Abstract
Picornaviral RNA replication utilizes a small virus-encoded protein, termed 3B or VPg, as a primer to initiate RNA synthesis. This priming step requires uridylylation of the VPg peptide by the viral polymerase protein 3Dpol, in conjunction with other viral or host cofactors. In this study, we compared the viral specificity in 3Dpol-catalyzed uridylylation reactions between poliovirus (PV) and human rhinovirus 16 (HRV16). It was found that HRV16 3Dpol was able to uridylylate PV VPg as efficiently as its own VPg, but PV 3Dpol could not uridylylate HRV16 VPg. Two chimeric viruses, PV containing HRV16 VPg (PV/R16-VPg) and HRV16 containing PV VPg (R16/PV-VPg), were constructed and tested for replication capability in H1-HeLa cells. Interestingly, only PV/R16-VPg chimeric RNA produced infectious virus particles upon transfection. No viral RNA replication or cytopathic effect was observed in cells transfected with R16/PV-VPg chimeric RNA, despite the ability of HRV16 3Dpol to uridylylate PV VPg in vitro. Sequencing analysis of virion RNA isolated from the virus particles generated by PV/R16-VPg chimeric RNA identified a single residue mutation in the VPg peptide (Glu6 to Val). Reverse genetics confirmed that this mutation was highly compensatory in enhancing replication of the chimeric viral RNA. PV/R16-VPg RNA carrying this mutation replicated with similar kinetics and magnitude to wild-type PV RNA. This cell culture-induced mutation in HRV16 VPg moderately increased its uridylylation by PV 3Dpol in vitro, suggesting that it might be involved in other function(s) in addition to the direct uridylylation reaction. This study demonstrated the use of chimeric viruses to characterize viral specificity and compatibility in vivo between PV and HRV16 and to identify critical amino acid residue(s) for viral RNA replication.
Human rhinoviruses (HRV) are members of an extensive genus of the Picornaviridae family that are most frequently associated with viral infections causing symptoms of the common cold. They are small, nonenveloped positive-stranded RNA viruses possessing genomes of approximately 7,200 nucleotides. Like all members of the Picornaviridae, the rhinovirus genome encodes a single polyprotein that is posttranslationally cleaved by viral proteases. In addition to these characteristics, rhinoviruses possess a long 5′ untranslated region, a shorter 3′ untranslated region, a poly(A) tail at their 3′ termini, and a small covalently bound virus-encoded protein termed VPg (3B) at their 5′ termini (6, 11, 22, 42).
Rhinovirus replication is believed to follow the same basic strategy outlined for the prototypic picornavirus poliovirus (PV). This replication strategy involves transcription of the viral genome into a cRNA (negative strand) followed by synthesis of new genomic RNA strands which template from the negative strand. Central to this process is the viral RNA-dependent RNA polymerase (RdRp) 3Dpol which is responsible for the synthesis of both strands. VPg has been shown to be essential for protein-primed initiation of both the positive and negative RNA strand replication (42).
Perhaps the most well studied rhinovirus serotype is HRV14, and many details of its structure (2, 7, 10, 36, 38) and function have been elucidated (24, 25, 29, 35). However, HRV14 represents a less significant target molecule for antiviral drug discovery since it belongs to the rhinovirus subgroup A, which generally lacks pathogenicity and circulates infrequently in human populations (1, 21). Phylogenetic tree analysis of picornaviruses suggests that HRV14 is more divergent than many other rhinoviruses (1). HRV16, on the other hand, is included in the more pathogenic subgroup B and is more closely related to other circulating rhinoviruses (21). At the present time, no effective therapeutics are available for prevention or treatment of rhinovirus infections. Current development of rhinovirus antivirals has focused on the inhibition of viral capsid function or 3C protease activity (reviewed in reference 40). The RdRp 3Dpol of rhinovirus represents another target for the development of antiviral compounds since viral RdRp is a key enzyme in viral RNA replication. The corresponding polymerase from PV has been the subject of many investigations, including those describing three-dimensional structure and potential replication mechanism(s) (14, 15, 28, 29); however, fewer of these details are known for HRV16. To this end, a recent report from Hung and coworkers (18) has shed light onto some of the detailed biochemical properties of bacterially expressed HRV16 3D RdRp.
The initiation of picornaviral RNA replication is regulated predominantly by the interaction between 3Dpol, VPg, an RNA template, and other viral and cellular cofactors (5, 20, 28, 32). This reaction has been reconstituted in vitro by using an enzymatic assay in which synthetic VPg peptides become uridylylated by 3Dpol in the presence of a poly(A) template and UTP (12, 28). A more sophisticated uridylylation assay has also been developed (12, 29) which relies on interactions between 3Dpol, VPg peptides, a cis-acting replication element (cre) within the coding sequence of rhinovirus or PV, and the precursor viral protein 3CDpro. Results from in vitro uridylylation assays with PV type 1 (Mahoney strain) 3Dpol suggested that other picornaviral VPg gene products are interchangeable with those of PV (29). To extend these observations, we investigated the interchangeability in VPg uridylylation in vitro between HRV16 and PV 3Dpol and their effects on in vivo viral RNA replication by using PV and rhinovirus VPg chimeric viruses. Our data suggested that in vitro uridylylation is essential but not sufficient for in vivo replication activity. Furthermore, a key amino acid residue (glutamate) at position 6 of the HRV16 VPg peptide was virus specific and did not support the replication of chimeric PV carrying HRV16 VPg. Mutation of the glutamate residue to a hydrophobic valine residue drastically enhanced the replication capability of the chimeric virus.
MATERIALS AND METHODS
Plasmids, cells, peptides, and enzymes.
A full-length HRV16 infectious cDNA clone was kindly provided by Wai-Ming Lee (University of Wisconsin). The HRV sequence was cloned into a modified pUC19 vector and was used as the template for PCR amplification of HRV16 3Dpol. An infectious cDNA clone of a PV type 1 (M) strain and its purified 3Dpol were obtained from Craig Cameron (13). H1-HeLa cells were purchased from the American Type Culture Collection and propagated in minimal essential medium supplemented with 10% fetal bovine serum. The expression vector pET26-Ub was a modified version of Novagen's pET26b (13) and contained the Saccharomyces cerevisiae ubiquitin gene immediately upstream of the SacII cloning site. Plasmid pCG1 contained the sequence from the yeast ubiquitin C-terminal-specific protease Ubp1 (4, 13). DNA oligonucleotides were purchased from Sigma/Genosys (The Woodlands, Tex.). Escherichia coli BL21(DE3) competent cells were obtained from Novagen for protein expression. VPg peptides were purchased from Alpha Diagnostics International (San Antonio, Tex.). All restriction enzymes were purchased from New England Biolabs (Beverly, Mass.).
Expression and purification of HRV 16 3Dpol protein.
The expression of HRV16 3Dpol with an authentic glycine amino terminus was accomplished by use of a pET26b-Ub vector (13). The HRV16 3Dpol fragment was PCR amplified from the infectious cDNA plasmid with HRV16-3DF and HRV16-3DR oligonucleotides and cloned into the pET26Ub vector via SacII and XhoI sites (pET26Ub-3Dpol). The HRV16 3Dpol construct possessed a GS diresidue linker followed by a hexahistidine tag at the carboxyl terminus. pET26Ub-3Dpol was cotransformed into BL21(DE3) competent cells along with the pCG1 plasmid and were selected on agar plates containing 25 μg of kanamycin/ml and 34 μg of chloramphenicol/ml. Co- and/or posttranslational cleavage of the Ub-3Dpol fusion protein was accomplished by the Ubp1 protease encoded on the pCG1 plasmid. Using this system, the 3Dpol protein is left with a perfect N-terminal glycine residue. Confirmation of the N-terminal amino acid residues of the purified 3Dpol product was performed by an Edman degradation mass spectroscopy method (University of California at Irvine). Expression and purification of HRV16 3Dpol were accomplished by using the procedures outlined below. BL21(DE3) cells containing pET26Ub-3Dpol and pCG1 plasmids were grown overnight at 37°C in 10 ml of Luria-Bertani medium containing 50 μg of kanamycin/ml and 34 μg of chloramphenicol/ml. For 1-liter cultures, 2 ml of the overnight seed culture were added to fresh Luria-Bertani medium containing kanamycin-chloramphenicol and shaken till the cellular density reached an optical density at 600 nm of 0.6. The temperature of the incubator was then lowered to 24°C. Protein expression was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 2 mM, after which time the culture was grown for an additional 4 h at 24°C. Cells were harvested by centrifugation, and the pellet was resuspended in 20 ml of buffer 1 (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 10% glycerol, and 5 mM β-mercaptoethanol). The cell suspension was frozen at −80°C for at least 1 h, thawed, supplemented with 100 μl of 0.1 M phenylmethylsulfonyl fluoride (ICN Biomedicals), and sonicated on ice with 4 2-min cycles. The crude cell lysate was centrifuged at 100,000 × g for 1 h in a Beckman Ti 45 rotor at 4°C. The supernatant was loaded at a rate of 1 ml/min onto a nickel(II) Sepharose column that had been equilibrated with buffer 1. The column was washed with 5 column volumes (CV) of 10% buffer 2 (50 mM Tris [pH 8.0], 100 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol, and 0.35 M imidazole) and 90% buffer 1 at a rate of 2 ml/min. Bound proteins were eluted with a 10 to 60% linear gradient of buffer 2 over 5 CV. Fractions that contained eluted 3Dpol were pooled and then diluted 1:2 with buffer A (50 mM Tris [pH 8.0], 10% glycerol, and 1 mM dithiothreitol [DTT]). Diluted proteins were subsequently loaded onto a heparin column at a rate of 1 ml/min and then washed with 5 CV of 10% buffer B (50 mM Tris [pH 8.0], 10% glycerol, 1 mM DTT, and 1 M NaCl) and 90% buffer A at a rate of 2 ml/min. The protein was eluted with 15 CV of a gradient of 10 to 60% buffer B at a rate of 2 ml/min. Peak fractions with greater than 95% purity as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) were pooled and supplemented with DTT to a concentration of 5 mM. 3Dpol protein was concentrated initially by use of a YM-10 membrane in an Amicon 8400 apparatus followed by a YM-10 Centriprep to a concentration of approximately 10 mg/ml.
Primer-dependent elongation using sym/sub RNA.
A symmetrical primer template substrate (sym/sub) (3, 9) was used to assess nucleotide incorporation by purified HRV16 3Dpol. The sym/sub RNA oligo was 5′ 33P-labeled and annealed as described by Arnold and Cameron (3). A standard nucleotide incorporation assay contained 50 mM HEPES (pH 7.3), 50 mM NaCl, 5 mM MgCl2, 10 mM β-mercaptoethanol, 1 U of RNasin (Promega), 100 μM ATP (and 100 μM UTP in some reactions), 2 μM sym/sub, and 2 μM 3Dpol and was incubated for 5 to 30 min at 30°C. Reactions were terminated by the addition of EDTA to a final concentration of 50 mM, extracted with phenol-chloroform, and ethanol precipitated. The precipitated pellets were air dried, heated at 85°C for 4 min in 2× sample loading buffer, and then electrophoresed at 15 W of constant power on a 25% PAGE-6 M urea gel. The undried gel was exposed and analyzed by phosphorimaging.
VPg uridylylation assay.
To assess the ability of HRV16 3Dpol to uridylylate VPg peptides, we followed a basic procedure established previously for PV 3Dpol (28). Briefly, VPg uridylylation assays were set up in 20-μl reaction volumes containing 50 mM HEPES buffer (pH 7.3), 10% glycerol, 3.5 mM magnesium acetate, 0.5 μg of poly(A) RNA (∼200 nucleotides long), 4 μg of VPg peptide, 3 μg of HRV16 3Dpol (or 0.25 μg of PV 3Dpol) and 2 μCi of [α-33P]UTP (3,000 Ci/mmol). Extension of uridylylated VPg products was determined by the addition of 100 μM unlabeled UTP. Reaction mixtures were incubated at 33°C for 60 min, and reactions were terminated by the addition of EDTA to a concentration of 70 mM. Products were analyzed by electrophoresis on 10 to 20% Tricine gels (Bio-Rad). The gels were vacuum dried prior to exposure to a phosphorimaging screen.
Construction of chimeric PV/R16-VPg and R16/PV-VPg clones.
To obtain a chimeric PV containing the HRV16 VPg gene (PV/R16-VPg), we used an overlap extension PCR strategy (17). PV infectious cDNA plasmids were used for PCR amplification of two separate reactions. The upstream PCR product was produced by the forward primer P1 2273F and the reverse primer P1/R16. The downstream PCR product was generated with forward primer R16/P1 and reverse primer P1 6076R. Annealed PCR product was double digested with NheI and KasI, subcloned into pBlueScript KS(+) vector (Stratagene), removed with the same enzymes, and finally ligated back into equally digested PV1 (M) infectious clone cDNA. Chimeric rhinoviruses incorporating a PV1 (M) VPg in place of HRV16 VPg (R16/PV-VPg) were constructed by a sequential QuikChange mutagenesis (Stratagene) strategy (41) with primer pairs R16-P1S-R16-P1AS, R16-P2S-R16-P2AS, and R16-P3S-R16-P3AS and then subcloned back into the HRV16 infectious clone by using a ClaI/SacI fragment. All mutagenized constructs were sequenced to confirm the VPg switch and lack of PCR-induced mutations with an ABI automated sequencer. Primer sequences are given in Table 1.
TABLE 1.
Oligonucleotides used for cloning and mutagenesis
| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| R16-3DF*a | TCCCCGCGGTGGAGGCCAAATTCAAATCTC |
| R16-3DR*a | CCGCTCGAGCGGCTAGTGGTGGTGGTGGTGGTGAGATCCGAATTTTTCATACCATTCATGTC |
| P1 2273F | TGGATTAGCAACACCACG |
| P1 6076R | GAAAGCACTGGGTTCAAGC |
| P1/R16 | CTCGGGAACTTTTGTCTTAGGTTTGGGCTCCCCCGAGTAAGGGCCCTGGTGTCCACAAACAGT |
| R16/P1 | GAGCCCAAACCTAAGACAAAAGTTCCCGAGAGAAGAGTGGTAGCTCAAGCCAGGTTCGATTACGC |
| R16-P1S | ATAAGCTCTTTTGCTCTCTACAGGGCGCTTACACCGGGTTGCCCAACGGTCCAGAAGAAGAATTTGGAAT |
| R16-P1AS | ATTCCAAATTCTTCTTCTGGACCGTTGGGCAACCCGGTGTAAGCGCCCTGTAGAGAGCAAAAGAGCTTAT |
| R16-P2S | GCGCTTACACCGGGTTGCCCAACAAAAAGCCAAACGTTCCCACGATTGGTCCAGAAGAAGAATTTGGAAT |
| R16-P2AS | ATTCCAAATTCTTCTTCTGGACCAATCGTGGGAACGTTTGGCTTTTTGTTGGGCAACCCGGTGTAAGCGC |
| R16-P3S | AAGCCAAACGTTCCCACGATTCGAACGGCAAAGGTACAAGGTCCAGAAGAAGAATTTGGAAT |
| R16-P3AS | ATTCCAAATTCTTCTTCTGGACCTTGTACCTTTGCCGTTCGAATCGTGGGAACGTTTGGCTT |
| P1 4730F | ATGGCATCCCTGGAGGAGAAAG |
| P1 6159R | ATTGCCTCCTCAAAGTCTG |
| P/R16 E6V S | AGGGCCCTTACTCGGGGGTGCCCAAACCTAAGACAA |
| P/R16 E6V AS | TTGTCTTAGGTTTGGGCACCCCCGAGTAAGGGCCCT |
The underlined sequences in R16-3DF and R16-3DR represent restriction sites.
In vitro transcription, RNA electroporation, and Northern blot analysis.
Chimeric or infectious cDNA clones were linearized with either SacI (HRV16 backbone) or EcoRI (PV1 backbone) and then used in the T7 MegaScript transcription kit (Ambion) according to the manufacturers' guidelines. For RNA electroporation, H1-HeLa cell monolayers at approximately 75% confluency were harvested, washed once in phosphate-buffered saline, and resuspended at a concentration of 2.2 × 106 to 3 × 106 cells/ml in phosphate-buffered saline. Fifteen micrograms of transcript RNA were mixed with the cell suspension and then electroporated on a Bio-Rad Gene Pulser II at a setting of 980 V with a capacitance of 25 μF at maximum resistance (24). Cells were then immediately diluted into fresh growth medium and plated onto six-well tissue culture plates. To collect virus particles from the electroporated cells, medium was harvested, snap-frozen, thawed, and then centrifuged (Allegra 6R; Beckman Coulter) at 2,000 rpm for 10 min at 4°C. Supernatants were collected and passed through a 0.45-μm-pore-size low-protein-binding filter and used as viral seed stock. Detection of viral RNA from transfected cells was accomplished by Northern blot analysis with a fragment from the 3Dpol gene of PV1(M) or HRV16 as a probe. Similarly, a probe for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cellular RNA was used to monitor the expression level of this endogenous gene. Northern blotting was done according to procedures described in the Northern Max kit (Ambion).
Serial passage and plaque assay of VPg chimeric viruses.
Chimera virus stocks were sequentially passaged 10 times in H1-HeLa cells by adding 0.5 ml of viral inoculum to 70% confluent cell monolayers in T-75cm2 flasks. The virus was permitted to bind the cells for 1 h at 37°C (PV) or 34°C (HRV), after which time the viral inoculum was removed and the cells were replenished with fresh growth medium. Upon detection of a cytopathic effect (CPE) (24 to 48 h postinfection), the supernatant was removed, snap-frozen once, and then prepared as described above. To determine virus titers and plaque morphology, H1-HeLa monolayers were infected with the viral stocks. After the attachment period, the virus-containing medium was removed and the cells were overlaid with equal volumes of a 2% SeaPlaque agarose (FMC) solution and a 2× growth medium. At 72 h postinfection, plaques were fixed with an acetic acid-ethanol-formaldehyde solution and stained with crystal violet.
Isolation of virion RNA for sequencing analysis.
To identify potential mutations in viruses derived from electroporated cells, virus-containing culture medium from the infected H1-HeLa cells (7 ml) was loaded onto a 30% sucrose cushion and centrifuged at 25,000 rpm at 15°C for 4 h (Beckman SW28 rotor). The virus pellet was resuspended in LS buffer (37), and viral RNA was isolated with the QiAMP viral RNA kit (Qiagen). Reverse transcription (RT) was performed using the OmniScript RT kit (Qiagen), viral RNA, and oligonucleotide P1 6159R. Briefly, PCR product was generated with one-fourth of the RT reaction mixture, Pfu polymerase (Stratagene), 4.5% dimethyl sulfoxide, and oligonucleotides P1 4730F and P1 6076R. PCR fragments were ligated into pCR-TOPO-Blunt vectors (Life Technologies) and transformed into XL-10 Gold competent cells. Ten successful transformants of each type were fully sequenced across the viral cDNA insert.
Construction of consensus E6V mutation into PV/R16-VPg.
Site-directed mutagenesis (QuikChange) was used to introduce the identified compensatory mutation E6V into the PV/R16-VPg chimeric backbone. Mutagenesis was initially performed on the PV/R16-VPg infectious cDNA clone using oligonucleotides P/R16 E6VS and P/R16 E6VAS. Transformants were sequenced and then digested with BglII and NheI for subcloning back into an identically cut PV/R16-VPg construct. Intact constructs were used to generate infectious viral RNA (T7 MegaScript) for electroporation into H1-HeLa cells.
RESULTS
Expression and purification of HRV16 3Dpol with an authentic N terminus.
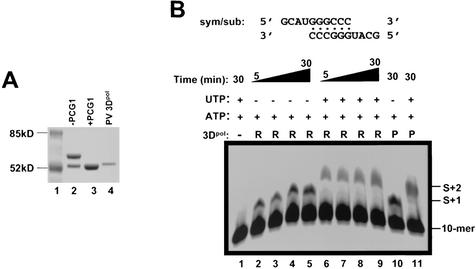
It was previously shown that the N-terminal region of the PV 3Dpol was essential for optimal enzymatic activity as the N terminus was reportedly wrapped around into the active site of the polymerase (15). Removal of a nonviral translation initiation amino acid (methionine) at the N terminus dramatically increased the enzymatic activity of PV 3Dpol (13, 31). Based on these observations, we adopted a similar binary expression strategy for HRV16 3Dpol in E. coli (13) to ensure that the expressed protein contained the authentic glycine residue at the N terminus. One plasmid (pET26Ub-3Dpol) produced HRV16 3Dpol as a fusion protein with ubiquitin at the N terminus and the other (pCG1) produced the ubiquitin C-terminal-specific protease Ubp1 (4, 13). Expression of Ubp1 in the same cells as ubiquitin-3Dpol resulted in cleavage of the fusion protein and release of 3Dpol with the authentic glycine residue at the N terminus (Fig. 1A, compare lanes 2 and 3). The expressed HRV16 3Dpol protein was purified to homogeneity as described in Materials and Methods. The authenticity and integrity of the N terminus of the purified protein was confirmed by Edman degradation/mass spectroscopy analysis.
FIG. 1.
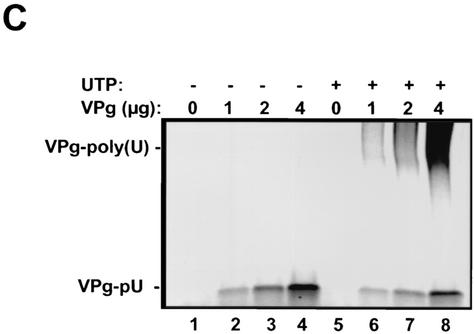
Purified HRV16 3Dpol is functionally active. (A) HRV16 3Dpol expressed from BL21(DE3) cells was purified on sequential nickel(II) and heparin chromatography columns prior to visualization on a sodium dodecyl sulfate-4 to 20% PAGE gel. Proteins were purified from BL21 cells transformed either with pET26Ub-3Dpol alone, which produced an ubiquitin-3Dpol fusion protein (lane 2), or together with the ubiquitinase-carrying pCG1 plasmid, which produced the fully cleaved HRV16 3Dpol (lane 3). Lane 4 contained the purified PV 3Dpol. (B) Nucleotide incorporation catalyzed by the purified HRV16 3Dpol with a duplexed symmetrical RNA template (sym/sub). Each reaction mixture contained 2 μM end-labeled, preannealed sym/sub RNA, 2 μg of HRV16 3Dpol (R), and 100 μM ATP (lanes 2 to 5) or 100 μM ATP and UTP (lanes 6 to 9). Purified PV 3Dpol (P) was also shown to be active in this assay (lanes 10 and 11). (C) Uridylylation of VPg peptide by HRV16 3Dpol. A 21-mer synthetic peptide corresponding to the wild-type HRV16 VPg was uridylylated (to become VPg-pU) by increasing amounts of VPg in the presence of HRV16 3Dpol, poly(A) RNA, and [α-33P]UTP (lanes 1 to 4). The addition of unlabeled UTP (100 μM) to the reaction mixture enabled elongation of the uridylylated UTP primer and produced elongated VPg-poly(U) (lanes 5 to 8).
Purified HRV16 3Dpol was enzymatically active in both primer-dependent elongation and VPg uridylylation assays.
Active picornaviral 3Dpol proteins are capable of catalyzing primer-dependent RNA replication and uridylylation of VPg, the protein primer required for initiation of RNA replication. To demonstrate that the purified HRV16 3Dpol was enzymatically active, we tested the protein in two assays designed to measure these activities. One assay used a duplexed, symmetrical RNA (sym/sub) template to assess primer-dependent RNA elongation activity (Fig. 1B). The other assay measured the ability of the purified HRV16 3Dpol to catalyze uridylylation of VPg peptide (Fig. 1C and 2). As shown in Fig. 1B, the HRV16 3Dpol was able to use sym/sub RNA as a template and catalyze nucleotide incorporation in a template-dependent fashion, an activity similar to that of PV 3Dpol. In the VPg uridylylation assay (Fig. 1C), the HRV16 3Dpol catalyzed, in the presence of poly(A), the covalent addition of radiolabeled UMP to a synthetic peptide corresponding to the VPg of HRV16 and produced the labeled product VPg-pU (Fig. 1C, lanes 2 to 4). Note that more HRV16 3Dpol protein was used in order to achieve comparable activity to PV 3Dpol in these assays (see Materials and Methods). In the presence of additional cold UTP, the labeled VPg-pU was further extended to the high-molecular-weight species VPg-poly(U) (Fig. 1C, lanes 6 to 8). These results demonstrated that the HRV16 3Dpol was enzymatically active and capable of RNA-dependent RNA polymerization as well as VPg uridylylation.
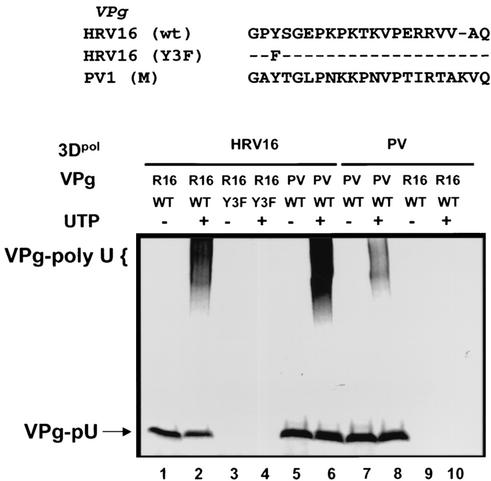
FIG. 2.
Uridylylation potential of HRV16 and PV 3Dpol toward heterologous VPg peptides. HRV16 3Dpol successfully uridylylated, at similar efficiencies, HRV16 VPg (lanes 1 and 2) and PV VPg peptides (lanes 5 and 6). PV 3Dpol uridylylated PV VPg peptide (lanes 7 and 8) but was unable to utilize HRV16 VPg peptide (lanes 9 and 10). In addition, the mutant VPg (Y3F) of HRV16, in which the tyrosine residue at position 3 had been changed to a phenylalanine residue, was inactive for uridylylation by HRV16 3Dpol (lanes 3 and 4). Odd lanes contained [α-33P]UTP alone; even lanes contained both [α-33P]UTP and 100 μM unlabeled UTP for further elongation.
Cross-uridylylation of VPg peptides by HRV16 3Dpol and PV 3Dpol.
The results in Fig. 1C showed that the purified HRV16 3Dpol possessed the ability to uridylylate its VPg peptide in vitro, an activity that is believed to be essential for the initiation of viral RNA replication. As reported previously for PV VPg uridylylation (28), the HRV16 VPg peptide lost its uridylylation capacity when the tyrosine residue at position 3 was changed to a phenylalanine (Y3F) (Fig. 2, lanes 3 and 4), confirming this tyrosine residue as the site of uridylylation. To assess potential viral specificity of 3Dpol-catalyzed uridylylation, we analyzed the cross-uridylylation activity of PV and HRV16 3Dpol by using opposite VPg peptides. As shown in Fig. 2, HRV16 3Dpol was able to uridylylate PV VPg as efficiently as it did with its natural substrate (R16 VPg) (Fig. 2, lanes 5 and 6). In contrast, no significant uridylylation of HRV16 VPg peptide was observed for PV 3Dpol (Fig. 2, lanes 9 and 10). This result suggested that different viral specificity existed between these two related polymerase proteins. PV 3Dpol seemed to possess a higher specificity than HRV16 3Dpol (or HRV2 3Dpol) (12, 30) in the selection of VPg substrate. This finding was somewhat surprising considering that HRV14 VPg was capable of acting as a uridylylation substrate for PV 3Dpol in vitro (29). This difference in uridylylation efficiency by PV 3Dpol was likely due to the sequence variations in VPg peptides between HRV14 and HRV16 (∼62% identity).
Replication capability of PV/R16-VPg and R16/PV-VPg chimeric RNA in H1-HeLa cells.
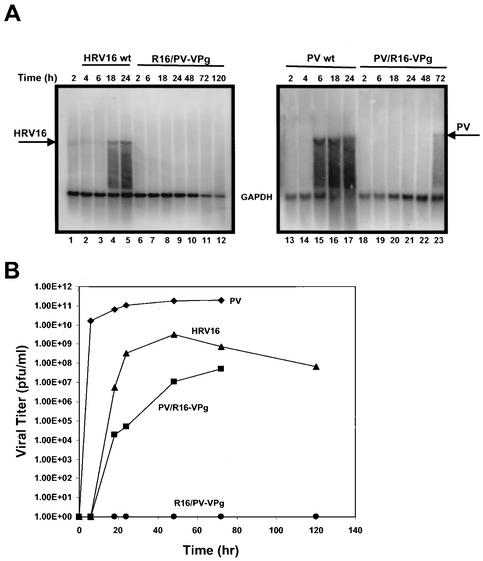
The previous observations that HRV16 3Dpol was capable of uridylylating PV VPg whereas PV 3Dpol was unable to uridylylate HRV16 VPg prompted us to investigate further whether such viral specificity existed in vivo. To this end, two chimeric clones were constructed. One was the PV containing the HRV16 VPg gene in place of its own VPg (PV/R16-VPg); the other was HRV16 containing PV VPg gene (R16/PV-VPg) (Fig. 3). The chimeric viral RNAs maintained the residues flanking the cleavage sites at the 3A-3B (Q/GA) and 3B-3C (Q/GP) junctions, which would likely be recognized by respective 3C proteinases. RNA derived from the chimeric clones was transfected into H1-HeLa cells, and RNA replication, virus production, and the appearance of CPE were monitored. It was found that PV/R16-VPg produced significant CPE approximately 48 to 72 h posttransfection, a rate much slower than that of wild-type PV (approximately 6 h). Interestingly, no CPE was observed for R16/PV-VPg in H1-HeLa cells up to 5 days posttransfection when incubated at 34°C (or 37°C) (data not shown). Northern blotting and plaque assay confirmed that cells transfected with PV/R16-VPg chimeric RNA, but not that of R16/PV-VPg, supported viral RNA synthesis and produced infectious viral particles (Fig. 4). This result was somewhat surprising since it did not correlate with the in vitro cross-uridylylation activities between PV and HRV16 3Dpol. As shown earlier, HRV16 3Dpol was able to uridylylate PV VPg as efficiently as its own VPg, but PV 3Dpol could not uridylylate HRV16 (Fig. 2). These results indicated that additional constraints might be affected in the R16/PV-VPg chimeric RNA. These constraints may include, but are not limited to, proteolytic processing at the chimeric junctions (3A-3B and 3B-3C, in this case), and interactions between the VPg primer and 3Dpol or VPg and the cis-acting RNA element in the context of other viral and cellular factors. Any defect in these important functions may abolish the ability of the chimeric RNA to replicate. We are currently in the process of identifying and characterizing such replication defect(s) in R16/PV-VPg chimeric RNA. Preliminary in vitro translation and processing experiments using HeLa S10 extracts were inconclusive due to the extremely inefficient translation from both R16/PV-VPg and HRV16 wild-type RNA templates in this system (data not shown) (5, 26, 27, 39).
FIG. 3.
Schematic representation of VPg (3B) chimeric virus constructs in a PV backbone (PV/R16-VPg) (left) or an HRV16 backbone (R16/PV-VPg) (right).
FIG. 4.
Viability of chimeric viral RNAs in transfected H1-HeLa cells. (A) PV/R16-VPg and R16/PV-VPg chimeric RNA, as well as wild-type (wt) PV and HRV16 RNA, were electroporated into H1-HeLa cells. Total cellular RNA was extracted from the transfected cells at various time intervals posttransfection and subjected to Northern blot analysis. Virus-specific probes from either HRV16 3Dpol (left panel) or PV 3Dpol (right panel) and GAPDH were used in the hybridization. (B) Production kinetics of infectious virus particles released from transfected cells as determined by plaque assay. Medium from the cells was collected over time and assayed for PFU as described in Materials and Methods. Infectious virions derived from the PV/R16-VPg chimera first appeared in the electroporated cultures between 6 to 18 h. No infectious virus was detected for the R16/PV-VPg chimera even after 120 h of incubation.
Sequence analysis of viral RNA isolated from the recovered PV/R16-VPg chimeric viruses after serial passages and identification of a mutation in HRV16 VPg.
To further characterize the viruses produced by H1-HeLa cells upon transfection with PV/R16-VPg RNA, medium from the transfected cells was serially passaged up to 10 times through naive H1-HeLa cells. Plaque assay was performed to determine virus titers and plaque morphology of the passaged chimeric viruses. As shown in Fig. 5A, a gradual increase in plaque size was observed during the extended passages, suggesting that cell culture adaptation(s) occurred with the chimeric virus.
FIG. 5.
Plaque morphology of serially passaged PV/R16-VPg chimeric viruses and sequence alteration in the VPg region. (A) Virus particles generated from H1-HeLa cells transfected with PV/R16-VPg chimeric (P0) were further passaged 10 times (P1 to P10). A plaque assay was then performed to determine changes in plaque phenotype. Shown are typical plaques from P0, P1, and P10 of the chimeric virus and from wild-type PV1(M) for comparison. All plaques were developed over a period of 72 h at 37°C. (B) Sequence analysis of the VPg gene in the chimeric viral RNA at various passages. A consensus mutation was identified at position 6 (Glu to Val or Ala) of the VPg gene. This mutation became fixed on E6V after the P0 generation. No other sequence changes were noted in the chimeric backbone.
To determine whether the HRV16 VPg sequence was conserved during the passaging and to identify any possible compensatory or adaptive mutation(s), viral RNA was extracted from the recovered viral particles at different passages and the region spanning the VPg gene was amplified by RT-PCR. At least 10 independent clones were sequenced for each passage. Sequencing analysis showed that HRV16 VPg was found in all the chimeric isolates, confirming that the virus particles were indeed produced from the chimeric viral RNA PV/R16-VPg. More interestingly, a single nucleotide mutation (A to T) was observed at amino acid position 6 of the HRV16 VPg sequence, consequently altering a negatively charged glutamate residue to a valine residue (E6V) in virion RNA of all passages (Fig. 5B). In the initial cells transfected with the chimeric RNA (P0), the glutamate residue (E) at this position was changed to either an alanine (E6A) or a valine (E6V). However, only the change to valine (E6V) was further propagated in the subsequent passages (P1 to P10). This result indicated that the presence of the negatively charged glutamate residue in HRV16 VPg was detrimental to the efficient initiation of RNA replication for the chimeric PV virus. Changing of the glutamate residue to a noncharged hydrophobic reside, preferably a valine residue, significantly improved the replication capability of the chimeric virus. It is noteworthy to mention that a hydrophobic leucine residue is present at this position in the natural PV VPg, suggesting that the hydrophobic residue at this position is preferred in interactions with PV 3Dpol during initiation of viral RNA replication.
Based on these observations, we concluded that the initial PV/R16-VPg chimeric RNA was capable of low-level RNA replication and that PV 3Dpol quickly created compensatory mutations in the HRV16 VPg region (E6V or E6A). Further improvement in plaque phenotype, however, was likely due to the appearance of additional compensatory mutations in other regions of the viral genome.
E6V mutation in HRV16 VPg is highly compensatory in enhancing chimeric PV/R16-VPg RNA replication.
To demonstrate that the E6V mutation is indeed compensatory for replication of the chimeric RNA, site-directed mutagenesis was performed to introduce the mutation into the chimeric clone. RNA transcribed from the modified clone was transfected into H1-HeLa cells, along with the unmodified chimeric RNA as well as the wild-type PV RNA. Aliquots of culture medium from the transfected cells were collected at various time intervals after transfection, and the production of virus particles was determined by plaque assay. As shown in Fig. 6A, the E6V mutation enabled the chimeric RNA to produce infectious virus particles much sooner than the original chimeric RNA (PV/R16-VPg). The viruses that resulted from transfection of modified chimeric RNA (PV/R16-VPg/E6V) replicated with similar kinetics and magnitude to wild-type PV RNA. The unmodified PV/R16-VPg RNA showed a 14-h delay in initiation of virus production, though it reached a similar virus titer at much later time points. This result demonstrated that the E6V mutation in the HRV16 VPg gene significantly enhanced the replication capability of the chimeric viral RNA at the initiation stage. On the other hand, this result also implied that the negatively charged residue at position 6 of HRV16 VPg was detrimental to PV replication. This observation was consistent with the sequence analysis of the recovered chimeric virus particles where the E6V change occurred quite early after RNA transfection (P0) and the requirement for amplifying such a change might account for the time delay in infectious virus production. The plaque sizes for the PV/R16-VPg chimera and the E6V revertant virus (72 h postelectroporation) were judged to be smaller than those of the PV1(M) plaques, suggesting that these altered viruses suffered from a decrease in viral replication robustness (Fig. 6B).
FIG. 6.
The E6V mutation within VPg of PV/R16-VPg chimeric RNA significantly enhanced its replication capability in transfected cells. (A) Growth kinetics of wild-type (wt) or chimeric viral RNA upon transfection of H1-HeLa cells. Supernatants from the electroporated cells were plaque assayed to determine virus titers. (B) Plaque morphologies from supernatants collected 72 h posttransfection.
E6V mutation in HRV16 VPg moderately enhanced its substrate activity for uridylylation by PV 3Dpol.
To determine whether the cell culture-induced compensatory mutation identified from culturing the PV/R16-VPg chimeric virus enhanced the uridylylation activity of HRV16 VPg by PV 3Dpol, synthetic peptides were made and tested in the in vitro uridylylation assay. One peptide representing HRV16-VPg contained the E6V modification while a second peptide PV-VPg, with an L6E modification (note that the corresponding position in PV VPg is a leucine), served to demonstrate the impact of altering the VPg position 6 to a negatively charged residue. As shown in Fig. 7, the HRV16 E6V VPg mutation enabled a partial recovery of VPg uridylylation compared with wild-type VPg (Fig. 7, compare lanes 7 and 8 to lanes 5 and 6). This result suggested that the cell culture-induced adaptive mutation, while only moderately improving the uridylylation of the HRV16 VPg by PV 3Dpol, might be involved in additional functions critical for efficient initiation of viral RNA replication. Alternatively, it is possible that the VPg uridylylation reaction is not the rate-limiting step in the initiation of PV RNA replication. The E6V compensatory mutation in R16-VPg rendered enough substrate activity (despite being suboptimal) for PV 3Dpol and resulted in efficient RNA replication. Inclusion of the L6E mutation in PV VPg drastically reduced the ability of PV 3Dpol to use this peptide as a uridylylation substrate (Fig. 7, compare lanes 3 and 4 to lanes 1 and 2). Consistent with this, PV RNA carrying the L6E mutation in the VPg gene was defective in replication in transfected H1-HeLa cells (data not shown). These results highlighted the significance of the VPg amino acid residue at position 6 for successful in vitro uridylylation and virus replication in vivo.
FIG. 7.
PV 3Dpol requires a hydrophobic residue at position 6 of VPg peptide for in vitro uridylylation. Wild-type (WT) PV VPg (lanes 1 and 2), mutant PV VPg/L6E (lanes 3 and 4), WT HRV16 VPg (lanes 5 and 6), and mutant HRV16 VPg/E6V (lanes 7 and 8) were tested for in vitro uridylylation by PV 3Dpol. The E6V mutation in HRV16 VPg rendered partial recovery in uridylylation activity by PV 3Dpol.
DISCUSSION
RNA-dependent RNA polymerases are key players in the replication of positive-stranded RNA viruses such as the human rhinoviruses. In the case of the Picornaviridae family, the interaction and subsequent uridylylation of the protein primer, VPg, by the viral polymerase 3Dpol is crucial to the successful initiation of RNA replication. Indeed, it is generally thought that uridylylation of VPg represents the initial reaction in minus-strand RNA synthesis (12). Although many details regarding the identification of amino acid residues essential for PV or HRV 3Dpol-directed in vitro VPg uridylylation have been reported (12, 28-30, 33), we sought to extend these studies by using chimeric PV/HRV16 in order to identify critical residues in VPg necessary for assessing combined uridylylation and in vivo replication potentials.
In this study, we compared the cross-uridylylation activities of the HRV16 and PV 3Dpol polymerases. It was found that HRV16 3Dpol was able to uridylylate PV VPg, but PV 3Dpol was unable to uridylylate HRV16 VPg. This finding was consistent with a previous report that HRV2 3Dpol, which is approximately 90% homologous to HRV16 3Dpol, was capable of uridylylating PV VPg (12) but less consistent with another report where PV 3Dpol uridylylated VPg from HRV14 (29). A major difference between HRV14 and HRV16 VPg peptides is the presence of a negatively charged residue (Glu) at position 6 of HRV16 VPg, which is absent in HRV14 VPg. It is conceivable that the presence of a negatively charged residue at this location perturbs the highly adapted molecular interaction between the PV 3Dpol and VPg primer, hence preventing efficient uridylylation. It is noteworthy to mention that an uncharged polar residue is present at this position in VPg of HRV14 (Asn), and this might account in part for the uridylylation activity of HRV14 VPg but not of HRV16 VPg by PV 3Dpol. This hypothesis was further supported by the in vivo replication study of a chimeric PV (PV/R16-VPg) whose VPg gene was replaced by that of HRV16. A compensatory mutation (E6V) at position 6 was identified in progeny virion RNA, which resulted in significant enhancement of viral replication. Therefore, it is apparent that the amino acid residue at position 6 of the VPg peptide plays a key role in determining viral specificity during uridylylation, and the subsequent initiation of viral RNA replication. Based on the observations in this study, we predict that PV 3Dpol would prefer a hydrophobic residue at this position while HRV16 3Dpol can accommodate a negatively charged residue. In fact, E6V modification in HRV16 VPg peptide did not affect its uridylylation potential by HRV16 3Dpol (data not shown). This residue is likely involved in direct interaction with either the viral polymerase protein or the incoming UTP molecule, or both, during the uridylylation reaction. Interestingly, a PV containing a methionine residue at VPg position 6 has been reported to be replication competent (8, 19), supporting the notion that PV prefers an amino acid with a nonpolar hydrophobic side chain at this location. Another report indicated that a PV/coxsackievirus VPg chimera, which contained a valine residue at VPg position 6 (plus four other alterations at positions 9, 12, 16, and 18) was viable but less fit than wild-type PV (34). In addition, a PV construct containing tandemly repeated VPg sequences which differed only at VPg position 6 (either Leu or Met) generated viable virus with a complete deletion of one VPg sequence (8). The VPg sequence maintained in the virus always derived from the VPg adjacent to the 3A sequence regardless of VPg type. The authors stated that selection was unlikely to be based on the VPg position 6 sequence but rather may have occurred through a homologous recombination or intramolecular deletion event.
A recent report by Paul et al. (30) demonstrated the viability of a PV/HRV14 VPg chimera 48 h posttransfection (compared with 20 h for wild-type PV) with the occurrence of a compensatory mutation (L12P) in the HRV14 VPg. This single nucleotide mutation restored the proline residue found at the comparable position 11 in the wild-type PV VPg. Curiously, no viable progeny virions were obtained if the rhinovirus VPg derived from serotypes 2 or 89 (subgroup B viruses) was used. Both HRV2 and HRV89 VPg contain only 2 different amino acid residues from that of HRV16 (positions 13 and 20), yet they possess identical glutamate residues at positions 6 and 15. It is unclear why E6V reversion mutations identified with our PV/HRV16 VPg chimeras could not have developed with PV/HRV2 and PV/HRV89 VPg chimeras in vivo. It is conceivable that the unique residues at positions 13 and 20 of HRV2 and HRV89 VPg completely abolished compatibility with PV polymerase and subsequent initiation of viral replication. Therefore, adaptive mutations did not occur for these chimeras due to the lack of viral RNA replication. In comparison, PV/HRV16 VPg chimera may still allow at least low levels of viral RNA replication that created the required VPg mutation for virus growth.
Little is known regarding the precise location on 3Dpol where VPg binds or the temporal regulation of this interaction. Mutational analysis of PV VPg has led to the identification of several key amino acid residues, including Tyr3, Gly5, Lys9, Lys10, and Arg17, which are essential for both uridylylation and viral growth (12, 28, 30). Exactly how these residues participate with the 3Dpol in the uridylylation of VPg remains unknown. In PV, several residues in 3Dpol have been implicated in binding the precursor protein 3AB, including F377, R379, E328, and V391 (16, 23, 43). These residues are believed to form part of a 3Dpol surface binding site for 3AB, and mutation of any of these amino acids leads to both decreased VPg (3B) uridylylation and binding to 3Dpol. Mutational analysis in the flanking regions identified residues that were less critical for successful 3AB binding yet important for uridylylation (M394T, R358A, R359A, and K395A). Collectively, these data represent the current proposed model of physical and functional interactions between the two proteins without consideration of how the RNA template (cre) might further influence these interactions. A complexed structural model of protein RNA will be necessary to define this influence. Information concerning protein-protein interactions between 3Dpol and VPg are not yet available for other related picornaviruses, including HRV16. Based on sequence alignment, similar residues are also present in HRV16 3Dpol. It is likely that additional residues in the polymerase protein are also involved in binding to VPg, and these residues may account for the different preference in substrate specificity.
Acknowledgments
This work was supported by Ribapharm, Inc.
We thank Craig Cameron for kindly providing the PV infectious cDNA clone, purified PV polymerase, and expression vectors pCG1 and pET26-Ub; Wai-Ming Lee for the HRV infectious clone; David Barton for providing the HeLa S10 extract for in vitro translation reactions; and Agnes Henschen for protein sequencing. We also thank Vicky C. H. Lai, Shahul Nilar, and Nanhua Yao for helpful discussions.
REFERENCES
- 1.Andries, K., B. Dewindt, J. Snoeks, L. Wouters, H. Moereels, P. J. Lewi, and P. A. J. Janssen. 1990. Two groups of rhinoviruses revealed by a panel of antiviral compounds present sequence divergence and differential pathogenicity. J. Virol. 64:1117-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, E., J. W. Erickson, G. Shay Fout, E. A. Frankenberger, H.-J. Hecht, M. Luo, M. G. Rossmann, and R. R. Rueckert. 1984. Virion orientation in cubic crystals of human common cold virus HRV14. J. Mol. Biol. 177:417-430. [DOI] [PubMed] [Google Scholar]
- 3.Arnold, J. J., and C. E. Cameron. 2000. Poliovirus RNA-dependent RNA polymerase (3Dpol). Assembly of stable elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub). J. Biol. Chem. 275:5329-5336. [DOI] [PubMed] [Google Scholar]
- 4.Baker, R. T., S. A. Smith, R. Marano, J. McKee, and P. G. Board. 1994. Protein expression using cotranslational fusion and cleavage of ubiquitin. J. Biol. Chem. 269:25381-25386. [PubMed] [Google Scholar]
- 5.Barton, D. J., E. P. Black, and J. B. Flanegan. 1995. Complete replication of poliovirus in vitro: preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J. Virol. 69:5516-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butterworth, B. E., R. R. Grunert, B. D. Korant, K. Longberg-Holm, and F. H. Yin. 1976. Replication of rhinoviruses. Arch. Virol. 51:169-189. [DOI] [PubMed] [Google Scholar]
- 7.Callahan, P. L., S. Mizutani, and R. J. Colonno. 1985. Molecular cloning and complete sequence determination of the RNA genome of human rhinovirus type 14. Proc. Natl. Acad. Sci. USA 82:732-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, X., R. J. Kuhn, and E. Wimmer. 1993. Replication of poliovirus RNA containing two VPg coding sequences leads to a specific deletion event. J. Virol. 67:5572-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crotty, S., D. Maag, J. J. Arnold, W. Zhong, J. Y. N. Lau, Z. Hong, R. Andino, and C. E. Cameron. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6:1375-1379. [DOI] [PubMed] [Google Scholar]
- 10.Erickson, J. W., E. A. Frankenberger, M. G. Rossmann, G. Shay Fout, K. C. Medappa, and R. R. Rueckert. 1983. Crystallization of a common cold virus, human rhinovirus 14: “isomorphism” with poliovirus crystals. Proc. Natl. Acad. Sci. USA 80:931-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flanegan, J. B., and D. Baltimore. 1977. A poliovirus-specific primer-dependent RNA polymerase able to copy poly(A). Proc. Natl. Acad. Sci. USA 74:3677-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerber, K., E. Wimmer, and A. V. Paul. 2001. Biochemical and genetic studies of the initiation of human rhinovirus 2 RNA replication: purification and enzymatic analysis of the RNA-dependent RNA polymerase 3Dpol. J. Virol. 75:10969-10978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gohara, D. W., C. S. Ha, S. K. B. Ghosh, J. J. Arnold, T. J. Wisniewski, and C. E. Cameron. 1999. Production of “authentic” poliovirus RNA-dependent RNA polymerase (3Dpol) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr. Purif. 17:128-138. [DOI] [PubMed] [Google Scholar]
- 14.Gohara, D. W., S. Crotty, J. J. Arnold, J. D. Yoder, R. Andino, and C. E. Cameron. 2000. Poliovirus RNA-dependent RNA polymerase (3Dpol). J. Biol. Chem. 275:25523-25532. [DOI] [PubMed] [Google Scholar]
- 15.Hansen, J. L., A. M. Long, and S. C. Schultz. 1997. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5:1109-1122. [DOI] [PubMed] [Google Scholar]
- 16.Hope, D. A., S. E. Diamond, and K. Kirkegaard. 1997. Genetic dissection of interaction between poliovirus 3D polymerase and viral protein 3AB. J. Virol. 71:9490-9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horton, R. M., Z. L. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8:528-535. [PubMed] [Google Scholar]
- 18.Hung, M., C. S. Gibbs, and M. Tsiang. 2002. Biochemical characterization of rhinovirus RNA-dependent RNA polymerase. Antiviral Res. 56:99-114. [DOI] [PubMed] [Google Scholar]
- 19.Kuhn, R. J., H. Tada, M. F. Y. Wong, J. J. Dunn, B. L. Semler, and E. Wimmer. 1988. Construction of a “mutagenesis cartridge” for poliovirus genome-linked viral protein: isolation and characterization of viable and nonviable mutants. Proc. Natl. Acad. Sci. USA 85:519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lama, J., A. V. Paul, K. S. Harris, and E. Wimmer. 1994. Properties of purified recombinant poliovirus protein 3AB as substrate for viral proteinases and as a cofactor for RNA polymerase 3Dpol. J. Biol. Chem. 269:66-70. [PubMed] [Google Scholar]
- 21.Lee, W.-M., W. Wang, and R. R. Rueckert. 1994. Complete sequence of the RNA genome of human rhinovirus 16, a clinically useful common cold virus belonging to the ICAM-1 receptor group. Virus Genes 9:177-181. [DOI] [PubMed] [Google Scholar]
- 22.Lee, Y. F., A. Nomoto, B. M. Detjen, and E. Wimmer. 1977. A protein covalently linked to poliovirus genome RNA. Proc. Natl. Acad. Sci. USA 74:59-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyle, J. M., A. Clewell, K. Richmond, O. C. Richards, D. A. Hope, S. C. Schultz, and K. Kirkegaard. 2002. Similar structural basis for membrane localization and protein priming by an RNA-dependent RNA polymerase. J. Biol. Chem. 277:16324-16331. [DOI] [PubMed] [Google Scholar]
- 24.McKnight, K. L., and S. M. Lemon. 1996. Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J. Virol. 70:1941-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKnight, K. L., and S. M. Lemon. 1998. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA 4:1569-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 27.Pathak, H. B., S. K. B. Ghosh, A. W. Roberts, S. D. Sharma, J. D. Yoder, J. J. Arnold, D. W. Gohara, D. J. Barton, A. V. Paul, and C. E. Cameron. 2002. Structure-function relationships of the RNA-dependent RNA polymerase from poliovirus (3Dpol). J. Biol. Chem. 277:31551-31562. [DOI] [PubMed] [Google Scholar]
- 28.Paul, A. V., J. H. van Boom, D. Filippov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature (London) 393:280-284. [DOI] [PubMed] [Google Scholar]
- 29.Paul, A. V., E. Reider, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paul, A. V., J. Peters, J. Mugavero, J. Yin, J. H. van Boom, and E. Wimmer. 2003. Biochemical and genetic studies of the VPg uridylylation reaction catalyzed by the RNA polymerase of poliovirus. J. Virol. 77:891-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plotch, S. J., O. Palant, and Y. Gluzman. 1989. Purification and properties of poliovirus RNA polymerase expressed in Escherichia coli. J. Virol. 63:216-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plotch, S. J., and O. Palant. 1995. Poliovirus protein 3AB forms a complex with and stimulates the activity of the viral RNA polymerase, 3Dpol. J. Virol. 69:7169-7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reider, E., A. V. Paul, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74:10371-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reuer, Q., R. J. Kuhn, and E. Wimmer. 1990. Characterization of poliovirus clones containing lethal and nonlethal mutations in the genome-linked protein VPg. J. Virol. 64:2967-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohll, J. B., D. H. Moon, D. J. Evans, and J. W. Almond. 1995. The 3′ untranslated region of picornavirus RNA: features required for efficient genome replication. J. Virol. 69:7835-7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossmann, M. G., E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H.-J. Hecht, J. E. Johnson, G. Kamer, M. Luo, A. G. Mosser, R. R. Rueckert, B. Sherry, and G. Vriend. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145-153. [DOI] [PubMed] [Google Scholar]
- 37.Sherry, B., and R. Rueckert. 1985. Evidence for at least two dominant neutralization antigens on human rhinovirus 14. J. Virol. 53:137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stanway, G., P. J. Hughes, R. C. Mountford, P. D. Minor, and J. W. Almond. 1984. The complete nucleotide sequence of a common cold virus: human rhinovirus 14. Nucleic Acids Res. 12:7859-7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Todd, S., J. S. Towner, and B. L. Semler. 1997. Translation and replication properties of the human rhinovirus genome in vivo and in vitro. Virology 229:90-97. [DOI] [PubMed] [Google Scholar]
- 40.Turner, R. B. 2001. The treatment of rhinovirus infections: progress and potential. Antiviral Res. 49:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, W., and B. A. Malcolm. 1999. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange site-directed mutagenesis. BioTechniques 26:680-682. [DOI] [PubMed] [Google Scholar]
- 42.Wimmer, E. 1982. Genome-linked proteins of viruses. Cell 28:199-201. [DOI] [PubMed] [Google Scholar]
- 43.Xiang, W., A. Cuconati, D. Hope, K. Kirkegaard, and E. Wimmer. 1998. Complete protein linkage map of poliovirus P3 proteins: interaction of polymerase 3Dpol with VPg and with genetic variants of 3AB. J. Virol. 72:6732-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]