angle of 0.008 Å and 1.53°, respectively. The average isotropic temperature factors were 18.1 Å2 for protein atoms and 19.5 Å2 for inhibitor atoms. All amino acids lie in the allowed Ramachandran regions according to PROCHECK (19). Coordinates have been deposited with the Protein Data Bank (accession no. 1O55).
Figure preparation.
Figure 1B was prepared with XFIT and Raster3D (24). Figure 2B was made with Insight II (Molecular Simulations, 1998). Figure 2A, 3, and 5A were made with Molscript (17), and Raster3D. Figure 5B was made with GRASP (28). For Fig. 4, homology assessment was done by using BK NS5B to search the pFAM database (Washington University). Only full-length HCV NS5B sequences contained in the viral_RdRp family were used for this analysis.
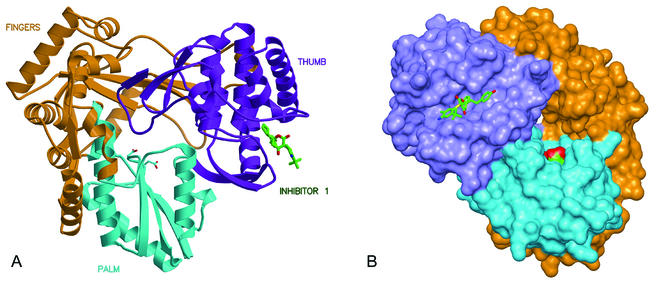
FIG. 2.
Overall views of HCV NS5B polymerase with bound inhibitor 1. (A) Ribbon representation of NS5B with domains colored according to thumb (purple; residues 371 to 563), palm (blue; residues 188 to 227 and 287 to 370), and fingers (orange; residues 1 to 187 and 228 to 286). Active site aspartic acids 318 and 319 are visible at the center of the molecule in the palm domain. (B) Molecular surface of NS5B with bound inhibitor 1, rotated ∼90° from view in panel A.
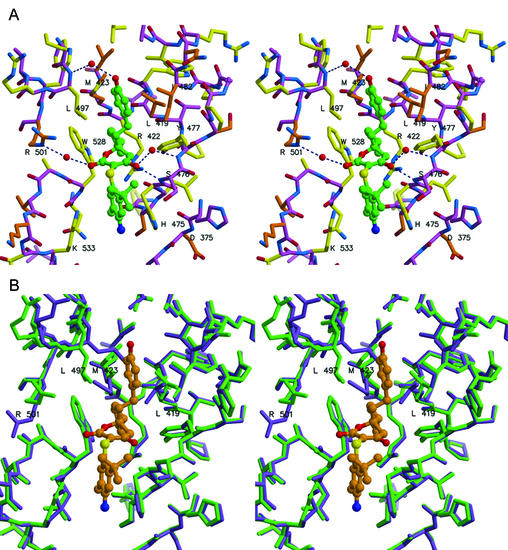
FIG. 3.
Stereo views of inhibitor 1 bound to NS5B. (A) Emphasis on amino acid conservation among HCV genotypes, with yellow side chains for highly conserved residues (>99% occurrence) and orange side chains for type-conserved residues. The inhibitor's carbon atoms are colored green. (B) Adjustments at the binding site upon inhibitor binding. Superposition of NS5B before (purple) and after (green) inhibitor binding. Inhibitor carbon atoms are colored orange. Several side chains are found to rearrange (see labeled residues), and main chain atoms at residues 496 to 499 shift by ∼1 Å.
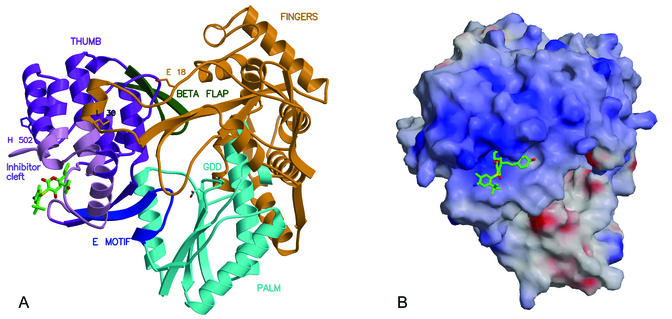
FIG. 5.
(A) Location of several motifs proposed to play roles in NS5B function: The E motif (blue) connects the thumb and palm domains; β-loop (positions 443 to 454; green) is a flap covering the active site; one finger loop (orange) extends across to interact tightly with the thumb, particularly at Leu 30. Residues defining the thumb's inhibitor-binding cleft (light purple) either contact these motifs directly or are linked to them through secondary structure elements. In addition, inhibitor 1 lies near His 502, which is reported to play a role in functional dimerization of NS5B (along with Glu 18). (B) Electrostatic potential surface for NS5B (blue, positive; red, negative), showing an extensive region of conserved positive charge adjacent to the thumb's inhibitor binding site. If this region plays a role in the binding of oligonucleotide and/or nucleotides during RNA elongation, then the presence of a bound inhibitor may interfere sterically or electrostatically.
FIG. 4.
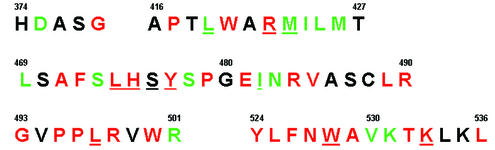
Residues defining the long cleft on the NS5B thumb in which inhibitor 1 is bound. The sequence shown is for the HCV BK isolate. Amino acids in red are >99% identical across 222 HCV genotypes. Residues in green are type conserved among HCV sequences (e.g., Ile, Leu, or Val at that position). Underlined residues have at least one atom within 4.2 Å of bound inhibitor 1.
RESULTS AND DISCUSSION
Inhibitor identification.
High-throughput screening of the Pfizer proprietary compound archive identified a novel racemic NS5B inhibitor whose chemical structure is shown in Fig. 1A. Compound 1 demonstrated inhibition of NS5B polymerization activity with a calculated IC50 of 0.93 μM (Fig. 1C). Subsequent biochemical experiments demonstrated that the Km for GTP did not vary significantly with inhibitor concentration, while the observed Vmax did (Fig. 1D). In addition, isothermal titration calorimetry analysis indicated specific binding of the compound to the purified NS5B enzyme, with a measured Kd of 0.14 μM and an inhibitor-enzyme stoichiometry close to 1:1 (Fig. 1E). Collectively, these data are consistent with specific, reversible binding of compound 1 to NS5B in a manner that noncompetitively inhibits the enzyme's activity with respect to GTP. We therefore initiated efforts to elucidate crystallographic details of the enzyme-inhibitor complex.
Crystallization strategy and structure determination.
Critical to obtaining crystals of the NS5B-inhibitor 1 complex was a strategy of site-directed mutagenesis aimed at improving the crystallization properties of the enzyme, starting from a 21-amino-acid C-terminal deletion of wild-type NS5B (BK isolate). Initial mutagenesis focused on changing nonconserved surface lysines to arginine or glutamine, since the latter are favored in crystal lattice contacts over lysine (10). Single mutations of K106Q and K114R led to an enhancement of crystal quality for uncomplexed NS5B, and the X-ray structures for these mutants were solved (see Materials and Methods). Additional mutations were then designed to further improve properties of the protein, based on the SM crystal structures. Phe 101 and Leu 47 were selected for modification based on the exposure of their hydrophobic side chains to solvent. A TM construct, K114R/L47Q/F101Y, ultimately proved most successful in NS5B inhibitor cocrystallization trials. For the TM NS5B construct, polymerase activity and inhibition by compound 1 (IC50 = 1.2 μM) were not significantly different from those found with wild-type enzyme. Importantly, none of the mutated TM residues are located near where inhibitor 1 was subsequently shown to bind to the enzyme.
We determined the structure of the TM NS5B-inhibitor 1 complex by molecular replacement using our unligated structures of SM NS5B and then refined the complex at 2.2-Å resolution (see Materials and Methods). The TM NS5B enzyme is very similar to the already reported NS5B structures and to the SM structures (superimposed Cα r.m.s.d. of 0.54 and 0.35 Å, respectively). The observed electron density for inhibitor 1 (Fig. 1B) is consistent with the enol tautomer of the compound (i.e., sp2 hybridization of the dihydropyran-2-one carbon atom bonded to the molecule's sulfur moiety). However, due to the crystallographic similarity between oxygen and carbon atoms at this resolution, the electron density alone cannot establish which, if any, enantiomer of compound 1 (transposition of enol and ester functional groups) is preferentially bound to NS5B.
Description of bound inhibitor.
The location of inhibitor 1 with respect to the entire NS5B protein is depicted in Fig. 2. Unexpectedly, the compound is found partially buried within a long cleft that extends nearly the entire length of the thumb's surface and eventually connects with the finger domain. This cleft is approximately 30 Å long, 10 Å wide, and 10 Å deep. The complete surface of the cleft can be defined by NS5B residues 374 to 378, 416 to 427, 469 to 490, 493 to 501, and 524 to 536. The inhibitor occupies the central portion of the extended cleft, and detailed interactions between inhibitor and enzyme are shown in Fig. 3A. The interface consists primarily of nonpolar surfaces along with several hydrogen bonds. The most intimate interaction involves the compound's cyclopentyl ring, which fits into a small, complementary, hydrophobic pocket defined by main chain and side chain atoms of Met 423, Trp 528, Leu 419, Tyr 477, and Arg 422 (alkyl portion of side chain). One edge of the hydroxyphenyl ring of the inhibitor rests on side chains of Met 423, Leu 419, Leu 489, and Val 485 (the “floor” of the cleft), while the ring's faces are surrounded by side chains of Leu 497 and Ile 482 (the “walls” of the cleft). The compound's phenol moiety forms a water-mediated hydrogen bond to the backbone amide of Leu 497. The enol oxygen of the central dihydropyran-2-one ring makes a direct hydrogen bond to the backbone amide of Ser 476 (distance, 2.8 Å) and a water-mediated hydrogen bond to the amide of Tyr 477. The carbonyl oxygen of the inhibitor's dihydropyran-2-one forms a second water-mediated hydrogen bond with Arg 501. The phenylsulfanyl ring of compound 1 is observed to make few interactions with the NS5B protein (e.g., parallel stacking with the imidazole of His 475). The above description of protein-ligand interactions also applies if the other enantiomer of compound 1 is considered (transposition of enol and ester functional groups).
Inhibitor-induced changes.
The structures for inhibitor-bound TM NS5B and unbound TM NS5B derived from the same crystal form are very similar. Superposition of the two structures reveals no interdomain shifts, minor rearrangement of a few thumb amino acid side chains (Leu 497, Met 423, Leu 419, and Arg 501) surrounding the inhibitor, and a small shift (∼1 Å) in residues 496 to 499, which lead into α-helix residues 497 to 513 (Fig. 3B). Thumb domain Cα atoms (residues 371 to 563) of the complexed and uncomplexed enzymes superimpose with an r.m.s.d. of only 0.29 Å. Thus, in contrast to the relatively dramatic structural distortions effected by the binding of nonnucleoside inhibitors to the human immunodeficiency virus (HIV) reverse transcriptase (RT) enzyme (reviewed in reference 15), interaction of compound 1 with HCV NS5B produces minimal changes in overall protein architecture.
Conservation of binding site.
The possible existence of the inhibitor-binding cleft on NS5B proteins from other HCV genotypes was evaluated by homology analysis. The sequences of amino acids comprising the BK construct's extended cleft are shown in Fig. 4. Of these, about 50% are invariant among 222 HCV polymerase sequences studied, and this number increases to 66% if conservation of amino acid type is considered. Examination of residues immediately surrounding bound inhibitor 1 reveals an especially high number of invariant or conserved amino acids (Fig. 3A). Therefore, this inhibitor binding site may retain a high degree of three-dimensional similarity across NS5B enzymes derived from known HCV genotypes. Consistent with this possibility, we have found similar activity for inhibitor 1, using NS5B enzymes derived from several HCV genotypes other than type 1b (data not shown). No analogous cleft exists in the thumb domain of other polymerases (cellular or viral) for which three-dimensional structures are known. This is primarily due to the smaller size of the thumb domain in related polymerases, as well as a different arrangement of secondary structure. Thus, NS5B inhibitors that interact with the described binding site have the potential to function as selective, broad-spectrum anti-HCV agents.
Mechanism of inhibition.
Several hypotheses can be offered to explain the inhibition of NS5B by compound 1 in the absence of gross structural changes induced by ligand binding. First, the presence of a molecule bound in the thumb's cleft may perturb dynamic properties of the thumb domain, or of interdomain contacts, that are necessary for normal enzymatic function. Possibilities include the following: (i) the thumb β-loop (residues 443 to 454), which is proposed to move and/or interact with RNA during elongation (2, 6, 20); (ii) the E motif (residues 360 to 370) and several subsequent residues located at the thumb-palm junction, which may contact the nascent primer strand (20) and may serve as a hinge for rotations of the thumb domain analogous to those in RdRp from rabbit hemorrhagic disease virus (27) or in C-terminal deletion constructs of NS5B (1); and (iii) the finger-thumb interface created by extended finger domain loops (primarily 11 to 45, but also 139 to 160), the disturbance of which by mutation of Leu 30 reduces enzymatic activity (18). In each case, the noted NS5B motif or interface is linked to secondary structural elements that form part of or lie adjacent to the inhibitor-binding cleft (Fig. 5A).
Second, recent studies suggest that NS5B can form oligomers in the absence or presence of RNA (18, 31, 34) and that this property is important for enzyme function in vitro and in vivo. At least two protein residues have been identified as necessary for homomeric association of NS5B: Glu 18 and His 502 (31). His 502 lies along one wall of the thumb's inhibitor-binding cleft (Fig. 5A). If a complementary intermolecular interface is indeed required for enzyme function, then the presence of a small molecule bound on the surface near that interface might perturb the association.
Third, a bound compound may interfere with important protein-RNA interactions, should these occur on the outer thumb region. The amino acid content of the thumb is unusually rich in arginines and lysines, and mutagenesis experiments have suggested a functional role for several arginines in the range of 500 to 505 (35). The spatial arrangement of basic side chains on the thumb has the appearance of a patch of positive charge that lies parallel and adjacent to the inhibitor-binding cleft described here (Fig. 5B). If this patch plays a role in the binding of polynucleotide, then a bound small molecule may interfere with such interactions. Interestingly, the patch includes a low-affinity but specific rGTP binding site (estimated Kd of 0.25 to 0.45 mM) proposed to be an allosteric regulatory site responsible for the enhancement of NS5B RNA synthesis at high rGTP concentrations (5). Part of this rGTP site forms a wall near one end of the thumb's extended cleft, about 10 to 15 Å beyond the phenol moiety of inhibitor 1. While none of the residues defining the rGTP site are in contact with inhibitor 1, several are involved in small structural shifts that accommodate the inhibitor's binding (Pro 496 through Val 499; Fig. 3B). Therefore, the inhibitor's presence might perturb features of the adjacent site that are necessary for interaction with rGTP. However, at the low GTP concentrations of our assay (∼1 μM), rGTP allosteric regulation is unlikely to contribute significantly to the measured NS5B polymerase activity. Thus, perturbation of the rGTP site does not readily explain the observed NS5B inhibition by compound 1.
Finally, as in other NS5B structures with a 21-amino-acid C-terminal deletion (1, 2, 20), residues 545 to 563 of our NS5B structure are found to occupy the enzyme's putative RNA-binding cleft (Fig. 2A) and presumably must reposition to allow polymerase function. The presence of residues 545 to 570 has been shown to have an inhibitory effect on the activity of purified NS5B (1). Since residues 524 to 536 upstream of the C terminus participate in defining the binding site for inhibitor 1, it is conceivable that the small molecule acts indirectly to stabilize the C terminus at the active site. However, this explanation appears improbable, because we find equivalent inhibition with compound 1 when using a 55-amino-acid C-terminal deletion construct of NS5B (data not shown), the crystal structure of which reveals no occlusion of the enzyme's central cavity by the C terminus (1, 6).
Following the discovery of compound 1, we determined the cocrystal structures of numerous other NS5B inhibitors (both chemically related and unrelated to compound 1) with the TM enzyme. These additional inhibitors are observed to bind NS5B at a position similar to that of compound 1, and the character of ligand-protein interactions correlates well with biochemical and biophysical data. We therefore believe that, in analogy to the strategy previously developed for nonnucleoside HIV RT inhibitors, a structure-based approach for the design of compounds that noncompetitively inhibit HCV NS5B through binding at the described thumb site offers the potential to deliver novel, enzyme-specific agents for HCV therapy.
ADDENDUM IN PROOF
Recently, crystal structures were published for HCV NS5B in complex with inhibitors that bind noncompetitively to the enzyme in the same general location as compound 1 (M. Wang et al., J. Biol. Chem. 278:9489-9495, 2003).
Acknowledgments
We thank D. Matthews for a critical reading of the manuscript.
Note that publication of the information contained in this article shall not be construed as granting or conveying an assignment, license, or right under any patent owned, controlled, or licensed by Pfizer, Inc., or its subsidiaries.
REFERENCES
- 1.Adachi, T., H. Ago, N. Habuka, K. Okuda, M. Komatsu, S. Ikeda, and K. Yatsunami. 2002. The essential role of C-terminal residues in regulating the activity of hepatitis C virus RNA-dependent RNA polymerase. Biochim. Biophys. Acta 1601:38-48. [DOI] [PubMed] [Google Scholar]
- 2.Ago, H., T. Adachi, A. Yoshida, M. Yamamoto, N. Habuka, K. Yatsunami, and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure 7:1417-1426. [DOI] [PubMed] [Google Scholar]
- 3.Beaulieu, P. L., and M. Llinas-Brunet. 2002. Therapies for hepatitis C infection: targeting the non-structural proteins of HCV. Curr. Med. Chem. 1:163-176. [Google Scholar]
- 4.Boyer, F. E., J. V. Vara Prasad, J. M. Domagala, E. L. Ellsworth, C. Gajda, S. E. Hagen, L. J. Markoski, B. D. Tait, E. A. Lunney, A. Palovsky, D. Ferguson, N. Graham, T. Holler, D. Hupe, C. Nouhan, P. J. Tummino, A. Urumov, E. Zeikus, G. Zeikus, S. J. Gracheck, J. M. Sanders, S. VanderRoest, J. Brodfuehrer, K. Iyer, M. Sinz, and S. V. Gulnik. 2000. 5,6-Dihydropyran-2-ones possessing various sulfonyl functionalities: potent nonpeptidic inhibitors of HIV protease. J. Med. Chem. 43:843-858. [DOI] [PubMed] [Google Scholar]
- 5.Bressanelli, S., L. Tomei, F. A. Rey, and R. De Francesco. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brünger, A. 1992. XPLOR v3.1. A system for X-ray crystallography and NMR. Yale University Press, New Haven, Conn.
- 8.Butcher, S. J., J. M. Grimes, E. V. Makeyev, D. H. Bamford, and D. I. Stuartt. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235-240. [DOI] [PubMed] [Google Scholar]
- 9.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from blood-borne non-A, non-B viral hepatitis. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 10.Dasgupta, S., G. H. Iyer, S. H. Bryant, C. E. Lawrence, and J. A. Bell. 1997. Extent and nature of contacts between protein molecules in crystal lattices and between subunits of protein oligomers. Proteins 28:494-514. [DOI] [PubMed] [Google Scholar]
- 11.Dhanak, D., K. J. Duffy, V. K. Johnston, J. Lin-Goerke, M. Darcy, A. N. Shaw, B. Gu, C. Silverman, A. T. Gates, M. R. Nonnemacher, D. L. Earnshaw, D. J. Casper, A. Kaura, A. Baker, C. Greenwood, L. L. Gutshall, D. Maley, A. DelVecchio, R. Macarron, G. A. Hofmann, Z. Alnoah, H.-Y. Cheng, G. Chan, S. Khandekar, R. M. Keenan, and R. T. Sarisky. 2002. Identification and biological characterization of heterocyclic inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 277:38322-38327. [DOI] [PubMed] [Google Scholar]
- 12.Ferrari, E., J. Wright-Minogue, J. W. S. Fang, B. M. Baroudy, J. Y. N. Lau, and Z. Hong. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagedorn, C. H., E. H. Van Beers, and C. De Staercke. 2000. Hepatitis C virus RNA-dependent RNA polymerase (NS5B polymerase). Curr. Top. Microbiol. Immunol. 242:225-260. [DOI] [PubMed] [Google Scholar]
- 14.Ivashkina, N., B. Wölk, V. Lohmann, R. Bartenschlager, H. E. Blum, F. Penin, and D. Moradpour. 2002. The hepatitis C virus RNA-dependent RNA polymerase membrane insertion sequence is a transmembrane segment. J. Virol. 76:13088-13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonckheere, H., J. Anne, and E. De Clercq. 2000. The HIV-1 reverse transcription (RT) process as target for RT inhibitors. Med. Res. Rev. 20:129-154. [DOI] [PubMed] [Google Scholar]
- 16.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 18.Labonte, P., V. Axelrod, A. Agarwal, A. Aulabaugh, A. Amin, and P. Mak. 2002. Modulation of hepatitis C virus RNA-dependent RNA polymerase activity by structure-based site-directed mutagenesis. J. Biol. Chem. 277:38838-38846. [DOI] [PubMed] [Google Scholar]
- 19.Laskowski, R. J., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 20.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 21.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Love, R. A., X. Yu, W. Diehl, M. J. Hickey, H. E. Parge, J. Gao, and S. Fuhrman. 2002. Hepatitis C virus (HCV) NS5B RNA polymerase and mutants thereof. European patent application EP 1256628.
- 23.McRee, D. E. 1992. XtalView: a visual protein crystallographic system for X11/Xview. J. Mol. Graph. 10:44-47. [Google Scholar]
- 24.Merrit, E. A., and D. J. Bacon. 1997. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 25.Morris, R. J., A. Perrakis, and V. S. Lamzin. 2002. ARP/wARP's model-building algorithms. I. The main chain. Acta Crystallogr. Sect. D 58:968-975. [DOI] [PubMed] [Google Scholar]
- 26.Navaza, J. 1994. AMoRe: an automated package for molecular replacement. Acta Crystallogr. Sect. A 50:157-163. [Google Scholar]
- 27.Ng, K. K. S., M. M. Cherney, A. L. Vazquez, A. Machnin, J. M. M. Alonso, F. Parra, and M. N. G. James. 2002. Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase. J. Biol. Chem. 277:1381-1387. [DOI] [PubMed] [Google Scholar]
- 28.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski, Z., and W. Minor. 1997. Processing X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 30.Prasad, J. V., F. E. Boyer, J. M. Domagala, E. L. Ellsworth, C. Gajda, H. W. Hamilton, S. E. Hagen, L. J. Markoski, B. A. Steinbaugh, B. D. Tait, C. Humblet, E. A. Lunney, A. Pavlovsky, J. R. Rubin, D. Ferguson, N. Graham, T. Holler, D. Hupe, C. Nouhan, P. J. Tummino, A. Urumov, E. Zeikus, G. Zeikus, S. J. Gracheck, J. W. Erickson et al. 1999. Nonpeptidic HIV protease inhibitors possessing excellent antiviral activities and therapeutic indices. PD 178390: a lead HIV protease inhibitor. Bioorg. Med. Chem. 7:2775-2800. [DOI] [PubMed] [Google Scholar]
- 31.Qin, W., H. Luo, T. Nomura, N. Hayashi, T. Yamashita, and S. Murakami. 2002. Oligomeric interaction of hepatitis C virus NS5B is critical for catalytic activity of RNA-dependent RNA polymerase. J. Biol. Chem. 277:2132-2137. [DOI] [PubMed] [Google Scholar]
- 32.Rosenberg, S. 2001. Recent advances in the molecular biology of hepatitis C virus. J. Mol. Biol. 313:451-464. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt-Mende, J., E. Bieck, T. Hugle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276:44052-44063. [DOI] [PubMed] [Google Scholar]
- 34.Wang, Q. M., M. A. Hockman, K. Staschke, R. B. Johnson, K. A. Case, J. Lu, S. Parsons, F. Zhang, R. Rathnachalam, K. Kirkegaard, and J. M. Colacino. 2002. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76:3865-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamashita, T., S. Kaneko, Y. Shirota, W. Qin, T. Nomura, K. Kobayashi, and S. Murakami. 1998. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 273:15479-15486. [DOI] [PubMed] [Google Scholar]