Abstract
Small interfering RNAs (siRNAs) can induce potent gene silencing by degradation of cognate mRNA. However, in dividing cells, the silencing lasts only 3 to 7 days, presumably because of siRNA dilution with cell division. Here, we investigated if sustained siRNA-mediated silencing of human immunodeficiency virus type 1 (HIV-1) is possible in terminally differentiated macrophages, which constitute an important reservoir of HIV in vivo. CCR5, the major HIV-1 coreceptor in macrophages, and the viral structural gene for p24 were targeted either singly or in combination. When transfected 2 days prior to infection, both CCR5 and p24 siRNAs effectively reduced HIV-1 infection for the entire 15-day period of observation, and combined targeting of both genes abolished infection. To investigate whether exogenously introduced siRNA is maintained stably in macrophages, we tested the kinetics of siRNA-mediated viral inhibition by initiating infections at various times (2 to 15 days) after transfection with CCR5 and p24 siRNAs. HIV suppression mediated by viral p24 siRNA progressively decreased and was lost by day 7 posttransfection. In contrast, viral inhibition by cellular CCR5 knockdown was sustained even when transfection preceded infection by 15 days, suggesting that the continued presence of target RNA may be needed for persistence of siRNA. The longer sustenance of CCR5 relative to p24 siRNA in uninfected macrophages was also confirmed by detection of internalized siRNA by modified Northern blot analysis. We also tested the potential of p24 siRNA to stably silence HIV in the setting of an established infection where the viral target gene is actively transcribed. Under these circumstances, long-term suppression of HIV replication could be achieved with p24 siRNA. Thus, siRNAs can induce potent and long-lasting HIV inhibition in nondividing cells such as macrophages.
RNA interference (RNAi) is an evolutionarily conserved posttranscriptional gene-silencing mechanism in which small interfering 21- to 23-mer double-stranded RNA (siRNA) mediates sequence-specific degradation of mRNA (22, 30). The recent discovery that exogenously delivered siRNA can trigger RNAi in mammalian cells raises the possibility of harnessing RNAi technology as a therapeutic tool against pathogenic viruses (11). Several studies have recently shown that siRNAs can suppress human immunodeficiency virus type 1 (HIV-1) replication in cell lines and proliferating CD4 T cells (6, 8, 13, 14, 17, 26, 28).
The advantage of using siRNA as a potential antiviral tool is that it is effective at concentrations that are several orders of magnitude lower than that required for other RNA-based antiviral gene therapies, such as antisense RNA or ribozymes (4, 10). However, the silencing effect of siRNAs in actively replicating cells peaks around 96 h but tapers off thereafter and is lost by 7 days (26, 33, 34). Because most studies have used actively dividing cell lines for siRNA studies, it is generally presumed that the siRNA effect is transient because of its dilution with cell division. However, whether siRNA can stably suppress gene expression in nondividing cells has not been tested. Terminally differentiated macrophages offer an ideal cell type with which to test this possibility. Macrophages constitute a significant reservoir for HIV-1 (23) and, being relatively immune to the cytopathic effects of HIV-1, survive for long periods after infection (2).
Previous studies of siRNA-directed protection against HIV-1 have generally targeted viral genes to suppress HIV-1 (6, 8, 13, 14, 17, 26). In an earlier study, we achieved viral suppression by targeting the cellular CD4 receptor (26). However, CD4 targeting may not be a feasible therapeutic approach because of its importance in immune function. On the other hand, CCR5, the major HIV-1 coreceptor for viral entry into macrophages, may be a potentially useful cellular target (16) since a 32-bp homozygous deletion of the gene, which abolishes its function, has no deleterious immunological consequences (25) but provides protection from HIV-1 infection (19).
In the present study, we aimed to achieve sustained and synergistic siRNA-mediated inhibition of HIV-1 replication in monocyte-derived macrophages (MDMs) by silencing the genes for cellular CCR5 and viral p24.
MATERIALS AND METHODS
Preparation of human MDMs.
Human monocytes were isolated from buffy coats prepared from healthy volunteer donors. Peripheral blood mononuclear cells isolated by Ficoll-Hypaque (Pharmacia Corporation, Peapack, N.J.) density gradient centrifugation were seeded at 2 × 106/ml in 24-well plates in RPMI 1640 medium (BioWhittaker, Inc., Walkersville, Md.) supplemented with 10% heat-inactivated human AB serum (Nabi, Boca Raton, Fla.), 50 U of penicillin per ml, 50 μg of streptomycin per ml, and 2 mM l-glutamine. After 5 days of culture, nonadherent cells were removed by repeated gentle washing with warm medium. More than 95% of the adherent cells obtained with this technique were CD14+ macrophages (data not shown).
Preparation of siRNAs.
All siRNAs, including the Cy5-labeled p24 siRNA, were synthesized at Dharmacon Research, Lafayette, Colo. The sequences of the sense and antisense strands of siRNAs were as follows: CCR5, 5′-P.CUCUGCUUCGGUGUCGAAAdTdT-3′ (sense) and 5′-P.UUUCGACACCGAAGCAGAGdTdT-3′ (antisense); p24, 5′-P.GAUUGUACUGAGAGACAGGCU-3′ (sense) and 5′-P.CCUGUCUCUCUCAGUACAAUCUU-3′ (antisense); green fluorescent protein (GFP), 5′-P.GGCUACGUCCAGGAGCGCACC-3′ (sense) and 5′-P.UGCGCUCCUGGACGUAGCCUU-3′ (antisense).
RNAs were deprotected and annealed in accordance with the manufacturer's instructions.
Transfection of siRNAs.
Adherent macrophages were generated by seeding peripheral blood mononuclear cells at 2 × 106/well in 24-well plates. Before transfection, the cells were washed and resuspended in 900 μl of RPMI 1640 medium. Cationic lipid complexes, prepared by incubating 1 μM siRNA duplexes with 3 μl of oligofectamine (Gibco-Invitrogen, Rockville, Md.) in 100 μl of RPMI 1640 medium, were added to the wells. The cells were washed after overnight incubation and resuspended in RPMI 1640 medium with serum for further experiments.
Flow cytometry.
To test CCR5 expression and HIV-1 infection, adherent MDMs were trypsinized at the times indicated and stained with biotin-conjugated CCR5 antibody (R&D Systems, Inc., Minneapolis, Minn.), followed by avidin-labeled streptavidin-phycoerythrin (BD Pharmingen, San Diego, Calif.). Where indicated, the cells were permeabilized with the Caltag Laboratories (Burlingame, Calif.) Fix and Perm kit and stained with fluorescein isothiocyanate (FITC)-labeled p24 monoclonal antibody (Beckman Coulter, Brea, Calif.). Cells were analyzed by flow cytometry on a FACScalibur instrument with CellQuest software (Becton Dickinson, Franklin Lakes, N.J.).
HIV-1 infection.
MDMs were infected with the R5 BAL strain of HIV-1 by using 50 ng of p24gag antigen per well. At the times indicated, HIV-1 replication in infected macrophages was evaluated by flow cytometric analysis of p24 expression. Cell-free viral production was measured by p24 antigen enzyme-linked immunosorbent assay (ELISA) of supernatants with the Alliance HIV-1 p24 ELISA kit (Perkin-Elmer Life Science Inc., Boston, Mass.).
In situ hybridization.
Infected macrophages cultured in slide chambers were evaluated for HIV-1 mRNA expression by using the ViroTect HIV-1 Cell Detection System (Invirion Inc., Frankfurt, Mich.). Cells were fixed, permeabilized, and hybridized with an FITC-labeled gag-pol oligonucleotide probe cocktail as described in the manufacturer's protocol. The cells were stained with Texas red-X phalloidin (Molecular Probes Inc., Eugene, Oreg.) and analyzed by epifluorescence microscopy. The fluorochromes were independently recorded at excitation wavelengths of 494 and 591 nm.
Reverse transcription PCR (RT-PCR).
Total RNA was extracted from macrophage cultures at the times indicated with an RNeasy Mini Kit (QIAGEN Inc., Valencia, Calif.), and cDNA was synthesized with a TaqMan reverse transcription kit (Applied Biosystems, Foster City, Calif.). Aliquots of cDNA were PCR amplified for CCR5 with the primers 5′-ATGGATTATCAAGTGTCAAGTCC-3′ and 5′-CCAGAATTGATACTGACTGTATGG-3′ and for γ-actin with the primers 5′-TCTGTCAGGGTTGGAAAGTC-3′ and 5′-AAATGCAAACCGCTTCCAAC-3′. Amplified PCR products were visualized on 1.2% agarose gels.
Modified Northern blot assay.
Total cellular RNA was extracted with Trizol reagent in accordance with the manufacturer's protocol (Invitrogen Life Technologies, Carlsbad, Calif.). After precipitation, the RNA pellets were washed in 70% ethanol, air dried, resuspended in nuclease-free H2O, and quantitated by UV absorption. Three micrograms of total RNA was loaded onto a Tris-borate-EDTA-15% urea gel and electrophoresed at 10 W. The gel was electrotransferred to Nytran Plus (Schleicher & Schuell, Inc., Keene, N.H.) for 2.5 h, UV cross-linked at 1,200 μF, and prehybridized for 30 min at 40°C in UltraHyb buffer (Ambion). One hundred picomoles of the sense strand of CCR5 or p24 siRNA was end labeled with [γ-32P]ATP (150 μCi). The labeled probe was purified on a G-25 MicroSpin Column (Amersham), heated to 65°C for 5 min, added to the prehybridization buffer, and hybridized overnight. Blots were washed at room temperature (2 × 5 min in 2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]-0.1% sodium dodecyl sulfate and 3 × 10 min in 0.1× SSC-0.1% sodium dodecyl sulfate) and analyzed on a phosphorimager (Molecular Dynamics, Carlsbad, Calif.).
RESULTS
Efficient uptake of labeled siRNA in transfected MDMs.
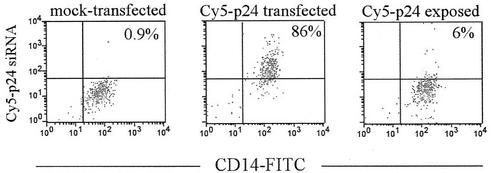
To determine the efficiency of siRNA delivery in MDMs, we transfected the cells with Cy5-labeled p24 siRNA. After 24 h of transfection, 86% of the CD14+ macrophages were Cy5+ by flow cytometry (Fig. 1), a transfection efficiency comparable to that observed with HeLa cells (∼90%; data not shown). The siRNA was not taken up efficiently by nonspecific phagocytosis because, in the absence of oligofectamine, <6% of the MDMs were Cy5+ (Fig. 1).
FIG. 1.
Efficient delivery of duplex siRNA into macrophages. MDMs were mock transfected or exposed to Cy5-labeled p24 siRNA in the presence or absence of transfection reagent. After 24 h of culture, the cells were removed by trypsinization, stained with anti-CD14-FITC, and analyzed by flow cytometry. The percentage of Cy5+ cells is indicated in each panel.
The CCR5 and p24 siRNAs inhibit HIV-1 replication in MDMs for prolonged periods of time, and when used in combination, they completely abolish infection.
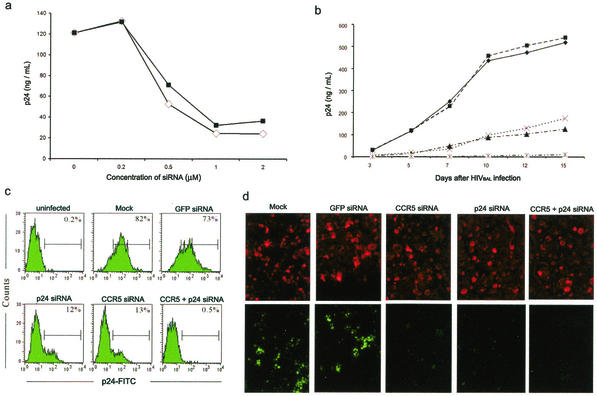
To determine the antiviral effects of siRNAs, MDMs were transfected with either CCR5 or p24 siRNA at a range of concentrations (0.2 to 2 μM) and challenged with the R5 BAL macrophagetropic virus strain 2 days later. After further culture for 7 days, the reduction of cell-free viral particle production was assessed by p24 ELISA. The p24 titers were reduced relative to those of mock-transfected cultures in both CCR5 and p24 siRNA-transfected MDMs, with a maximal sixfold inhibition at a 1 μM dose (Fig. 2a). No reduction in p24 levels was seen in MDMs transfected with 2 μM unrelated GFP siRNA (data not shown). To evaluate the stability of viral suppression, MDMs were transfected with CCR5 or p24 siRNA at a 1 μM concentration singly or in combination and similarly challenged with the R5 BAL virus. Periodic p24 ELISAs of culture supernatants revealed a sustained four- to sixfold reduction of p24 production in both CCR5 and p24 siRNA-transfected MDMs compared to those of mock-transfected and GFP siRNA-transfected controls for the total duration of the experiment (Fig. 2b). Similarly, flow cytometric analysis of p24 expression also demonstrated a sevenfold reduction in p24 expression for up to 15 days with either CCR5 or p24 siRNA compared to that of controls (Fig. 2c). Fluorescence in situ hybridization analysis of the cultures on day 7 after infection revealed a corresponding reduction in HIV-1 RNA in CCR5 or p24 siRNA-transfected MDMs (Fig. 2d). More importantly, cotransfection with both siRNAs was able to abolish HIV-1 infection throughout the 15-day period of observation (Fig. 2b to d). Thus, siRNAs can provide lasting protection against HIV-1 in macrophages.
FIG. 2.
CCR5 and p24 siRNAs inhibit HIVBAL infection in MDMs. (a) MDMs were transfected with the indicated doses of CCR5 (▪) or p24 (□) siRNA and infected 2 days later with HIVBAL. Cell-free virus production was measured on day 7 postinfection by p24 ELISA. (b) MDMs were either mock transfected (♦) or transfected with the GFP (▪), p24 (▴), or CCR5 (×) siRNA or with the p24 and CCR5 siRNAs (*) and infected after 2 days with HIVBAL, and virus production was measured by p24 ELISA at the indicated times postinfection. (c) The siRNA-transfected cells described in panel b were stained with anti-p24-FITC 15 days after infection and examined by flow cytometry. The percentage of p24+ cells is shown in each panel. (d) siRNA-transfected and HIVBAL-infected MDMs were probed for HIV-1 RNA by in situ hybridization with a fluorescein-labeled HIV-1 gag-pol oligonucleotide probe cocktail 7 days after infection. Fluorescence microscopy (magnification, ×200) was used to evaluate fluorescence signals for HIV-1 RNA (bottom). At the top are the same cells counterstained with Texas red-X phalloidin.
Differential persistence and antiviral kinetics of CCR5 and p24 siRNA in MDMs.
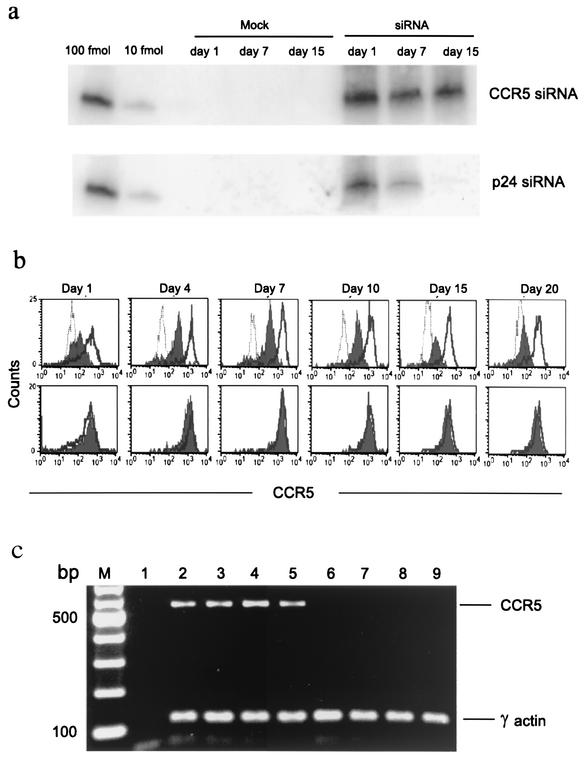
Our results suggest that siRNA has the potential to prevent HIV-1 infection. However, to realize this potential, it is important that siRNAs persist in cells for long periods of time before infection. To address this issue, we examined the intracellular persistence of transfected CCR5 and p24 siRNA in uninfected MDMs. Retention of exogenously introduced p24 and CCR5 siRNAs in uninfected MDMs was evaluated by modified Northern blot analysis of RNA samples at different time points after transfection by using γ-32P-labeled sense strands of CCR5 or p24 siRNA as probes. To quantitate siRNAs, lanes were also loaded with titrated concentrations of single-stranded antisense siRNA. Small RNA species with the relative mobility of 20- to 23-bp oligonucleotides were visualized at a 10-fmol level of sensitivity (Fig. 3a) The hybridization signals for CCR5 and p24 were similar in intensity on day 1. However, by day 7, the p24 signal became weaker and was completely lost by day 15, whereas the CCR5 signal remained robust throughout. No signal was detected in mock-transfected samples used as controls. These results suggested that p24 siRNA may be relatively short-lived in the absence of infection, while CCR5 siRNA persists under these conditions. Stability of CCR5 siRNA-mediated gene silencing was also assessed by parallel flow cytometric analysis of endogenous CCR5 expression over time. Compared to mock-transfected cells, CCR5 siRNA-transfected cells maintained a uniform 70 to 80% reduction in the mean fluorescence intensity of CCR5 staining from 1 to 20 days after transfection (Fig. 3b). Moreover, CCR5 mRNA was undetectable by RT-PCR analysis on days 1, 4, 7, and 15 after CCR5 siRNA transfection (Fig. 3c).
FIG. 3.
CCR5, but not p24, siRNA persists in uninfected MDMs. (a) Modified Northern blot analysis showing levels of internalized CCR5 and p24 siRNAs in MDMs on the indicated days after transfection. Before loading, samples were normalized for total RNA content. The sense strand of each siRNA was end labeled with γ-32P and used as a probe. Lanes loaded with graded amounts of the antisense strand of siRNA and mock-transfected samples served as positive and negative controls, respectively. (b) CCR5 (top) and GFP (bottom) siRNA-transfected MDMs were examined for CCR5 expression over time. Overlay histograms of CCR5-stained mock-transfected (open solid line), control immunoglobulin-stained (open dotted line), and siRNA-transfected (filled) cells are shown. (c) RT-PCR for CCR5 and γ-actin mRNA expression was performed with mock-transfected (lanes 2 to 5) and CCR5 siRNA-transfected (lanes 6 to 9) cells on days 1 (lanes 2 and 6), 4 (lanes 3 and 7), 7 (lanes 4 and 8), and 15 (lanes 5 and 9) after transfection (M, molecular weight marker; lane 1, negative control). CCR5 mRNA was not detected, even after an additional 25 cycles of PCR amplification (data not shown).
To test if these differences in intracellular persistence are also reflected in their differential abilities to sustain HIV-1 suppression, we transfected MDMs with CCR5 or p24 siRNA and initiated infections at increasing intervals of time after transfection. Viral replication was measured by flow cytometric analysis of p24 expression 10 days after infection. Consistent with long-term suppression of CCR5 expression, CCR5 siRNA provided protection whether the cells were infected 2 or 15 days after transfection (Fig. 4). On the other hand, p24 siRNA provided maximal protection when the cells were infected within 5 days of transfection, but the protection level gradually declined when the interval between transfection and infection was extended further. Because the gene for CCR5 is an endogenously expressed gene while that for p24 is expressed only after infection, we interpret these results to suggest that to sustain silencing, siRNA may need the presence of intracellular target mRNA.
FIG. 4.
CCR5, but not p24, siRNA confers sustained and uniform protection when MDMs are infected at increasing intervals after transfection. MDMs were transfected with GFP (top), p24 (middle), or CCR5 (bottom) siRNA and infected with HIVBAL at the indicated times after transfection. Cells were analyzed 10 days postinfection for p24 expression by flow cytometry. The percentage of p24+ cells is shown in each panel.
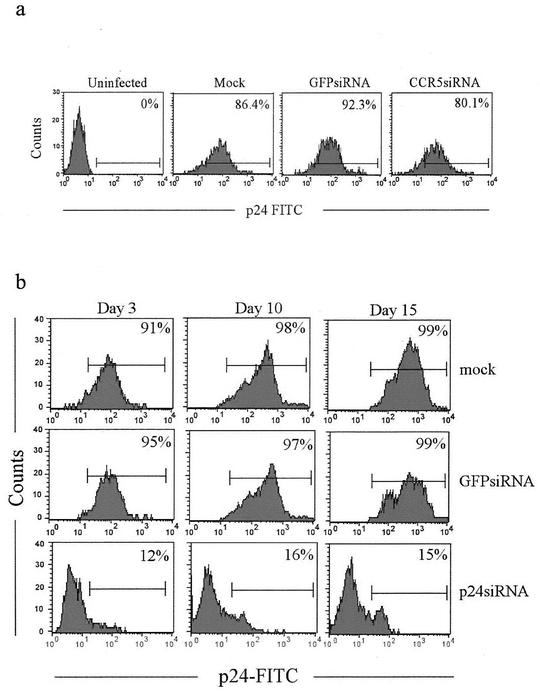
p24 siRNA can stably suppress HIV-1 replication in MDMs with an established infection.
To test if siRNAs can stably suppress HIV-1 in previously infected MDMs and to further examine the hypothesis that the siRNA effect may be long lasting if the target mRNA is continually available, we tested the p24 and CCR5 siRNAs for the ability to suppress viral replication in the setting of an established infection. MDMs were transfected with CCR5 or p24 siRNA 16 days after infection with HIVBAL, at which time >90% of the MDMs were infected (data not shown). The suppression of viral replication after transfection was monitored over time by evaluation of intracellular p24 expression. As expected, CCR5 blockade did not significantly reduce virus replication in this setup (Fig. 5a). In contrast, p24 siRNA was able to reduce viral replication in infected cells by nearly 90% 3 days after transfection (Fig. 5b). More importantly, p24 siRNA transfected after infection was able to suppress viral replication throughout the 15-day period of observation. Thus, long-lasting viral suppression can be achieved with p24 siRNA in MDMs with an established infection.
FIG. 5.
p24, but not CCR5, siRNA suppresses HIV-1 replication in an established infection. (a) MDMs infected with HIVBAL for 16 days (>90% of the MDMs were p24+; data not shown) were transfected with CCR5 siRNA and examined for p24 expression 3 days later. The percentage of p24+ cells is shown in each panel. (b) MDMs infected for 16 days were transfected with p24 or control siRNA and examined for p24 expression on various days posttransfection.
DISCUSSION
Our results take RNAi-based therapeutics a significant step forward by showing that a single application of synthetic siRNA is able to achieve long-lasting suppression of HIV-1 in a physiologic setting. We show that in primary macrophages, siRNAs not only prevent infection but also suppress viral replication after an infection is established.
Macrophages represent a key target of HIV-1 in vivo. Although the absolute number of infected macrophages is relatively low compared to that of CD4 T cells, the unique dynamics of HIV-1 replication in these cells make them a formidable viral reservoir (2). Macrophages are relatively immune to the cytopathic effects of HIV-1 and can survive for long periods after infection. They replicate large amounts of virus in sequestered cytoplasmic vacuoles, with a plateau of virus production lasting as long as 60 days (1). They are also the primary targets of HIV-1 in the nervous system (2). HIV-1-infected macrophages are commonly found in the blood and tissues of seropositive patients receiving highly active antiretroviral therapy (32), suggesting that virus production in macrophages may not be effectively controlled by currently available antiviral therapy. Thus, it is significant that this recalcitrant cell type is particularly amenable to sustained siRNA-mediated viral inhibition.
Why is the siRNA effect sustained in macrophages? Previous studies in which actively dividing cells were used to demonstrate siRNA-mediated gene silencing have found that the effect in mammalian cells is transient, lasting for only 4 to 7 days (7, 26, 33, 34). This has been attributed to siRNA dilution with cell division. However, even in nondividing macrophages, Northern blot analysis revealed that CCR5, but not p24, siRNA was stably present for a prolonged period after transfection. p24 siRNA persisted for only up to 7 days after transfection in uninfected cells, suggesting that siRNA degradation may contribute to loss of siRNA. Consistent with the lack of sustenance of p24 siRNA in uninfected MDMs, the level of viral inhibition declined significantly when the interval between transfection and infection was prolonged beyond 7 days. While p24 siRNA appears to be rapidly degraded in uninfected cells, it effectively suppressed viral replication for prolonged periods in previously infected cells (Fig. 5b), where target mRNA is continually synthesized. These results suggest that the presence or absence of target mRNA may determine whether siRNA is sustained or degraded. Moreover, CCR5 siRNA was able to suppress the expression of the endogenous CCR5 gene, as well as prevent viral entry for long periods, even when transfection preceded infection by 15 days (Fig. 4). Thus, continued presence of target RNA may be required for intracellular sustenance of siRNAs. These results are also consistent with the finding, in Caenorhabditis elegans, that RNAi directed against GFP did not result in detectable siRNA unless the gene for GFP was simultaneously expressed in the target cells (27). However, although this finding would imply that the antisense strand of the siRNA would be preferentially maintained in the cells, no bias toward longer persistence or higher concentration of the antisense strand relative to the sense strand of CCR5 siRNA was observed in modified Northern blot analysis (data not shown).
Another mechanism that can extend the effectiveness of siRNA is its amplification. In lower species, such as C. elegans, it is well documented that siRNAs can prime target mRNA to generate new siRNAs by using an RNA-dependent RNA polymerase (15). Whether siRNA amplification occurs in mammalian cells is unclear, and no direct evidence of RNA-dependent RNA polymerase activity exists (7 29a). Moreover, the silencing effect of siRNA rapidly fades in dividing mammalian cell lines, even in the presence of target mRNAs (10, 30, 33). These issues underscore the need to elucidate the fundamental mechanisms of RNAi in mammalian cells. Nondividing MDMs may provide an ideal cell type for such mechanistic studies because of the absence of complicating dilutional effects.
Only one of the two siRNA sequences that we used in our initial screening was effective at silencing CCR5 expression in primary macrophages (data not shown). Similar results have been reported in a recent study by Qin et al., who used two sequences distinct from the ones that we used and found that only one of them was highly effective at ablating CCR5 expression in primary CD4 T cells (29). Surprisingly, the siRNA that we found to be completely ineffective at silencing CCR5 expression in MDMs is identical in sequence to the one used by Martinez et al. for silencing CCR5 expression in the U87-CD4-CCR5 cell line in a recent study (20). However, in agreement with their data, we also observed a significant decrease in CCR5 expression in the U87-CD4-CCR5 cell line transfected with the same siRNA (data not shown). This brings up the intriguing possibility of cell type-specific differences in effectiveness of siRNAs and underscores the importance of testing their effectiveness in primary CD4 T cells and macrophages, which are the physiologically relevant targets of HIV-1.
Given the high mutation rate of HIV-1, it may be desirable to target highly conserved regions of the viral genome and to use combinations of siRNAs. In this regard, host cellular gene targets that can interrupt the HIV-1 life cycle may offer particularly attractive targets because endogenous genes are not under immune pressure to generate escape mutations. CCR5 is a feasible target since homozygous mutations of the gene are well tolerated and reduce susceptibility to infection (19, 25). Combinations of siRNAs targeting two different steps in the viral life cycle had a strong synergistic antiviral effect. Combined treatment with the CCR5 and p24 siRNAs completely eliminated HIV-1 replication in macrophages, probably by interrupting two steps in the viral life cycle, with CCR5 siRNA blocking viral entry and p24 siRNA destroying the virus that slipped through other coreceptors, or by passive uptake (24). Thus, it may be possible to develop optimal RNAi-based therapeutic strategies that target multiple aspects of the virus life cycle akin to drug cocktails used in highly active antiretroviral therapy.
Efficient in vivo delivery of siRNAs into CD4 T cells and macrophages remains the major bottleneck in gene therapeutic approaches to HIV-1. Viral vectors capable of generating siRNA under the control of an RNA polymerase III promoter have been used effectively in cell lines (3, 5, 32a). Qin et al. have demonstrated the feasibility of using a lentivirus-based vector to introduce siRNAs against the HIV-1 coreceptor CCR5 into primary cells (29). However, the use of these vector systems in vivo is fraught with potential dangers of retroviral recombination and malignant transformation due to random insertion within the host genome. Two out of nine children receiving retroviral gene therapy for X-linked severe combined immunodeficiency developed a leukemia-like disease in a recent clinical trial (12). In this regard, synthetic siRNA administration may be preferable if it can provide reasonably long-lasting protection, as appears to be the case for macrophages. However, efficient in vivo delivery methods need to be developed. In fact, several groups, including ours, have successfully administered duplex siRNAs systemically to mice (4, 18, 21, 31). Given the rapid strides being made in the field, the use of siRNA as an antiviral therapeutic approach in humans appears to be promising.
Acknowledgments
The first two authors contributed equally to this work.
We thank Z. Xu and V. François-Borgarçon for technical assistance. The R5 BAL strain of HIV-1 was provided by S. Gartner, M. Popovic, and R. Gallo and was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
This work was supported by National Institutes of Health grants AI49792, AI45306 (P.S.), AI46566 (N.M.), and AI42519 (J.L.), R37-GM34277 (P.A.S.), and F32-AI10523 (C.N.), NCI grant PO1-42063 (P.A.S.), and amfAR fellowship grant 70589-32RF (S.-K.L.).
REFERENCES
- 1.Aquaro, S., P. Bagnarelli, T. Guenci, A. De Luca, M. Clementi, E. Balestra, R. Calio, and C. F. Perno. 2002. Long-term survival and virus production in human primary macrophages infected by human immunodeficiency virus. J. Med. Virol. 68:479-488. [DOI] [PubMed] [Google Scholar]
- 2.Aquaro, S., R. Calio, J. Balzarini, M. C. Bellocchi, E. Garaci, and C. F. Perno. 2002. Macrophages and HIV infection: therapeutical approaches toward this strategic virus reservoir. Antiviral Res. 55:209-225. [DOI] [PubMed] [Google Scholar]
- 3.Barton, G. M., and R. Medzhitov. 2002. Retroviral delivery of small interfering RNA into primary cells. Proc. Natl. Acad. Sci. USA 99:14943-14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertrand, J., M. Pottier, A. Vekris, P. Opolon, A. Maksimenko, and C. Malvy. 2002. Comparison of antisense oligonucleotides and siRNAs in cell culture and in vivo. Biochem. Biophys. Res. Commun. 296:1000-1004. [DOI] [PubMed] [Google Scholar]
- 5.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2:243-247. [DOI] [PubMed] [Google Scholar]
- 6.Capodici, J., K. Kariko, and D. Weissman. 2002. Inhibition of HIV-1 infection by small interfering RNA-mediated RNA interference. J. Immunol. 169:5196-5201. [DOI] [PubMed] [Google Scholar]
- 7.Chiu, Y. L., and T. M. Rana. 2002. RNAi in human cells: basic structural and functional features of small interfering RNA. Mol. Cell 10:549-561. [DOI] [PubMed] [Google Scholar]
- 8.Coburn, G. A., and B. R. Cullen. 2002. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 76:9225-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devroe, E., and P. A. Silver. 2002. Retrovirus-delivered siRNA. BMC Biotechnol. 2:15-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 11.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15:188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hacein-Bey-Abina, S., A. Fischer, and M. Cavazzana-Calvo. 2002. Gene therapy of X-linked severe combined immunodeficiency. Int. J. Hematol. 76:295-298. [DOI] [PubMed] [Google Scholar]
- 13.Hu, W., C. Myers, J. Kilzer, S. Pfaff, and F. Bushman. 2002. Inhibition of retroviral pathogenesis by RNA interference. Curr. Biol. 12:1301-1311. [DOI] [PubMed] [Google Scholar]
- 14.Jacque, J. M., K. Triques, and M. Stevenson. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418:435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ketting, R. F., S. E. Fischer, E. Bernstein, T. Sijen, G. J. Hannon, and R. H. Plasterk. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15:2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitabwalla, M., and R. M. Ruprecht. 2002. RNA interference—a new weapon against HIV and beyond. N. Engl. J. Med. 347:1364-1367. [DOI] [PubMed] [Google Scholar]
- 17.Lee, N. S., T. Dohjima, G. Bauer, H. Li, M. J. Li, A. Ehsani, P. Salvaterra, and J. Rossi. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20:500-505. [DOI] [PubMed] [Google Scholar]
- 18.Lewis, D. L., J. E. Hagstrom, A. G. Loomis, J. A. Wolff, and H. Herweijer. 2002. Efficient delivery of siRNA for inhibition of gene expression in postnatal mice. Nat. Genet. 32:107-108. [DOI] [PubMed] [Google Scholar]
- 19.Liu, R., W. A. Paxton, S. Choe, D. Ceradini, S. R. Martin, R. Horuk, M. E. MacDonald, H. Stuhlmann, R. A. Koup, and N. R. Landau. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367-377. [DOI] [PubMed] [Google Scholar]
- 20.Martinez, M. A., A. Gutierrez, M. Armand-Ugon, J. Blanco, M. Parera, J. Gomez, B. Clotet, and J. A. Este. 2002. Suppression of chemokine receptor expression by RNA interference allows for inhibition of HIV-1 replication. AIDS 16:2385-2390. [DOI] [PubMed] [Google Scholar]
- 21.McCaffrey, A. P., L. Meuse, T. T. Pham, D. S. Conklin, G. J. Hannon, and M. A. Kay. 2002. RNA interference in adult mice. Nature 418:38-39. [DOI] [PubMed] [Google Scholar]
- 22.McManus, M. T., and P. A. Sharp. 2002. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 3:737-747. [DOI] [PubMed] [Google Scholar]
- 23.Meltzer, M. S., D. R. Skillman, P. J. Gomatos, D. C. Kalter, and H. E. Gendelman. 1990. Role of mononuclear phagocytes in the pathogenesis of human immunodeficiency virus infection. Annu. Rev. Immunol. 8:169-194. [DOI] [PubMed] [Google Scholar]
- 24.Moore, J. P., and M. Stevenson. 2000. New targets for inhibitors of HIV-1 replication. Nat. Rev. Mol. Cell. Biol. 1:40-49. [DOI] [PubMed] [Google Scholar]
- 25.Nansen, A., J. P. Christensen, S. O. Andreasen, C. Bartholdy, J. E. Christensen, and A. R. Thomsen. 2002. The role of CC chemokine receptor 5 in antiviral immunity. Blood 99:1237-1245. [DOI] [PubMed] [Google Scholar]
- 26.Novina, C. D., M. F. Murray, D. M. Dykxhoorn, P. J. Beresford, J. Riess, S. K. Lee, R. G. Collman, J. Lieberman, P. Shankar, and P. A. Sharp. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8:681-686. [DOI] [PubMed] [Google Scholar]
- 27.Plasterk, R. H. 2002. RNA silencing: the genome's immune system. Science 296:1263-1265. [DOI] [PubMed] [Google Scholar]
- 28.Pomerantz, R. J. 2002. RNA interference meets HIV-1: will silence be golden? Nat. Med. 8:659-660. [DOI] [PubMed] [Google Scholar]
- 29.Qin, X. F., D. S. An, I. S. Chen, and D. Baltimore. 2003. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. USA 100:183-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Schwarz, D. S., G. Hutvagner, B. Haley, and P. D. Zamore. 2002. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell 10:537-548. [DOI] [PubMed] [Google Scholar]
- 30.Sharp, P. A. 2001. RNA interference—2001. Genes Dev. 15:485-490. [DOI] [PubMed] [Google Scholar]
- 31.Song, E., S. K. Lee, J. Wang, N. Ince, N. Ouyang, J. Min, J. Chen, P. Shankar, and J. Lieberman. 2003. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat. Med. 9:347-351. [DOI] [PubMed] [Google Scholar]
- 32.Sonza, S., H. P. Mutimer, R. Oelrichs, D. Jardine, K. Harvey, A. Dunne, D. F. Purcell, C. Birch, and S. M. Crowe. 2001. Monocytes harbour replication-competent, non-latent HIV-1 in patients on highly active antiretroviral therapy. AIDS 15:17-22. [DOI] [PubMed] [Google Scholar]
- 32a.Stewart, S. A., D. M. Dykxhoorn, D. Palliser, H. Mizuno, E. Y. Yu, D. S. An, D. M. Sabatini, I. S. Chen, W. C. Hahn, P. A. Sharp, R. A. Weinberg, and C. D. Novina. 2003. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9:493-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuschl, T. 2002. Expanding small RNA interference. Nat. Biotechnol. 20:446-448. [DOI] [PubMed] [Google Scholar]
- 34.Ullu, E., A. Djikeng, H. Shi, and C. Tschudi. 2002. RNA interference: advances and questions. Philos. Trans. R. Soc. Lond. B Biol. Sci 357:65-70. [DOI] [PMC free article] [PubMed] [Google Scholar]