Abstract
Entry of Epstein-Barr virus (EBV) into B lymphocytes requires the binding of viral glycoprotein 42 (gp42), a C-type lectin family member, to HLA class II. Recently, the structure of the gp42:HLA-DR1 complex was determined. In order to confirm the interaction as determined in the structural study and to identify other potential interactive residues, a mutational analysis of HLA class II was performed. A secreted form of gp42 (sgp42) reacted with a conformation-specific monoclonal antibody and blocked EBV infection. The binding of sgp42 and EBV entry to two sets of HLA class II mutants were tested. The first set of mutants were based on the known interaction of the C-type lectin Ly49A with HLA class I, and the second set of mutants were based on the identified interface in the gp42:HLA-DR1 complex. As expected, none of the mutants that would be predicted to interfere with the interaction of Ly49A with class I affected the interaction of gp42 with HLA class II, whereas mutants in amino acids identified in the gp42:HLA-DR1 structure inhibited sg42 binding to class II. In general, sgp42 binding correlated with efficient entry of EBV, as demonstrated by the necessity of glutamic acid 46 or arginine 72 in class II molecules. Furthermore, other HLA class II residues buried within the interface of gp42 and HLA class II when mutated had either no effect or a decrease in both binding and entry and implicate a region of class II important in stabilizing the interaction with gp42. These studies provide insight into the entry and fusion processes of the critical interaction between gp42 and HLA class II.
Epstein-Barr virus (EBV), a gammaherpesvirus, causes infectious mononucleosis and is associated with a variety of malignancies, such as Burkitt's lymphoma and nasopharyngeal carcinoma (18). The host range of EBV in vitro is largely restricted to B cells. Entry of the virus occurs through multiple interactions of viral glycoproteins with receptors found on B cells. The initial entry step is the interaction of the major viral glycoprotein 350/220 (gp350/220) with the complement receptor type 2 molecule CD21 (CR2/CD21) (15, 25). This interaction is thought to bring the virus closer to the B-cell membrane, allowing for attachment of the viral glycoprotein gp42 with B-cell surface HLA class II molecules. Fusion of the viral envelope with the B-cell membrane requires gp42 along with the concerted action of the EBV-encoded glycoproteins gB, gH, and gL (8). Furthermore, the association of gp42 and HLA class II seems to be the key signal for triggering virus entry and fusion into B cells as well as tropism. A virus lacking gp42 is not able to infect B cells, and the amount of gp42 present on the virion determines the cell type that EBV infects (2, 28).
The HLA class II molecule, HLA-DR, was first shown to interact with gp42 in an expression library screen for proteins that bound a soluble gp42-Fc construct (23). HLA class II molecules are comprised of two distinct gene products, α and β, which noncovalently heterodimerize to form mature molecules of approximately 62 kDa. The α1 and β1 together create a peptide binding groove formed by an eight-stranded β-pleated sheet supporting two α-helices. The biological function of these molecules is to bind foreign peptide antigens and to form complexes that are recognized by antigen-specific T lymphocytes. The site that gp42 binds does not involve any direct peptide contacts (13), though antigen presentation to T cells is inhibited by the binding of a soluble gp42-Fc fusion protein (23). Subsequent studies demonstrated that the gp42 interaction with HLA-DR is crucial for EBV infection in B cells, since monoclonal antibodies to both gp42 and HLA-DR can inhibit the infection of B cells in vitro (11).
HLA-DR is only one isotype of the HLA class II antigen family. The genetic locus encodes three different pairs of α and β chains referred to as HLA-DR, -DP, and -DQ. These antigens exhibit restricted patterns of expression and are found predominantly but not exclusively on specialized antigen-presenting cells, such as B cells, macrophages, and dendritic cells (1). The HLA class II loci are extremely polymorphic and encode numerous alleles. Not surprisingly, EBV also uses the other two HLA class II isotypes, HLA-DP and -DQ, to gain entry into B cells (7).
The HLA class II receptor binding protein gp42, a member of the C-type lectin family, is not conserved across the herpesvirus family homologues, being found only in closely related gammaherpesviruses that infect animals. This illustrates the evolution or acquisition of specific viral proteins that have evolved to allow EBV to specifically infect B cells. gp42 is most closely related to natural killer (NK) receptors such as Ly49A and shares the functional characteristic of binding to major histocompatibility complex (MHC) superfamily members (14). By taking advantage of the high degree of polymorphism in HLA class II molecules, it was established that a glutamic acid at residue 46 of the HLA class II β chain is necessary for EBV entry (9). At that time, it was noted that the domain surrounding residue 46 is homologous to a site on MHC class I that interacts with the murine NK receptor Ly49A (26). Furthermore, despite amino acid sequence differences, MHC class I and HLA class II molecules are structurally very similar, suggesting overall that the complex of gp42 and HLA-DR might be similar to the Ly49A and class I supercomplex (1, 3). Recently, the crystal structure of gp42 bound to HLA-DR1 was solved and was found to have a distinct interaction site in comparison to NK receptor supercomplexes (13). Although the overall structure of gp42 was similar to that of Ly49A, reflecting the fact that they both have conserved features of the C-type lectin family, gp42 interacted with HLA class II in an entirely different manner. The homodimerization domain within Ly49A corresponds to the interaction site of gp42 with HLA-DR. As a result of the binding site differences, gp42 binds much more highly and closely to the peptide binding groove of the HLA class II molecule.
To completely understand protein-protein associations, both structural and functional analyses should be performed to assess important domains and/or specific residues within the interaction interface. Thus far, functional information is still lacking on the participation of class II interface residues. To investigate further the gp42 and HLA class II interaction, site-specific mutagenesis of the HLA-DQ molecule was performed and tested for binding to a soluble form of gp42 (sgp42) and the ability to mediate EBV entry. The HLA-DQ molecule was chosen to be mutated to determine if gp42 interacts with HLA-DQ in the same way that gp42 interacts with HLA-DR. The biochemical results here confirm and support the crystal structure and have identified two additional residues (all HLA residues refer exclusively to the β chain), lysine 65 and arginine 72, that upon mutation dramatically reduce sgp42 binding and EBV entry. Thus, identification of these residues as important for EBV infectivity provides new information on the mechanism that gp42 utilizes to serve as a trigger to initiate viral membrane fusion by the concerted action of gB, gH, and gL.
Expression and purification of sgp42.
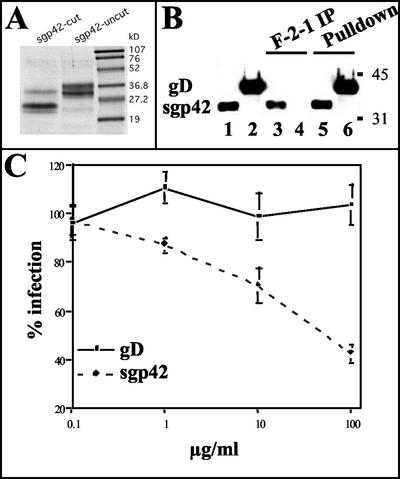
To generate sufficient quantities of gp42 for structural and functional studies, a secreted form of gp42 (sgp42) was made by utilizing a recombinant baculovirus. The expression construct was generated by subcloning DNA coding for residues 33 to 223 of gp42 (lacking the transmembrane domain) into the baculovirus pBACgus-3 transfer vector (Novagen), which contains a baculovirus gp64 signal sequence as well as His and S tags upstream of an enterokinase cleavage site. After purification, the resulting sgp42 was of high purity and migrated as a doublet with molecular masses of ∼34 and 36 kDa in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 1A, lane 2). Treatment of the protein with enterokinase (cleavage of the His and S tags) resulted in the expected shift in the digested protein on SDS-PAGE gels (Fig. 1A, lanes 1 and 2). The two bands of the sgp42 fusion protein may represent heterogeneity either in the glycosylated state of the protein or in the C-terminal section of the polypeptide. There are four potential N-linked glycosylation sites in the soluble gp42 protein, which may not be modified homogeneously.
FIG. 1.
Characterization of sgp42. (A) Supernatants of infected Trichoplasia ni insect cells were harvested, and sgp42 was purified by gel filtration. The purified protein was run on an SDS-PAGE gel. Both the enterokinase cleaved and uncleaved forms of the protein are shown. (B) sgp42 is recognized by the gp42-specific monoclonal antibody F-2-1. Media from cells infected with the gD or sgp42 baculoviruses were immunoprecipitated (IP) with either F-2-1 antibodies (lanes 3 and 4) or Ni-agarose, which binds the histidine tag (lanes 5 and 6) present in both constructs. Following immunoprecipitation, proteins were resolved by SDS-PAGE and were probed in immunoblots with the antihistidine antibodies. Purified gD or sgp42 is controls (lanes 1 and 2). Molecular mass standards are in kilodaltons. (C) sgp42 blocks infection of Daudi cells with EBfaV-GFP. Daudi cells were first incubated with either purified sgp42 or gD in the amounts indicated and were then infected with EBfaV-GFP. Cells infected were then determined 2 days postinfection by using flow cytometry. These experiments were performed a minimum of three times with similar results obtained.
Next, before both structural and functional studies were performed, antibodies directed against gp42 and the His tag were used to test the immunoreactivity and conformation of sgp42. The herpes simplex virus gD-expressing baculovirus, kindly provided by Gary Cohen and Roslyn Eisenberg, was used as a control, as it also contains a His tag (20). As expected, the secreted and uncleaved forms of both gD and sgp42 were readily precipitated with Ni-agarose (Fig. 1B, lanes 5 and 6), whereas only sgp42 was detected in an F-2-1 (gp42 monoclonal antibody [24]) immunoprecipitation (Fig. 1B, lanes 3 and 4). The antihistidine antibodies react with both purified gD and sgp42 added to the gel directly (Fig. 1B, lanes 1 and 2). These experiments demonstrate that expression of sgp42 in baculovirus is reactive with at least one gp42 conformation-specific monoclonal antibody, suggesting that the protein is properly folded.
Inhibition of EBV entry by sgp42.
Since the interaction of class II and gp42 is necessary for EBV to infect B cells and since a previously described Fc-gp42 fusion was able to block EBV entry, the effect of sgp42 on entry of EBV was tested (29). Various amounts of purified sgp42 were incubated with the CD21-positive, HLA-DQ, -DP, and -DR-positive Burkitt's lymphoma cell line Daudi for 15 min at 4°C (7). Again, purified gD was used as a control. The reporter virus, EBfaV-GFP, was then added for 1 h at 4°C (22). Unbound virus was removed, and after 2 days green fluorescent protein (GFP) expression was monitored by flow cytometry. As shown in Fig. 1C, the purified sgp42 was able to block EBV infection, as measured by a decrease in GFP expression in a dose-dependent fashion, with maximal inhibition observed at the highest dose of 100 μg of the purified protein per ml. No inhibition was seen when purified gD was used as a control. The ability of the purified protein to block EBV infection of Daudi cells illustrates that the purified sgp42 is functional in regard to binding of HLA-DQ.
HLA class II glutamic acid 46 is necessary for binding to sgp42.
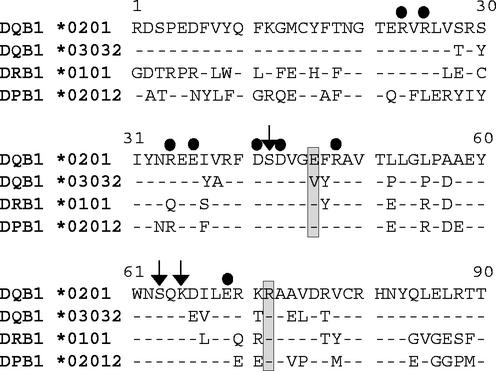
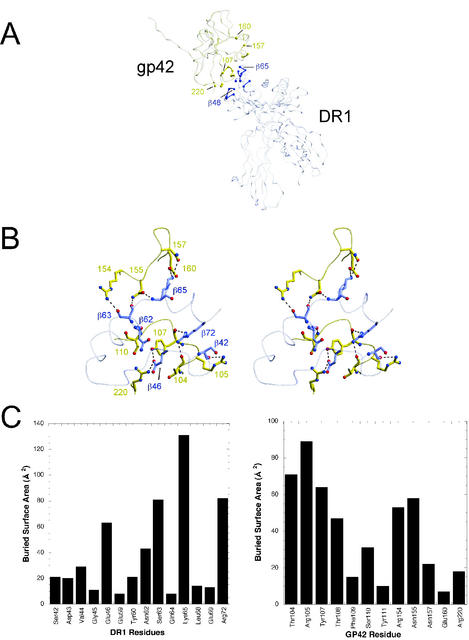
Previously, it was established that a glutamic acid or aspartic acid at amino acid 46 of the HLA class II β chain was absolutely required for EBV entry of B lymphocytes (9). All known HLA-DR and -DP alleles encode a glutamic acid residue at this position in the β chain, with the exception of a single change in an HLA-DR allele of glutamic acid to aspartic acid (Anthony Nolan database [http://www.anthonynolan.com/HIG/index.html]) (Fig. 2). Only a small subset of HLA-DQ β-chain alleles contain a glutamic acid, with the majority having a valine at this position, which does not mediate EBV entry (9). As expected, a more dramatic change of replacing a glutamic acid with a lysine in a HLA β chain that mediates EBV entry results in an HLA class II molecule that does not mediate EBV infection. Confirming the importance of the glutamic acid at position 46, an extensive interaction at this site with gp42 was discovered in the gp42:HLA-DR1 crystal structure (13). The glutamic acid at residue 46 in HLA class II forms a salt bridge with arginine 220 as well as a hydrogen bond with tyrosine 107 in gp42 (Fig. 3A and B).
FIG. 2.
Sequence comparison of the N-terminal residues of the HLA class II β chain is shown. The dots indicate the residues where mutations were made based on the Ly49A-class I homology interaction site. The arrows highlight amino acids identified as putative interaction sites determined by residues with large buried surface area. Boxes surround the two residues essential for the gp42-HLA-DQ interaction.
FIG. 3.
Interactions between gp42 and HLA-DR1. (A) Line structure of gp42 bound to HLA class II. Key residues are highlighted by a ball-and-stick formation with the yellow indicative of gp42 and the blue representative of HLA-DR1. (B) Close-up of the area highlighted in panel A. (C) Buried surface area of gp42 and HLA-DR1 residues.
To further characterize the requirements of gp42 binding to HLA class II for EBV entry, the ability of sgp42 to bind to class II molecules, both functional and nonfunctional in EBV entry, was determined. Transient transfection of 721.174 cells (6), a CD21-positive, HLA class II-negative lymphoblastoid cell line, was performed by electroporation of 107 cells with 40 μg of DNA (1:1 ratio of α and β chains) at 960-μF capacitance and 0.210 kV. Twenty-four hours after electroporation, sgp42 was added to 106 721.174 cells and was rotated for 20 min at 4°C. The cells were washed and were then analyzed by two-color flow cytometry. To detect HLA-DQ expression, a biotin-conjugated monoclonal HLA-DQ antibody (Ia3; ICN Biochemicals) with a streptavidin-allophycocyanin secondary antibody was used, and sgp42 binding was detected by using a polyclonal rabbit gp42 antibody (PB1114) followed by a fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G. The polyclonal gp42 antibody was generated by immunizing rabbits with purified sgp42 by using standard methodologies. Cells expressing HLA class II and able to bind to gp42 are thus observed in the upper right quadrant. At the same time that binding was tested, the entry of EBV was tested by utilizing our recombinant virus expressing GFP. Briefly, 106 transiently transfected 721.174 cells were exposed to the reporter virus EBfaV-GFP under constant agitation at 37°C. Thirty-six hours after exposure to EBfaV-GFP, cells were analyzed by flow cytometry for expression of HLA class II and infection with EBV by monitoring GFP expression and HLA-DQ expression as described above and previously (7). The relative infection rate was calculated by the percentage of GFP-positive cells divided by the percentage of total HLA-DQ-positive cells in relation to the wild-type allele DQ 2 set at 1. These numbers are shown in more detail in Table 1.
TABLE 1.
EBV infection with HLA class II mutants
| Mutant | Relative infection avga | No. of infections |
|---|---|---|
| DQ 2 | 1 (NA)c | 5 |
| R 23 D | 0.59 ± 0.11 | 4 |
| R 25 D | 0.71 ± 0.16 | 3 |
| R 34 E | 0.94 ± 0.23 | 3 |
| E 36 K | 0b | 3 |
| D 41 A | 0.79 ± 0.03 | 4 |
| D 43 A | 0.53 ± 0.05 | 4 |
| R 48 E | 0.72 ± 0.27 | 5 |
| E 69 K | 0.57 ± 0.21 | 4 |
| S 42 A | 1.1 ± 0.20 | 4 |
| S 42 K | 0.8 ± 0.09 | 4 |
| E 46 K | 0.08 ± 0.06 | 3 |
| S 63 A | 0.72 ± 0.20 | 4 |
| S 63 K | 0.57 ± 0.37 | 3 |
| K 65 A | 1.09 ± 0.16 | 3 |
| K 65 E | 0.12 ± 0.10 | 4 |
| R 72 A | 0.26 ± 0.10 | 4 |
| R 72 E | 0.00 ± 0.01 | 4 |
This number is equal to the relative infection rate with the standard deviation for each mutant. For each experiment, the percentage of GFP-positive cells was divided by the percentage of HLA-DQ-positive cells in and compared to the ratio for wild-type DQ 2 when it was set at 1.
No surface expression was detected.
NA, not applicable.
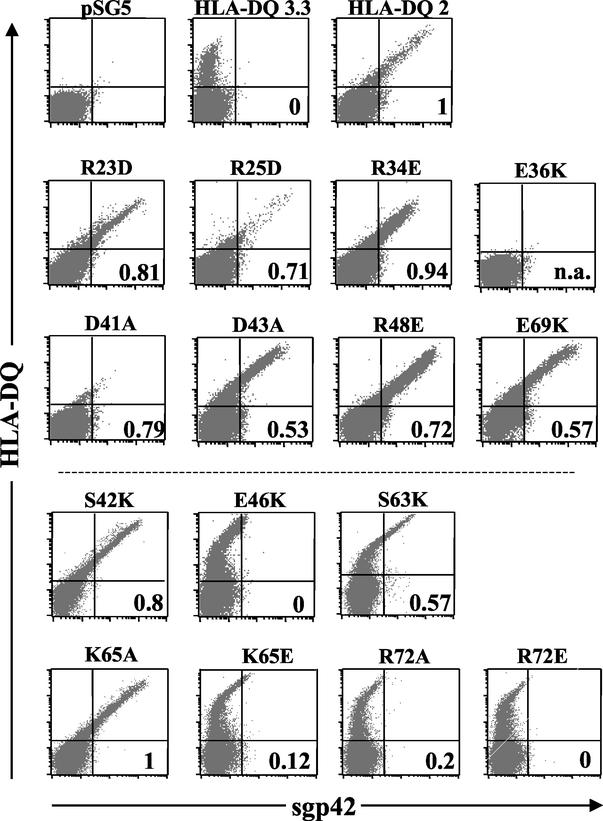
As expected, sgp42 was able to bind the HLA class II DQ 2 allele, which is able to mediate EBV entry as shown previously (Fig. 4, HLA-DQ 2, and Table 1) (9). Interestingly, sgp42 was unable to bind efficiently to the HLA class II DQ 3.3 allele, which does not mediate viral entry (Fig. 4, number in the lower right quadrant, or Table 1), although the receptor was abundantly expressed on the cell surface (Fig. 4, HLA-DQ 3.3). This observation provides an initial indication that the lack of entry-mediating ability of HLA-DQ 3.3 is a result of an inability to bind to gp42. We also tested a previously described glutamic acid 46-to-lysine mutation, which is unable to mediate viral entry, and as expected, this mutant was unable to efficiently bind sgp42 or mediate EBV entry (Fig. 4, E46K, and Table 1) (9). In cells transfected with vector, HLA class II was not expressed, and as expected, the cells did not bind sgp42 or mediate infection (Fig. 4, pSG5). This evidence suggests that sgp42 binds only to HLA-DQ alleles facilitating entry.
FIG. 4.
The HLA-gp42 interaction sites at class II β glutamic acid residue 46 and arginine 72 are necessary for EBV coreceptor activity. HLA class II expression and sgp42 binding were determined by flow cytometry by using a FacsCalibur (Becton Dickinson). Surface expression of HLA-DQ was detected by using the primary anti-HLA-DQ antibody Ia3 conjugated to biotin, followed by the secondary streptavidin antibody conjugated to allophycocyanin (Pharmingen). sgp42 was detected by using the polyclonal antibody PB1114, followed by a goat anti-rabbit secondary antibody conjugated to fluorescein isothyocyanate (Pharmingen). Entry of EBfaV-GFP was determined by flow cytometry analysis of GFP expression. The number in the lower right quadrant is the infection rate, with DQ 2 set at 1. The number was calculated by taking the percentage of GFP-positive cells divided by the number of HLA-DQ-positive cells. n.a., not applicable as cells were not positive for HLA-DQ expression. Dot plots contain 40,000 events. These experiments were performed a minimum of three times with similar results obtained.
The gp42:DR1 interaction is not similar to the Ly49A:class I interaction.
Due to the unique interaction site found in the gp42:HLA-DR structure when it was compared to those of other superfamily members, such as the interaction of Ly49A with MHC class I, and due to our desire to determine any differences in the interaction of gp42 with HLA-DQ and HLA-DR, additional point mutations were made in HLA-DQ to complement the structural analysis. The glutamic acid at residue 46 is located at a structural position similar to that of aspartic acid 137 on class I (26). This aspartic acid is part of a larger site containing multiple interactions that are important for Ly49A binding to class I. In the crystal structure of Ly49A bound to class I, two disparate sites on class I were identified as interaction sites; the site 2 interface contains the aspartic acid. Upon further analysis via alanine-scanning mutagenesis of class I, the functional binding site for Ly49A was determined to be site 2 (12, 27).
To verify that a similar region of class II was not important for binding gp42, as indicated by the structural studies, numerous point mutations were made in the homologous region important for binding Ly49A of HLA class II, including R23D, R25D, R34E, E36K, D41A, D43A, R48E, and E69K (26). By use of the QuikChange Site-Directed Mutagenesis Kit (Stratagene), point mutations were made in the DQ β0202 allele, an HLA-DQ allele that mediates EBV entry (shown above as HLA-DQ 2). The mutations were checked by sequence analysis and by unique restriction enzyme digest. The substitutions made in residues 23, 25, 34, 41, 43, 48, and 69 had no effect on sgp42 binding or EBV entry (Fig. 4, R23D, R25D, R34E, D41A, D43A, R48E and E69K, and Table 1). Lesser sgp42 binding with the HLA-DQ mutants R25D and D41A was consistent with lower cell surface expression of HLA-DQ. These two mutants still efficiently mediated entry of at least 70%, compared to wild-type DQ 2. E36K was not expressed on the cell surface, and as a result, sgp42 binding and GFP expression were also negative and zero, respectively (Fig. 4, E36K, and Table 1). None of the mutations interfered with the interaction of gp42 and HLA-DQ, confirming the structural study.
Arginine 72 in the HLA class II β1 chain is essential for sgp42 binding to HLA class II and EBV entry.
The crystal structure of the gp42:HLA-DR1 complex highlighted a second interaction of gp42 with class II at arginine 72 of the class II β1 chain (13). This arginine is completely conserved in HLA-DQ and HLA-DP sequences and is shown in Fig. 2 in a comparison with other HLA sequences. In HLA-DR sequences, the arginine is well conserved, with only substitutions of a glutamine and an alanine represented in only 2 of the 397 DRβ sequences in the HLA database (http://www.anthonynolan.com/HIG/index.html). Residues surrounding R72 are not as conserved (Fig. 2), but previously, a double mutation in residues 71 and 74 still mediated entry, negating the participation of these less conserved residues (9). An arginine at this position in HLA class II makes hydrogen bonds with the main chain atoms of threonine 104 and tyrosine 107 of gp42 (Fig. 3A and B). Individually an alanine and a glutamic acid replaced the arginine at this position and were tested as described above. The R72A and R72E mutants were expressed but were not able to bind sgp42 (Fig. 4, R72A and R72E). This inability correlated with the lack (R72E) or dramatic decrease (R72A) in entry of EBV mediated by these mutants (Fig. 4, R72A and R72E, and Table 1). These data confirm the importance of the extensive interaction of R72 with gp42 residues found in the gp42:HLA-DR1 crystal structure and establish this residue as essential in mediating the interaction.
Mutation of lysine 65 within the HLA-DR β chain can disrupt sgp42 binding and EBV entry.
Many protein-protein interactions display a critical role for two or three residues, while other interface residues contribute only small amounts to the overall binding energy. To help define the critical interaction residues of gp42 and HLA-DR required for gp42 binding and EBV entry, the buried, or solvent-inaccessible, surface area of HLA-DR1 residues and gp42 was determined (Fig. 3C). For HLA-DR1, lysine 65 stands out with the largest surface area (approximately 130 Å2) buried upon binding of gp42, an amount larger than that of the key residue 46 or 72, which is 62 or 80 Å2, respectively. To determine the importance of lysine 65 in sgp42 binding and EBV entry, lysine 65 was mutated to glutamic acid (K65E). When the lysine at amino acid 65 was replaced with a glutamic acid, there was a dramatic reduction in the binding of sgp42 to this class II mutant (Fig. 4, K65E). No significant difference is observed when this mutant is compared to E46K, R72A, or R72E. When infection with EBV was monitored, this mutant had a reduction in EBV entry and was similar to R72A, as entry was not completely inhibited (Fig. 4, K65E, and Table 1). In order to more definitively test this amino acid, since a dramatic substitution may alter the overall structure, an alanine replacement of the lysine at amino acid 65 was tested. The K65A both bound sgp42 and allowed for entry similar to that for the wild-type allele (Fig. 4, K65A, and Table 1). Since the alanine mutant was able to bind sgp42 as well as mediate entry efficiently, this suggests that the entire lysine side chain, including the positive charge, is not essential.
Other residues with large buried surface area do not play an essential role in the interaction of gp42 and class II.
We chose to examine two other residues based on buried surface area and their role in the class II-gp42 interaction. Serine 63 has an amount of buried surface area equal to that of R72, approximately 80 Å2, suggesting another potentially important interaction residue. On the opposite side of the interaction site, the largest buried surface area for gp42 is the residue arginine 105. This arginine appears to form hydrogen bonds with class II residue serine 42 (Fig. 3B). Therefore, both of these serines were independently mutated to alanine and lysine and were individually tested, in a manner similar to that for the mutants above. When either substitution is made in S42 or S63, the DQ mutants are still able to bind sgp42 and allow for entry of EBV (Fig. 4, S42K and S63K; data not shown). Serines 42 and 63 individually are not essential to the HLA class II-gp42 interaction.
Discussion.
The functional analysis of specific residues of class II molecules done in the present study was performed to complement the previous structural studies on the gp42:HLA-DR1 complex (13). In particular, we (i) confirmed that the interaction between gp42 and HLA class II was different from the interaction of MHC class I with Ly49A, (ii) established the functional roles of other residues in the interface contributing to the interaction, and (iii) determined that sgp42 binding correlates with efficient EBV entry.
To accomplish the first aspect, various point mutations were made in HLA class II in sites with homology to the previously identified HLA class I interaction sites with Ly49A (26). Despite gp42 being a member of the C-type lectin family, the interaction of gp42 with HLA class II in the crystal structure was distinct. In these studies, it was determined that, although gp42 interacted with a similar region of HLA class II, the overall interaction was entirely different from the interaction of Ly49A with MHC class I. In particular, gp42 used a surface site that is distinct from the canonical lectin and NK receptor ligand binding sites used by Ly49A to bind to MHC class I. By mutation of specific, relatively well-conserved amino acids contained within homologous regions of HLA class II that were shown to be important for Ly49A binding to MHC class I, we were able to convincingly show that this region is not important for gp42 binding to HLA class II.
By using the crystal structure of the gp42:HLA-DR1 complex, we sought to test the functional importance of residues observed to contribute to the interaction. These studies provide important verification of the crystal structure and identify the most critical interactions of gp42 with HLA class II. In particular, we demonstrated that, in addition to glutamic acid 46, arginine 72 in HLA class II, which makes hydrogen bonds with the main chain atoms of threonine 104 and tyrosine 107 of gp42, was absolutely required for sgp42 binding and viral entry. In addition, by further analysis of the gp42:HLA-DR1 complex, we were able to determine that lysine 65, which is buried in the gp42:HLA-DR1 interface and forms hydrogen bonds with asparagine 157 and glutamic acid 160, may also be important for efficient sgp42 binding and EBV entry. Mutation of lysine 65 to glutamic acid causes weak binding to HLA class II and a 10-fold reduction in the EBV entry-mediating ability compared to results for other working mutants. However, an alanine mutation behaves like the wild-type DQ 2 allele. These data suggest that the lysine 65 side chain is not absolutely required for EBV entry but that an electrostatic switch from a positively charged to a negatively charged side chain in the glutamic acid interferes with gp42 binding. This might result because of the close juxtaposition of glutamic acid 65 with glutamic acid 160 on gp42 or may result from an overall detrimental effect on the structure of HLA-DQ. Other contact residues, serines 42 and 63, may play a smaller role in entry and may be involved cooperatively in the interaction, as single mutations, either to alanine or more drastically to lysine, do not disrupt the interaction. In addition, these mutations would not be expected to interfere with main chain atom contacts that are observed in the interface. However, the slight difference in the sgp42 binding plots of S63 suggests that this residue may have a reduced binding affinity (compare the staining pattern of S63K with that of DQ 2 or other mutants that display wild-type binding of sgp42 in Fig. 4). Furthermore, most protein-protein interactions occur via a few key residues in maintaining the binding ability. This has been shown with hormone receptors as well as viral ligand-receptor complexes (4, 5). Similar to the interaction of the herpes simplex virus gD with its receptor(s), several key residues are required for viral entry and gD binding (5). Overall, the HLA-DQ residues do not contribute equally to gp42 binding. The variances in sgp42 binding shown above suggest a difference in the affinity for gp42 of different HLA class II molecules. Future optical biosensor experiments need to be performed to accurately examine the gp42's kinetics and affinity for binding to various mutant HLA class II molecules.
Of note is the conservation of residues between HLA alleles. The key contact residue, E46, is not completely conserved, though S42, S63, or K65 is found in all of the alleles sequenced to date and R72 is found in all but two. The evolution of EBV to infect B cells was likely through the acquisition of gp42 by the EBV progenitor virus. Did EBV infection select for individuals that may lack susceptibility to EBV infection? This is unlikely, since all individuals within the human population code for multiple HLA class II alleles, making it virtually impossible for an individual to express only class II alleles unable to mediate EBV entry. In addition, EBV has not typically been associated with a high mortality rate, which would be a requirement for a strong selection of HLA class II alleles defective for viral entry. The presence of HLA class II alleles that do not mediate entry is likely a result of random changes in the polymorphic class II alleles. EBV has also been associated with a variety of diseases, such as diabetes and nasopharyngneal carcinoma. In some cases, there is an association of disease with certain HLA class II alleles. There is some correlation between insulin-dependent diabetes mellitus and individuals expressing HLA-DQ alleles that mediate viral entry (10, 19). Further study is required to determine if susceptibility or lack of susceptibility to EBV-associated disease may be related to specific HLA class II alleles.
Little is known about the mechanism of fusion between the EBV envelope and the target membrane of susceptible cells. This lack of knowledge is also evident when one looks at the herpesvirus family in general. From our studies and from studies of other herpesviruses, it is known that a core group of viral glycoproteins are conserved and play an essential role in herpesvirus-induced membrane fusion. This core group of proteins, gB, gH, and gL, likely form the machinery that allows membrane fusion to occur (16, 17). Although gp42 is not conserved within the herpesvirus family, other members of the family contain proteins, which likely have similar function. For example, herpes simplex virus type 1 contains gD, a protein that also binds to specific cell surface receptors and also likely serves as a trigger for membrane fusion to occur following binding to specific receptors (21). To complement this study, mutagenesis of gp42 is being undertaken to further explore this interaction and, overall, the role of this interaction in fusion.
The entry model of EBV consists of the hypothesis that the binding of gp42 to class II causes a conformational change in the tripartite complex of gp42-gH-gL. The conformational change would lower the energy state and provide support for the model of this interaction being a fast-dissociating complex. None of the mutations tested in this study supports or negates this idea. Support for this idea would have been shown with a mutant displaying efficient binding of sgp42 though an inability or decreased entry capability. In order to more directly probe the possibility of the conformational change, we are in the process of solving the crystal structure of gp42 alone. Overall, our results from this study suggest that class II residues 46 and/or 72 may help in bringing other residues, specifically residue 65, closer together and in increasing the overall interaction.
Acknowledgments
We thank Roselyn Eisenberg and Gary Cohen for providing the gD-expressing baculovirus and Lindsey Hutt-Fletcher for providing the monoclonal antibody specific for gp42. Finally, we thank the members of the Longnecker, Jardetzky, and Spear laboratories for help in performing these studies.
R.L. is supported by Public Health Service grants CA62234, CA73507, and CA93444 from the National Cancer Institute and DE13127 from the National Institute of Dental and Craniofacial Research. R.L. is a Stohlman Scholar of the Leukemia and Lymphoma Society of America. T.S.J. is supported by grants from the NIH (GM61050, AI38972, and CA93444) and the Arthritis Foundation. T.S.J. is a Scholar of the Leukemia and Lymphoma Society of America and a recipient of a Cancer Research Institute Young Investigator Award. M.P.M. is supported by the training program in carcinogenesis (NCI/NIH T32 CA009560).
REFERENCES
- 1.Bjorkman, P. J., and P. Parham. 1990. Structure, function, and diversity of class I major histocompatibility complex molecules. Annu. Rev. Biochem. 59:253-288. [DOI] [PubMed] [Google Scholar]
- 2.Borza, C. M., and L. Hutt-Fletcher. 2002. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat. Med. 8:594-599. [DOI] [PubMed] [Google Scholar]
- 3.Brown, J. H., T. S. Jardetzky, J. C. Gorga, L. J. Stern, R. G. Urban, J. L. Strominger, and D. C. Wiley. 1993. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature 364:33-39. [DOI] [PubMed] [Google Scholar]
- 4.Clackson, T., and J. A. Wells. 1995. A hot spot of binding energy in a hormone-receptor interface. Science 267:383-386. [DOI] [PubMed] [Google Scholar]
- 5.Connolly, S. A., D. J. Landsburg, A. Carfi, D. C. Wiley, R. J. Eisenberg, and G. H. Cohen. 2002. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA (HVEM). J. Virol. 76:10894-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeMars, R., C. C. Chang, S. Shaw, P. J. Reitnauer, and P. M. Sondel. 1984. Homozygous deletions that simultaneously eliminate expressions of class I and class II antigens of EBV-transformed B-lymphoblastoid cells. I. Reduced proliferative responses of autologous and allogeneic T cells to mutant cells that have decreased expression of class II antigens. Hum. Immunol. 11:77-97. [DOI] [PubMed] [Google Scholar]
- 7.Haan, K. M., W. W. Kwok, R. Longnecker, and P. Speck. 2000. Epstein-Barr virus entry utilizing HLA-DP or HLA-DQ as a coreceptor. J. Virol. 74:2451-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haan, K. M., S. K. Lee, and R. Longnecker. 2001. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology 290:106-114. [DOI] [PubMed] [Google Scholar]
- 9.Haan, K. M., and R. Longnecker. 2000. Coreceptor restriction within the HLA-DQ locus for Epstein-Barr virus infection. Proc. Natl. Acad. Sci. USA 97:9252-9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kockum, I., R. Wassmuth, E. Holmberg, B. Michelsen, and A. Lernmark. 1993. HLA-DQ primarily confers protection and HLA-DR susceptibility in type I (insulin-dependent) diabetes studied in population-based affected families and controls. Am. J. Hum. Genet. 53:150-167. [PMC free article] [PubMed] [Google Scholar]
- 11.Li, Q., M. K. Spriggs, S. Kovats, S. M. Turk, M. R. Comeau, B. Nepom, and L. M. Hutt-Fletcher. 1997. Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J. Virol. 71:4657-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumoto, N., M. Mitsuki, K. Tajima, W. M. Yokoyama, and K. Yamamoto. 2001. The functional binding site for the C-type lectin-like natural killer cell receptor Ly49A spans three domains of its major histocompatibility complex class I ligand. J. Exp. Med. 193:147-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mullen, M. M., K. M. Haan, R. Longnecker, and T. S. Jardetzky. 2002. Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Mol. Cell 9:375-385. [DOI] [PubMed] [Google Scholar]
- 14.Natarajan, K., N. Dimasi, J. Wang, D. H. Margulies, and R. A. Mariuzza. 2002. MHC class I recognition by Ly49 natural killer cell receptors. Mol. Immunol. 38:1023-1027. [DOI] [PubMed] [Google Scholar]
- 15.Nemerow, G. R., C. Mold, V. K. Schwend, V. Tollefson, and N. R. Cooper. 1987. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complement fragment C3d. J. Virol. 61:1416-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pertel, P. E. 2002. Human herpesvirus 8 glycoprotein B (gB), gH, and gL can mediate cell fusion. J. Virol. 76:4390-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pertel, P. E., A. Fridberg, M. L. Parish, and P. G. Spear. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313-324. [DOI] [PubMed] [Google Scholar]
- 18.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2446. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, 3rd ed. Raven Press, New York, N.Y.
- 19.Sanjeevi, C. B., M. Landin-Olsson, I. Kockum, G. Dahlquist, and A. Lernmark. 2002. The combination of several polymorphic amino acid residues in the DQalpha and DQbeta chains forms a domain structure pattern and is associated with insulin-dependent diabetes mellitus. Ann. N. Y. Acad. Sci. 958:362-375. [DOI] [PubMed] [Google Scholar]
- 20.Sisk, W. P., J. D. Bradley, R. J. Leipold, A. M. Stoltzfus, M. Ponce de Leon, M. Hilf, C. Peng, G. H. Cohen, and R. J. Eisenberg. 1994. High-level expression and purification of secreted forms of herpes simplex virus type 1 glycoprotein gD synthesized by baculovirus-infected insect cells. J. Virol. 68:766-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spear, P. G., R. J. Eisenberg, and G. H. Cohen. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1-8. [DOI] [PubMed] [Google Scholar]
- 22.Speck, P., K. A. Kline, P. Cheresh, and R. Longnecker. 1999. Epstein-Barr virus lacking latent membrane protein 2 immortalizes B cells with efficiency indistinguishable from that of wild-type virus. J. Gen. Virol. 80:2193-2203. [DOI] [PubMed] [Google Scholar]
- 23.Spriggs, M. K., R. J. Armitage, M. R. Comeau, L. Strockbine, T. Farrah, B. Macduff, D. Ulrich, M. R. Alderson, J. Müllberg, and J. I. Cohen. 1996. The extracellular domain of the Epstein-Barr virus BZLF2 protein binds the HLA-DR β chain and inhibits antigen presentation. J. Virol. 70:5557-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strnad, B. C., T. Schuster, R. Klein, R. F. Hopkins III, T. Witmer, R. H. Neubauer, and H. Rabin. 1982. Production and characterization of monoclonal antibodies against the Epstein-Barr virus membrane antigen. J. Virol. 41:258-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanner, J., J. Weis, D. Fearon, Y. Whang, and E. Kieff. 1987. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell 50:203-213. [DOI] [PubMed] [Google Scholar]
- 26.Tormo, J., K. Natarajan, D. H. Margulies, and R. A. Mariuzza. 1999. Crystal structure of a lectin-like natural killer cell receptor bound to its MHC class I ligand. Nature 402:623-631. [DOI] [PubMed] [Google Scholar]
- 27.Wang, J., M. C. Whitman, K. Natarajan, J. Tormo, R. A. Mariuzza, and D. H. Margulies. 2002. Binding of the natural killer cell inhibitory receptor Ly49A to its major histocompatibility complex class I ligand. Crucial contacts include both H-2Dd and beta 2-microglobulin. J. Biol. Chem. 277:1433-1442. [DOI] [PubMed] [Google Scholar]
- 28.Wang, X., and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus lacking glycoprotein gp42 can bind to B cells but is not able to infect. J. Virol. 72:158-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, X., W. J. Kenyon, Q. Li, J. Müllberg, and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J. Virol. 72:5552-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]