Abstract
Herpes simplex virus type 1 (HSV-1) is a DNA virus that acquires an envelope by budding into the inner nuclear membrane of an infected cell. Recombinant HSV-1 lacking the UL34 gene cannot undergo this event. UL34 and UL31, another viral protein, colocalize in an infected cell and are necessary and sufficient to target both proteins to the inner nuclear envelope. In order to define and characterize sequences of UL34 that are necessary for primary envelopment to occur, a library of 19 UL34 charged cluster mutants and a truncation mutant lacking the putative transmembrane domain (ΔTM) were generated. Mutants in this library were analyzed in a complementation assay for their ability to function in the production of infectious virus. Seven of the mutants failed to complement a UL34-null virus. The remainder of the mutants complemented at or near wild-type UL34 levels. Failure of a mutant protein to function might be the result of incorrect subcellular localization. To address this possibility, confocal microscopy was used to determine the localization of the UL34 protein in charged cluster mutants and ΔTM. In transfection-infection experiments, all of the functional UL34 mutants and four of the six noncomplementing mutants localized to the inner nuclear envelope in a manner indistinguishable from that of wild-type UL34. All of the noncomplementing UL34 mutants mediated proper localization of UL31. Charged clusters critical for UL34 function are dispersed throughout the protein sequence and do not correlate well with highly conserved regions of the protein. These data suggest that UL34 has at least one function in addition to mediating proper localization of UL31 in infected cells and provide further support for the role of UL34 in mediating proper localization of UL31 in infected cells.
Herpes simplex virus type 1 (HSV-1) undergoes primary envelopment, budding from the interior of the nucleus into the perinuclear space, as one of the first steps in egress from the cell (10, 29, 34). Herpesvirus egress is a complex process involving many virus-encoded proteins (2-4, 12, 13, 18, 21, 26), but only the UL34 protein and its homologs have been shown to be required for primary envelopment in HSV-1 (35), pseudorabies virus (22), herpes simplex virus type 2 (HSV-2) (39), and most recently in equine herpesvirus type 1 (28). The HSV-1 UL34 gene product is a 30-kDa membrane-associated phosphoprotein that is a substrate for the virus-encoded protein kinase US3 (31, 32). The UL34 sequence contains a putative transmembrane domain of 14 to 17 amino acids near the C terminus of the protein. While localization and membrane association of the UL34 protein suggest that UL34 contains a transmembrane anchor, biochemical tests have not been done to verify the existence of such a domain. In addition, the interaction of UL34 with another HSV-1 protein, the UL31 gene product, is necessary and sufficient to target both proteins to the nuclear membrane, where they are thought to form a complex (33). Colocalization of UL34 and UL31 at the nuclear membrane is also seen in HSV-2 and pseudorabies virus (16, 43). Since the UL34 protein coding sequence is well conserved in alphaherpesviruses for which sequences are available (11, 15, 40), it seems likely that UL34 protein function is also widely conserved among herpesviruses.
Efficient primary envelopment in alphaherpesviruses is linked to complete packaging of DNA into the capsid. Many viral mutants that synthesize capsids but fail to correctly package their DNA are unable to envelop the empty capsids (1, 6, 7, 19, 25, 30, 36). Once genome packaging is complete, the nucleocapsid must gain access to the inner nuclear membrane, and this, in turn, likely requires a mechanism to break down the nuclear lamina. It is currently unknown how HSV-1 gains access to the nuclear membrane, but several lines of evidence suggest that herpesvirus infection leads to alterations of nuclear envelope structure. Infection with HSV-1 has been reported to lead to an increase in soluble lamin A/C, suggesting at least limited disassembly of the lamina (38).
The UL34 homolog in murine cytomegalovirus, a betaherpesvirus, recruits cellular protein kinase C to the nuclear lamina and induces lamin phosphorylation (27). Because lamin phosphorylation leads to a breakdown of the nuclear lamina during the normal cell processes of mitosis and apoptosis (17), it is logical to hypothesize that cytomegalovirus exploits lamin phosphorylation to promote access of nucleocapsids to the inner nuclear membrane. Overexpressed UL34 alters the morphology of the nuclear membrane so that the inner and outer nuclear membranes are separated (45). This separation at least superficially resembles the effects of HSV-1 infection on nuclear membrane morphology. Experiments with equine herpesvirus type 1 show that at early times in infection UL34 is evenly distributed at the nuclear rim, but localization changes to a punctate and filamentous pattern later in infection, further indicating that UL34 may be affecting the structure of the nuclear envelope (28).
Having gained access to the inner nuclear membrane, the capsid becomes wrapped in the inner nuclear membrane in the perinuclear space (18). One simple mechanism for this process might entail direct or indirect interaction between a protein on the exterior of the capsid and a protein anchored in the inner nuclear membrane. The protein(s) that mediates direct binding of the capsid to the nuclear membrane has not been determined, but a recombinant UL34 fusion protein has been shown to interact with VP5, the major capsid protein of HSV-1 (45). Once the capsid has been wrapped in the inner nuclear membrane, membrane scission is necessary to form a free enveloped virion between the two nuclear membranes. The accumulation of partially enveloped capsids in cells infected with a UL11 deletion virus, along with the presence of a possible PPPY consensus motif in the UL11 protein, suggests that the UL11 protein may facilitate this event (24). In retroviruses, the PPPY motif in envelope proteins is thought to recruit proteins involved in vesicle scission to the site of budding (23). The involvement of UL11 in vesicle scission does not, however, rule out the possibility that other proteins that localize to the nuclear membrane during viral infection, such as UL34, could also be involved in this process.
Based on established properties of the UL34 protein and experimental data regarding early events in viral envelopment, UL34 may participate in primary envelopment in multiple ways. UL34, either alone or in a complex with UL31, may interact directly with capsids and with the inner nuclear membrane to initiate the wrapping of the capsid. Alternatively, UL34 could recruit other viral and/or cellular proteins necessary for binding of capsids to the nuclear envelope. UL34 may also play a role in the membrane scission event that completes primary envelopment. In addition, UL34 could be facilitating a disruption of the nuclear lamina, allowing the capsid access to the inner nuclear membrane. These hypotheses are not mutually exclusive.
In this study we constructed a library of UL34 mutants and analyzed them by a complementation assay and confocal microscopy.
MATERIALS AND METHODS
Cells and viruses.
Vero, 143/1099E (derived from human osteocarcinoma 143B cell line), and HEp-2 cells were maintained as previously described (33, 35). The properties of HSV-1(F) and vRR1072 (thymidine kinase positive) have also been described previously (33).
Plasmids.
Plasmid pRR1072rep, which was used as the parent vector for the charged cluster mutants and ΔTM, was constructed by replacing the NcoI-BspEI fragment of pRR1072 (35) containing the green fluorescent protein (GFP) gene with the NcoI-BspEI fragment from pRR1099 containing the wild-type UL34 sequence.
Three methods were used to introduce mutations into the UL34 coding sequence. Mutants CL01, CL02, CL05, and CL06 were created by PCR megapriming (37). This method uses the product of the first PCR to prime the second PCR. Mutants CL03, CL04, CL07, CL08, CL09, CL10, CL11, CL12, and CL16 were constructed by unique-site elimination mutagenesis (5) as described in the Transformer site-directed mutagenesis kit (Clontech). Mutants CL13, CL14, CL15, CL17, CL18, and CL19 were constructed by site-directed PCR mutagenesis. Primers flanking the UL34 gene along with primers in the middle of the gene that contained the mutation and introduced a restriction site were used to amplify the gene in two pieces. After cutting with a restriction enzyme, the two halves of the gene were ligated and amplified by primers flanking the gene. The primers used for UL34 mutagenesis, the restriction site introduced by mutagenesis, and the amino acids changed in the charged cluster mutants are shown in Table 1.
TABLE 1.
Primers used for site-directed mutagenesis of UL34a
| Charged cluster mutant | Primer sequence (5′→3′) | Restriction site introduced | Wild-type amino acid sequence | Mutant amino acid sequence |
|---|---|---|---|---|
| CL01 | GAACGAGACCCgCGAAGGCGgCACCTGGGTGGCC | HphIb | GDAFE | GAAFA |
| CL02 | GGGACGATAAGcgcAATTgcCTGAAGCAGACCCTCG | MfeI | QRIRL | QAIAL |
| CL03 | CGTCCCATCTACGTTGgccGGCGGGGcCGGGGAGGCG | NgoMIV | LRGGD | LAGGA |
| CL04 | ACGTTGCGGGGCGGGGcCGGGGctGCaGGCCCCTACTCTCCC | PstI | DGEAG | AGAAG |
| CL05 | GGGAAACGACgCGgCGGACCCGgCATGGCCATGAAAC | PleIb | DGSDE | AGSAA |
| CL06 | GCCCAGTCGTTCATAAGCgctAgcACATACgCGATGGGAAACG | NheI | IEYVL | IAYAL |
| CL07 | ATCGAGTATGTACTGgcGCTTATGAACGcgTGGGCCGAGGTCCCG | MluI | RLMND | ALMNA |
| CL08 | CTGTCCCTCGGCGcgCTaGcCACCATCAAGGGG | NheI | GDLDT | GALAT |
| CL09 | GGCGACCTGGcCACCATCgcGGGGCGGCTCG | MscI | LDTIK | LATIA |
| CL10 | CCTGGACACCATCgcGGGagcGCTCGGCCTGG | AfeI | IKGRL | IAGAL |
| CL11 | CCATCAAGGGGgcGCTCGGCCTGGcgGCCCGGCCGATG | HaeII | RLGLD | ALGLA |
| CL12 | GCGGCTCGGCCTGGcTGCggcGCCGATGATGGCCAG | KasI | LDARP | LAAAP |
| CL13 | R:ATCAGCTGCTTTGTGgcCATGCCtgcaGTGCAGCTCGCGTTT | PstI | VRMPR | VAMPA |
| L:CAAACGCGAGCTGCACtgcaGGCATGgcCACAAAGCAGCTGA | ||||
| CL14 | L:CAGGATCCGTCTCGTCgcTCCGGCtgCagCGGGGCCCATGAACCG | PstI | EDAGR | AAAGA |
| R:CGGTTCATGGGCCCCGctGcaGCCGGAgcGACGAGACGGATCCTG | ||||
| CL15 | R:CCCGAAGATGCCGGAgcGACtgcAgcGATCCTGTGCCGCGCCG | PstI | GRTGG | GATAA |
| L:CGGCGCGGCACAGGATCgcTgcaGTCgcTCCGGCATCTTCGGG | ||||
| CL16 | GCCGGACGGACGAGAgcGATCCTGTGCgcCGCCGCCGAGCAGGCT | BamHIb | RILCR | AILCA |
| CL17 | R:GAGCAGGCTATTACCgcTgcCgcTgcAACCCGGCGGTCCCGG | HpyCH4IV | RRRRT | RRSRE |
| L:CCGGGACCGCCGGGTTgcAgcGgcAgcGGTAATAGCCTGCTC | ||||
| CL18 | R:CGTCGCCGCCGAACCgcGgcGTCCgctGcaGCGTACGGGGCCGAG | PstI | RRSRE | AASAA |
| L:CCGGCCTCGGCCCCGTACGCtcGagcGGACgcCgcGGTTCGGCGGCG ACGGGTAA | ||||
| CL19 | R:GCCGGAACGGGTTTCgcGGCtgcaGGGGcCGGTTTTGGCCCGCTC | PstI | RARGD | AAAGD |
| L:GAGCGGGCCAAAACCGgCCCCtgcaGCCgcGAAACCCGTTCCGGC |
Lowercase letters indicate bases that differ from the wild-type HSV-1 sequence.
Restriction site was eliminated in the mutant.
The ΔTM mutant plasmid was constructed by inserting the double-stranded oligonucleotide produced by annealing ΔTM1 (5′-TTAAGCACCTACGGATTGGCCCCCCCGCGTAA-3′) and ΔTM2 (5′-TTAATTACGCGGGGGGGCCAATCCGTAGGTGC-3′) into the AflII site in pRR1072Rep.
UL34 complementation assay.
We transfected 24-well cultures of Vero cells at 70% confluence with 0.125 μg of pCMVβ, expressing the β-galactosidase gene, and 0.125 μg of wild-type or mutant UL34 plasmid with Lipofectamine as described by the manufacturer (Gibco-BRL) and incubated at 37°C overnight. The cells were then infected with 10 PFU of the UL34 deletion virus vRR1072 per cell and incubated at 37°C for 90 min. Monolayers were washed three times with Dulbecco's modified Eagle's medium and then washed three times with pH 3 sodium citrate buffer (50 mM sodium citrate, 4 mM potassium chloride, adjusted to pH 3 with hydrochloric acid) to eliminate residual virus. Cells were washed with V medium (Dulbecco's modified Eagle's medium, penicillin-streptomycin, 1% heat-inactivated calf serum) until the pH returned to a normal level (about two to three times).
One milliliter of V medium was added to each well, and after 18 h of incubation at 37°C, cell lysates were prepared by freezing and thawing followed by sonication for 20 s at power level 2 with a Fisher sonic dismembrator. The amount of infectivity in each lysate was determined by titration on 143/1009E cells as described previously (35). Part of each cell lysate was assayed for β-galactosidase expression as described below.
To assay for β-galactosidase activity, 100 μl of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM β-mercaptoethanol, adjusted to pH 7) containing 0.27% β-mercaptoethanol and 0.2% Triton X-100 was added to 100 μl of cell lysate, and the mixture was vortexed for 30 s. Cell debris was pelleted by centrifuging at 14,000 rpm in a microcentrifuge for 30 s. Then 160 μl of the supernatant was transferred to a 96-well plate and reacted with 40 μl of 4-mg/ml o-nitrophenyl-β-d-galactopyranoside (ONPG) stock at 30°C. The reactions were stopped by adding 80 μl of 1 M NaCO3, and the absorbance was read at 420 nm.
Western blotting.
HEp-2 cells were transfected with 0.25 μg of a charged cluster mutant plasmid and 0.25 μg of pCMVβ with Lipofectamine (as described above) and incubated at 37°C overnight. The cells were then infected with the UL34-null virus at a multiplicity of infection (MOI) of 5 for 16 h. For the charged cluster mutants, the total cellular protein was harvested with a small-scale, whole-cell, high-salt extraction procedure. Briefly, cells were washed with phosphate-buffered saline (PBS) and then resuspended in 500 μl of PBS and pelleted for 20 s at 14,000 rpm in a microcentrifuge. The pellet was resuspended in 50 μl of 20 mM HEPES-0.5% NP-40-2 mM MgCl2 and incubated on ice for 5 min. Then 5 M NaCl was added to a final concentration of 0.4 M, and the reaction mixes were incubated on ice for 30 min. After spinning for 10 min at 17,000 rpm, the supernatant was mixed with 4× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and heated at 70°C for 10 min.
For the ΔTM mutant, transfected, superinfected 10-cm2 cultures were washed once with PBS, scraped into PBS, and pelleted at 3,000 rpm in a microcentrifuge. Cells were resuspended in 75 μl of water and lysed by addition of 25 μl of 4× SDS-PAGE sample buffer. All samples were heated to boiling for 10 min and then separated by SDS-PAGE. Following electrophoresis, proteins were transferred to nitrocellulose and membranes were blocked overnight in 10% nonfat milk in T-TBS (20 mM Tris [pH 7.5], 500 mM NaCl, 0.05% Tween) with 10% BlokHen II (Aves Labs, Portland, Oreg.). Primary chicken anti-UL34 antibody was diluted 1:2,000 in GT-TBS (0.5 g of gelatin in 50 ml of T-TBS; 1:1,000 for the ΔTM blot) and detected with 1:1,000-diluted goat anti-chicken immunoglobulin G-horseradish peroxidase conjugate. Chemiluminescence was captured with the ECL Western blotting detection system (Amersham Biosciences, London, England).
Calculation of complementation indices.
A complementation index for each mutant was calculated with the formula [(PFU of charged cluster mutant/normalization factor)/(PFU of wild-type UL34/normalization factor)]. The normalization factor was used to correct for differences in transfection efficiency and was derived from the results of the β-galactosidase assay as follows. Absorbance values for each sample were divided by the lowest absorbance value for any sample in that experiment. For example, if sample A has a reading of 0.45 and sample B has a reading of 0.9, the normalization factor for sample A is 1 and for sample B is 2. This signifies that sample B had a transfection efficiency that was twice as high as that of sample A.
Indirect immunofluorescence.
Indirect immunofluorescence was performed as previously described (33). Briefly, cells were fixed with 2% formaldehyde for 20 min and washed with PBS. They were permeabilized by incubation with immunofluorescence buffer and washed in PBS. The cells were blocked in 1:10 BlokHen II (Aves Laboratories) in PBS and washed with PBS. The chicken anti-UL34 antibody was diluted 1:4,000 in immunofluorescence buffer and detected with Texas Red-conjugated donkey anti-chicken immunoglobulin antibody. For the experiments in which UL34 and UL31 were detected simultaneously, cells were fixed with cold methanol for 20 min and then incubated with immunofluorescence buffer. Cells were blocked with 10% human serum in 1% bovine serum albumin in PBS and washed. The UL34 antibody was used as described above. The UL31 antibody was used at a 1:10 dilution in immunofluorescence buffer and detected by fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (Jackson Immunoresearch Laboratories). Slofade II (Molecular Probes, Eugene, Oreg.) was used to mount coverslips on glass slides. All confocal microscopy was done with a Zeiss 510 confocal microscope.
RESULTS
Charged cluster mutations in the UL34 gene.
All of the known or proposed functions for the UL34 protein suggest that it interacts with other viral or cellular proteins. To address the role of the UL34 protein in viral envelopment, we developed a library of mutants carrying amino acid substitutions designed to disrupt protein-protein interactions. The UL34 protein does not contain any domains homologous to previously characterized protein-protein interaction domains, making it difficult to predict which regions of the protein are necessary for protein function and protein interactions. To circumvent this problem, we chose to mutagenize the protein by introducing selected charged cluster mutations. A charged cluster mutation is defined as two or more charged amino acids within a five-amino-acid stretch (42). Charged cluster mutations disrupt exposed areas of the protein, increasing the chance that protein interactions will be disrupted (8). These mutations avoid disrupting hydrophobic amino acids, and therefore disruption of the protein secondary structure is not predicted and a greater number of mutant proteins are expressed at substantial levels (9, 42). Charged cluster mutations are often used to locate positions of ligand contact on proteins (41). In addition, this type of mutagenesis often results in high percentages of temperature-sensitive mutations (14, 20).
In addition to the charged cluster mutants, we also created a mutant UL34 protein in which a stop codon was inserted immediately upstream of the putative transmembrane domain. The hypothesis that UL34 acts as an anchor for a number of proteins involved in primary viral envelopment and/or facilitates capsid interactions predicts that such a mutant should be unable to function in envelopment.
Complementation assay identifies UL34 charged clusters necessary for protein function.
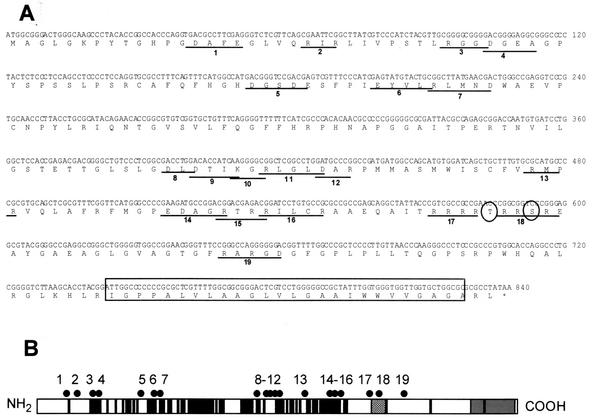
UL34 charged cluster mutants and a ΔTM mutant were created as described in Materials and Methods. Figure 1A shows the amino acid sequence of the UL34 gene, the location and name given to each charged cluster, the position of the proposed phosphorylation site, and the placement of the putative transmembrane domain. The schematic in Fig. 1B shows the spatial arrangement of the UL34 charged clusters, along with the putative transmembrane domain, the US3 phosphorylation consensus site, and amino acid residues of invariant regions for HSV-1, varicella-zoster virus, and equine herpesvirus type 1.
FIG. 1.
Locations of charged cluster mutations within the UL34 gene. (A) Nucleotide and amino acid sequences of the UL34 gene, showing the positions of charged cluster mutations. Each charged amino acid in a charged cluster was changed to an alanine. The location of a phosphorylation site recognized by US3 protein kinase is denoted by circles, and the putative transmembrane domain is indicated by the boxed region. (B) Schematic depiction of the sequence arrangement and location of charged clusters in the UL34 protein of HSV-1. The positions of the putative transmembrane domain (grey box), US3 phosphorylation consensus (hatched box), and residues invariant between HSV-1, varicella-zoster virus, and equine herpesvirus type 1 (black boxes) are indicated. Each of the black dots above the schematic represents the location of a charged cluster mutation within the gene. The mutations were numbered from 1 to 19, starting at the N terminus.
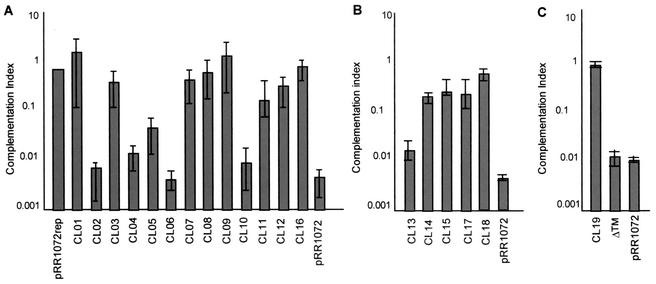
The 20 mutants were analyzed by a complementation assay designed to test the functionality of each mutant UL34 protein. Vero cells were transfected with plasmids encoding either wild-type or mutant UL34 proteins and pCMVβ (a plasmid carrying the β-galactosidase gene driven by the cytomegalovirus promoter) to normalize transfection efficiency. The monolayers were infected with a UL34-null virus, and after 18 h of infection, virus stock was prepared from each culture and titrated on complementing cells. A complementation index was calculated for each of the charged cluster mutants and pRR1072, a plasmid carrying a deletion of most of the UL34 gene. For each experiment, the complementation index for pRR1072rep (a plasmid carrying the wild-type UL34 sequence) was set equal to 1. Figure 2 shows the complementation indices for each of the UL34 charged cluster mutants and ΔTM (from three experiments). Mutants CL01, CL03, CL07, CL08, CL09, CL11, CL12, CL14, CL15, CL16, CL17, CL18, and CL19 were able to complement at levels not much different from that of wild-type UL34. Mutants CL02, CL04, CL06, CL10, CL13, and ΔTM complemented at levels equivalent to that of pRR1072, suggesting that a functional UL34 protein is not produced in these mutants. Only one of the mutants, CL05, functioned at an intermediate level, complementing 10-fold less efficiently than the wild-type protein.
FIG. 2.
Complementation indices of charged cluster mutants. Complementation index (log scale) versus charged cluster mutants. Vero cells were transfected with plasmids encoding pRR1072rep (wild-type UL34), UL34 charged cluster mutants (CL01 to CL19), ΔTM, or pRR1072 (a plasmid encoding GFP driven off the UL34 promoter) and then infected with a UL34-null virus. (A) Analysis of charged cluster mutants CL01 to CL12 and CL16. (B) Analysis of charged cluster mutants CL13 to CL15, CL17, and CL18. (C) Analysis of charged cluster mutant CL19 and ΔTM. Error bars represent the maximum and minimum complementation indices obtained from three independent experiments; the bar height represents the average complementation index.
All of the noncomplementing charged cluster mutant and ΔTM UL34 proteins are expressed at levels comparable to wild-type UL34.
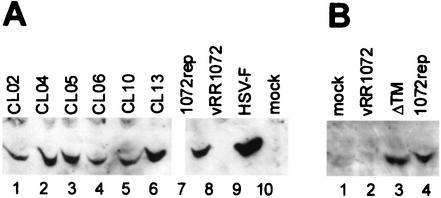
It is possible that some of the mutants were unable to complement a UL34-null virus because they failed to express mutant UL34 protein in sufficient amounts. In order to test for UL34 protein expression, 10-cm2 cultures of HEp-2 cells were transfected with plasmids encoding wild-type and mutant UL34 proteins and infected with the UL34-null virus. Cellular extracts were prepared, and proteins were separated by SDS-PAGE, blotted to nitrocellulose, and probed with antibody directed against UL34. As shown in Fig. 3, all of the noncomplementing charged cluster mutants produced full-length proteins at levels similar to that of pRR1072Rep, which encodes wild-type UL34. The ΔTM mutant expressed a protein that migrated slightly faster than wild-type UL34, consistent with loss of the putative transmembrane domain. The accumulation of mutant UL34 proteins in amounts similar to that of the wild-type protein suggests that differences exhibited by these mutants in the complementation assay are not due to differences in the level of protein expression but rather to defects in the intrinsic properties of the mutant proteins (i.e., either function, localization, or both).
FIG. 3.
Western blot of UL34 protein in noncomplementing cells transfected with charged cluster mutants or the ΔTM mutant. (A) For lanes 1 to 7, noncomplementing cells were transfected with plasmids encoding wild-type UL34 or the indicated charged cluster mutants and then infected with a UL34-null virus at an MOI of 5. In lanes 8 and 9, noncomplementing cells were infected with vRR1072 or HSV-1(F), respectively, at an MOI of 5. Protein was extracted as described in Materials and Methods. A Bradford assay was used to determine protein concentrations prior to gel loading; approximately 50 μg of protein was loaded in each lane. A β-galactosidase assay was used to determine relative transfection efficiencies, and sample amounts were normalized according to these results so that each lane contained equivalent amounts of transfected cells. (B) Lane 1 is a mock-infected control. For lane 2, noncomplementing cells were infected with vRR1072 at an MOI of 5. For lanes 3 and 4, noncomplementing cells were transfected with either ΔTM or pRR1072rep and then infected with a UL34-null virus at an MOI of 5. For these samples, cells were solubilized directly in SDS-PAGE sample buffer, and the entire sample was loaded on the gel. Panels A and B represent two independent experiments, in which the gels were run for different amounts of time, which accounts for the differences in size between the mutant UL34 proteins.
Some UL34 charged cluster mutants are able to localize correctly but fail to complement.
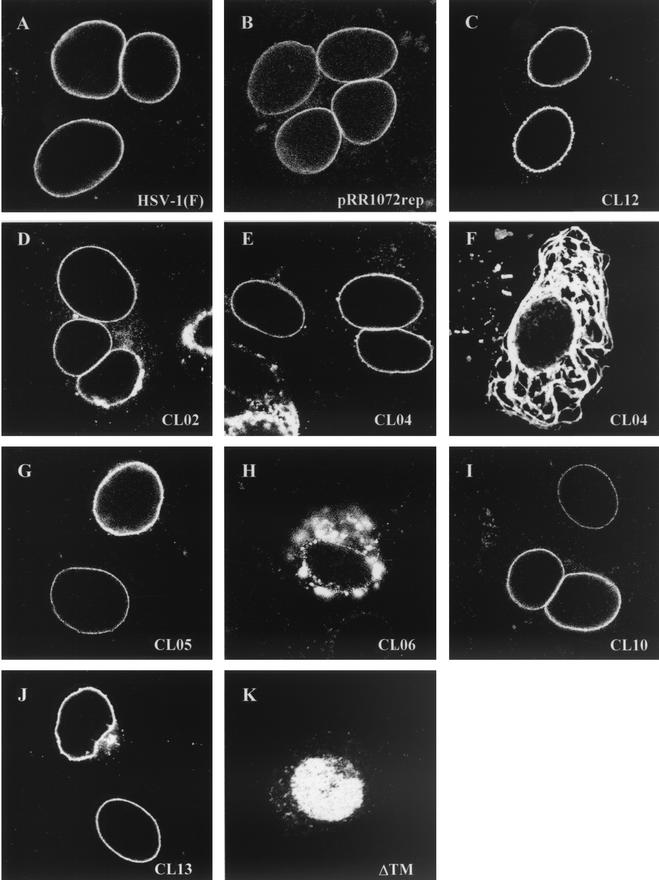
Proper localization of both the UL34 and UL31 proteins is dependent upon the expression of both proteins and their interaction in an infected cell (33). Nonfunctional UL34 mutants might fail to interact with UL31 as necessary for their localization to the nuclear envelope. Such mutant UL34 proteins should fail to localize properly in an infected cell. Alternatively, proper localization of a mutant UL34 protein might suggest that the protein interacts appropriately with UL31. To determine whether noncomplementing UL34 proteins localized correctly, Vero cells were transfected with plasmids encoding wild-type or mutant UL34 and then infected with the UL34-null virus vRR1072. Twelve hours later, the cells were fixed with formaldehyde and processed for immunofluorescence with an anti-UL34 antibody. The UL34 antibody was detected with a Texas Red-conjugated secondary antibody, and infected cells were identified by GFP expression from the UL34-null virus (Fig. 4).
FIG.4.
Digital confocal images showing the localization of wild-type and mutant UL34 proteins in transfected Vero cells infected with a UL34 deletion virus. (A) Vero cells were infected with wild-type HSV-1(F) at an MOI of 5. (B to K) Vero cells were transfected with plasmids encoding either the wild-type UL34 gene (B), charged cluster mutants (C to J), or the ΔTM mutant (K). After 24 h, cells were infected with a UL34-null virus at an MOI of 5. Panel F represents a cell that was only transfected, based on the absence of GFP expression, although other cells on the same coverslip were transfected and infected. For all panels, at 12 h postinfection the cells were fixed with formaldehyde, immunostained with chicken anti-UL34 antibody, and detected with donkey anti-chicken immunoglobulin-Texas Red conjugate or goat anti-chicken immunoglobulin-Alexa Red 524 conjugate. All experiments were performed independently a minimum of three times. The images shown are representative. (A) HSV-1(F), (B) pRR1072rep, (C) CL08, (D) CL02, (E and F) CL04, (G) CL05, (H) CL06, (I) CL10, (J) CL13, and (K) ΔTM.
All charged cluster mutants that showed wild-type complementation levels also localized to the inner nuclear membrane. Figures 4A and 4B show the localization pattern of wild-type UL34 in cells infected with HSV-1(F) and in cells transfected with the wild-type plasmid pRR1072rep and infected with vRR1072, respectively. Figure 4C shows CL12, which is representative of the group of mutants that complemented to wild-type levels. For all these mutants and for the wild-type control, many transfected-infected cells were seen showing a typical localization for UL34, in which the protein appears exclusively confined to the nuclear envelope. However, we also observed some transfected-infected cells in which a small percentage of the UL34 protein was distributed in the cytoplasmic structures typically seen in UL34-transfected cells. Thus, there was a range of UL34 localization in transfected-infected cells, which remained the same for both complementing and noncomplementing mutants. These results suggest that the influence of viral factors provided by infection on the localization of plasmid expressed UL34 is not always complete. This may depend upon the level of UL34 expression in the transfected cell.
Five of the six charged cluster mutants (CL02, CL04, CL05, CL10, and CL13) that failed to complement to wild-type levels were able to localize correctly in infected cells (Fig. 4D, E, G, I, and J, respectively). One of the charged cluster mutants that failed to complement, CL06, along with ΔTM, also failed to localize to the inner nuclear membrane (Fig. 4H and K). CL06 localized to large, irregular areas around the nucleus, but a tight ring around the nucleus never formed, and it was unclear whether UL34 was targeted to the nuclear membrane. There was also more cytoplasmic staining than was typically seen with wild-type UL34 localization. In contrast, ΔTM appeared to localize almost entirely inside the nucleoplasm. The localization of the ΔTM mutant did not depend on whether the transfected cell had been infected or not, suggesting that the intranuclear localization of this mutant protein was independent of viral factors. All of the mutant UL34 proteins accumulated to easily detectable levels in transfected-infected cells, confirming that failure to complement was not due to failure to accumulate UL34 protein.
Two of the noncomplementing mutants, CL04 and CL05, localized correctly in transfected-infected cells but did not exhibit a typical localization for UL34 in transfected uninfected cells. These mutant UL34 proteins were localized in extensive, rope-like structures that were present throughout most of the cytoplasm, as shown in Fig. 4F. Because these structures were present throughout the cell, we postulated that this pattern of UL34 staining might reflect attachment to a cytoskeletal element. However, the UL34 protein produced by these two mutants did not colocalize extensively with antitubulin, antivimentin, or phalloidin-stained cells, suggesting that these structures are not microtubules, microfilaments, or intermediate filaments (data not shown). In addition, UL34 localization did not coincide with similar rope-like structures formed by GFP-procaspase domain fusions (from caspases 1, 8, and 10), mitochondria, or endoplasmic reticulum structures in the cell (data not shown).
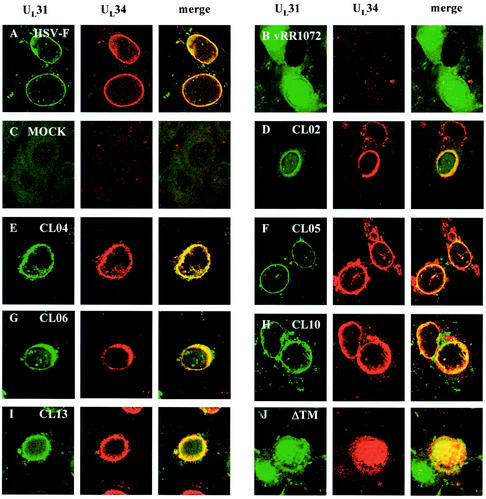
UL34 charged cluster mutants colocalize with UL31.
Because UL34 and UL31 are necessary and sufficient to target each other to the nuclear membrane (33), mutants that were able to localize to the inner nuclear envelope were assumed to be interacting with UL31. To determine whether noncomplementing UL34 mutants could mediate correct localization of UL31, Vero cells were transfected with plasmids containing wild-type or mutant UL34 and then infected with the UL34-null virus vRR1072. Twelve hours later, the cells were fixed with methanol and processed for immunofluorescence with both anti-UL31 and anti-UL34 antibodies. The anti-UL34 antibody was detected with a Texas Red-conjugated secondary antibody, and anti-UL31 antibody was detected with fluorescein isothiocyanate-conjugated secondary antibody (Fig. 5). All of the noncomplementing charged cluster mutants colocalized extensively with the UL31 protein, suggesting that the interaction between UL34 and UL31 is maintained in these mutants (Fig. 5D to J). UL34 proteins from all of the complementing charged cluster mutants also colocalized exclusively with UL31 (data not shown). UL34 and UL31 proteins in mutant CL06 localized to the same irregular areas around the nucleus, indicating that the association between these two proteins is likely maintained even though proper localization is not (Fig. 5G).
FIG. 5.
Digital confocal images showing colocalization of UL34 and UL31 in transfected Vero cells infected with a UL34-null virus. Vero cells were infected with wild-type HSV-1(F) (A), UL34 null virus (B), or mock-infected (C) at an MOI of 5. (D to J) Vero cells were transfected with plasmids encoding noncomplementing UL34 charged cluster mutations and infected after 24 h with a UL34-null virus at an MOI of 5. For all panels, at 12 h postinfection, the cells were fixed with cold methanol and immunostained with chicken anti-UL34 detected with goat anti-chicken immunoglobulin-Alexa 524 conjugate and rabbit anti-UL31 detected with goat anti-rabbit immunoglobulin-fluorescein isothiocyanate conjugate. Differences in morphological preservation between this figure and Fig. 4 are due to the different fixation techniques used. (A) HSV-1(F), (B) vRR1072, (C) mock infected, (D) CL02, (E) CL04, (F) CL05, (G) CL06, (H) CL10, (I) CL13, and (J) ΔTM.
DISCUSSION
The creation of a UL34 charged cluster mutant library was an effective strategy for generating stable mutant proteins; all 19 charged cluster mutants expressed nearly wild-type levels of UL34 protein in transfected and superinfected cells. Six of 19 charged cluster mutants were unable to fully complement a UL34 deletion virus in transfection-infection experiments, showing that this strategy effectively revealed residues of the UL34 protein that are important for function. Interestingly, all of the noncomplementing charged cluster mutations except CL06 occurred in regions of the UL34 protein coding sequence that are not conserved among alphaherpesviruses (Fig. 1). Conversely, many complementing charged cluster mutations were present in conserved areas of the UL34 protein, indicating that charged interactions in these areas of the protein are not necessary for protein function. These observations are consistent with two different hypotheses: (i) UL34 proteins in different herpesviruses may have separate, unique, essential functions in addition to a universal function conserved by all UL34 proteins, and (ii) different UL34 proteins may all have the same function, such as interaction with other viral proteins (e.g., UL31), but through coevolution with those other viral proteins, critical functions of UL34 are conserved without conservation of the sequence. Mutant CL06, the mutation of which is located in a highly conserved area of UL34 (Fig. 1), is also the only one of the noncomplementing charged cluster mutants that failed to correctly localize to the nuclear membrane (Fig. 4), possibly because the mutant UL34 protein can no longer interact with a conserved cellular protein(s) that is required for localization.
Confocal microscopy studies of the charged cluster library showed that the majority of mutants, whether they complemented or not, localized to the inner nuclear envelope. All of the mutants that complemented were able to correctly localize UL34 to the nuclear membrane; this suggests that correct localization is a prerequisite for complementation. A small amount of the UL34 protein from noncomplementing mutant CL06 was able to localize to the nuclear envelope, but most of the protein appeared in irregular areas within the cytoplasm (Fig. 4). The identity of these cytoplasmic structures was not determined.
Both complementing and noncomplementing charged cluster mutants were able to colocalize with UL31 to the inner nuclear envelope, suggesting that the interaction between UL34 and UL31 proteins may be maintained in these mutants.
We found that the ΔTM mutant was unable to complement and localized to the interior of the nucleus but was expressed in the transfection-infection experiment at levels equivalent to those of pRR1072rep (Fig. 2 to 4). Although this mutant failed to correctly localize to the nuclear envelope, colocalization between UL34 and UL31 in the nucleus was observed. However, since UL31 normally localizes to the nucleus (33), and the ΔTM mutant also localizes to the nucleus without infection (i.e., in the absence of UL31), it is possible that the ΔTM mutant and UL31 protein reach the interior of the nucleus independently of each other. Failure of the ΔTM UL34 protein to localize at the nuclear membrane was not surprising, since the putative transmembrane domain was removed and association with membranes was presumed to rely on the presence of this section of the protein.
More surprising was the failure of the ΔTM mutant to complement a UL34 deletion virus. Ye and Roizman (44) tested a similar mutation in the context of a recombinant HSV-1. Their mutant was able to grow almost as well as wild-type virus on noncomplementing cells, and the mutant protein accumulated to levels only slightly lower than those of the wild-type protein (44). Since these experiments used a recombinant virus to express the mutant UL34 protein, it is possible that the mutant UL34 was functional because compensatory secondary mutations were introduced elsewhere in the virus. Alternatively, the transfection-infection experiments used in this study may have produced mutant UL34 proteins with different expression kinetics than would be seen in the context of a viral infection.
The localization of two other charged cluster mutants, CL04 and CL05, in transfected uninfected cells revealed a previously unseen pattern of UL34 localization, rope-like chains of protein throughout the cytoplasm of cells. We attempted to identify these structures in the hope that they might reveal previously uncharacterized interactions of UL34 protein with other cellular factors. We did not see extensive colocalization of these structures with previously characterized cytoskeletal elements, membranous structures, or procaspase aggregates. These structures may represent ordered aggregates of UL34 protein alone. Interestingly, when procaspase-GFP fusion proteins were transfected into cells and infected with wild-type HSV-1(F), the rope-like chains were relocalized into the nucleus (data not shown). Perhaps the procaspases were interacting with other HSV-1 proteins.
The properties of the mutants analyzed in this study suggest that UL34 protein has at least one essential function in addition to its role in proper targeting of the essential UL31 protein and that the essential functions of UL34 protein are dependent upon its transmembrane domain. It seems likely that an additional UL34 protein function(s) is based on interactions with other viral or cellular proteins.
Acknowledgments
We thank Michael Choi for the gift of the procaspase-GFP fusion proteins, Wendy Maury for providing the vimentin antibody, and Mark Stamnes for providing the endoplasmic reticulum and mitochondrial antibodies. We also acknowledge Mark Stinski, Charles Grose, and members of the Roller laboratory for helpful discussions and reading of the manuscript.
These studies were supported by the University of Iowa and Public Health Service awards AI 41478 (R.J.R.) and GM 50740 (J.D.B.) and National Research Service Award F32 GM20448 (A.E.R.). J.K.K. was supported by the National Science Foundation site grant NSF DBI00-97361.
REFERENCES
- 1.al-Kobaisi, M. F., F. J. Rixon, I. McDougall, and V. G. Preston. 1991. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology 180:380-388. [DOI] [PubMed] [Google Scholar]
- 2.Baines, J. D., and B. Roizman. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 66:5168-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baines, J. D., P. L. Ward, G. Campadelli-Fiume, and B. Roizman. 1991. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J. Virol. 65:6414-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, M., A. R. MacLean, J. D. Aitken, and J. Harland. 1994. ICP34.5 influences herpes simplex virus type 1 maturation and egress from infected cells in vitro. J. Gen. Virol. 75:3679-3686. [DOI] [PubMed] [Google Scholar]
- 5.Carter, P. 1987. Improved oligonucelotide-directed mutagenesis with M13 vectors. Methods Enzymol. 154:382. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Y. E., C. Van Sant, P. W. Krug, A. E. Sears, and B. Roizman. 1997. The null mutant of the UL31 gene of herpes simplex virus 1: construction and phenotype in infected cells. J. Virol. 71:8307-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Church, G. A., and D. W. Wilson. 1997. Study of herpes simplex virus maturation during a synchronous wave of assembly. J. Virol. 71:3603-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunningham, B., and J. Wells. 1989. High resolution mapping of hGH-receptor interactions by alanine scanning mutagenesis. Science 244:1081-1085. [DOI] [PubMed] [Google Scholar]
- 9.Dao-Pin, S., D. B. Anderson, W. A. Baase, F. W. Dahlquist, and B. W. Matthews. 1991. Structural and thermodynamic consequences of burying a charged residue within the hydrophobic core of T4 lysozyme. Biochemistry 30:11521-11529. [DOI] [PubMed] [Google Scholar]
- 10.Darlington, R. W., and L. H. Moss. 1968. Herpesvirus envelopment. J. Virol. 2:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 12.Desai, P. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desai, P., G. Sexton, J. McAffrey, and S. Person. 2001. A null mutation in the gene encoding the herpes simplex virus type 1 UL37 polypeptide abrogates virus maturation. J. Virol. 75:10259-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diamond, S. E., and K. Kirkegaard. 1994. Clustered charged-to-alanine mutagenesis of poliovirus RNA-dependent RNA polymerase yields multiple temperature-sensitive mutants defective in RNA synthesis. J. Virol. 68:863-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerace, L., and G. Blobel. 1980. The nuclear envelope lamina is reversibly depolymerized during mitosis. Cell 19:277-287. [DOI] [PubMed] [Google Scholar]
- 18.Granzow, H., B. Klupp, W. Fuchs, W. Veits, N. Osterrieder, and T. Mettenleiter. 2000. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 73:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grose, C., R. Harson, and S. Beck. 1995. Computer modeling of prototypic and aberrant nucleocapsids of varicella-zoster virus. Virology 214:321-329. [DOI] [PubMed] [Google Scholar]
- 20.Hassett, D. E., and R. C. Condit. 1994. Targeted construction of temperature-sensitive mutations in vaccinia virus by replacing clustered charged residues with alanine. Proc. Natl. Acad. Sci. USA 91:4554-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutchinson, L., and D. C. Johnson. 1995. Herpes simplex virus glycoprotein K promotes egress of virus particles. J. Virol. 69:5401-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Blanc, I., M. Prevost, M. Dokhelar, and A. R. Rosenberg. 2002. The PPPY motif of human T-cell leukemia virus type 1 Gag protein is required in the budding process. J. Virol. 76:10024-10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loomis, J., B. Bowzard, R. Courtney, and R. Wills. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 75:12209-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mossman, K., R. Sherburne, C. Lavery, J. Duncan, and J. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854-857. [DOI] [PubMed] [Google Scholar]
- 28.Neubauer, A., J. Rudolph, C. Brandmüller, F. Just, and N. Osterrieder. 2002. The equine herpesvirus 1 UL34 gene product is involved in an early step in virus egress and can be efficiently replaced by a UL34-GFP fusion protein. Virology 300:189-204. [DOI] [PubMed] [Google Scholar]
- 29.Ni, S., C. Morgan, and H. M. Rose. 1968. Electron Microscopy of herpes simplex virus. II. Sequence of development. J. Virol. 2:517-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poon, A. P., and B. Roizman. 1993. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J. Virol. 67:4497-5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purves, F. C., D. Spector, and B. Roizman. 1991. The herpes simplex virus 1 protein kinase encoded by the US3 gene mediates posttranslational modification of the phosphoprotein encoded by the UL34 gene. J. Virol. 65:5757-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purves, F. C., D. Spector, and B. Roizman. 1992. UL34, the target of the herpes simplex virus US3 protein kinase, is a membrane protein which in its unphosphorylated state associates with novel phosphoproteins. J. Virol. 66:4295-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reynolds, A., B. Ryckman, J. Baines, Y. Zhou, L. Liang, and R. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 from a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roizman, B., and A. E. Sears. 1996. Herpes simplex viruses and their replication, p. 2231-2295. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Field's virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 35.Roller, R. J., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salmon, B., C. Cunningham, A. J. Davison, W. J. Harris, and J. D. Baines. 1998. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 72:3779-3788.,. [DOI] [PMC free article] [PubMed]
- 37.Sarkar, G., and S. S. Sommer. 1990. The “Megaprimer” method of site-directed mutagenesis. BioTechniques 8:404-407. [PubMed] [Google Scholar]
- 38.Scott, E., and P. O'Hare. 2001. Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infections. J. Virol. 75:8818-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shiba, C., T. Daikoku, F. Goshima, H. Takakuwa, O. Kowai, and Y. Nishiyama. 2000. The UL34 gene product of herpes simplex virus type 2 is a tail-anchored type II membrane protein that is significant for virus envelopment. J. Gen. Virol. 81:2397-2405. [DOI] [PubMed] [Google Scholar]
- 40.Telford, E. A., M. S. Watson, K. McBride, and A. J. Davison. 1992. The DNA sequence of equine herpesvirus-1. Virology 189:304-316. [DOI] [PubMed] [Google Scholar]
- 41.Wertman, K. F., D. G. Drubin, and D. Botstein. 1992. Systematic mutational analysis of the yeast ACT1 gene. Genetics 132:337-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiskerchen, M., and M. A. Muesing. 1995. Identification and characterization of a temperature-sensitive mutant of human immunodeficiency virus type 1 by alanine scanning mutagenesis of the integrase gene. J. Virol. 69:597-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamauchi, Y., F. Goshima, A. Nawa, T. Murata, and Y. Nishiyama. 2001. Herpes simplex virus 2 UL34 protein requires UL31 protein for its relocation to the internal nuclear membrane in transfected cells. J. Gen. Virol. 82:1423-1428. [DOI] [PubMed] [Google Scholar]
- 44.Ye, G., and B. Roizman. 2000. The essential protein encoded by the UL31 gene of herpes simplex virus 1 depends for its stability on the presence of UL34 protein. Proc. Natl. Acad. Sci. USA 97:11002-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye, G., K. Vaughan, R. Vallee, and B. Roizman. 2000. The herpes simplex virus type 1 UL34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J. Virol. 74:1355-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]