Abstract
The herpes simplex virus (HSV) single-stranded DNA-binding protein, ICP8, is required for viral DNA synthesis. Before viral DNA replication, ICP8 colocalizes with other replication proteins at small punctate foci called prereplicative sites. With the onset of viral genome amplification, these proteins become redistributed into large globular replication compartments. Here we present the results of immunocytochemical and biochemical analysis of ICP8 showing that various antibodies recognize distinct forms of ICP8. Using these ICP8-specific antibodies as probes for ICP8 structure, we detected a time-dependent appearance and disappearance of ICP8 epitopes in immunoprecipitation assays. Immunofluorescence staining of ICP8 in cells infected with different HSV mutant viruses as well as cells transfected with a limited number of viral genes demonstrated that these and other antigenic changes occur coincident with ICP8 assembly at intranuclear replication structures. Genetic analysis has revealed a correlation between the ability of various ICP8 mutant proteins to form the 39S epitope and their ability to bind to DNA. These results support the hypothesis that ICP8 undergoes a conformational change upon binding to other HSV proteins and/or to DNA coincident with assembly into viral DNA replication structures.
Changes in protein conformation can be critical to polypeptide function. Therefore, a complete understanding of how some proteins are regulated involves identifying changes in their tertiary structure. For example, structural changes in many transcription factors (e.g., OxyR, AP-1, Sp-1, NF-κB, and p53) (reviewed in references 28, 29, 34, 71, 74, and 91), the Escherichia coli replication protein dnaB (2, 3, 35), adenovirus 72K single-stranded DNA-binding protein (SSB) (17), bacteriophage T4 gene 32 SSB (85), and the eukaryotic SSB replication protein A (RP-A) (6, 25) are functionally related to the activity of these proteins. Changes in protein structure can be regulated by a variety of means, including binding to DNA (reviewed in references 25 and 73), interactions with ions (84, 88), interactions with other proteins (19, 47, 66), posttranslational modifications (46, 49; reviewed in reference 36), or even environmental conditions, such as redox potential (reviewed in references 5 and 13).
We have studied herpes simplex virus (HSV) DNA replication as a model system for the localization, maturation, and ordered assembly of protein complexes within the cell. HSV encodes seven proteins that are necessary for viral origin-dependent DNA synthesis (12, 39, 75, 87, 90). These include the SSB (ICP8), as well as a polymerase (UL30), its processivity factor (UL42), an origin-binding protein (UL9), and three proteins that form the viral helicase-primase complex (UL5, UL8, and UL52). ICP8 serves several functions during viral DNA synthesis (67). It functions as an SSB to stabilize displaced single-stranded DNA strands during HSV DNA replication and also stimulates the helicase activity of UL9 during initiation and that of the UL5/UL8/UL52 complex during elongation stages of DNA synthesis.
Within HSV-infected cells, the HSV DNA replication proteins assemble at specific intranuclear sites to form large globular replication compartments (8, 16, 26, 48, 51, 61, 64). If viral DNA synthesis is blocked during infection, many of these proteins are found in smaller prereplicative sites, which display a punctate distribution throughout the nucleus (8, 9, 16, 48, 51, 64). We and others have shown that, under natural infection conditions, three components of the viral replication machinery, the tripartite helicase-primase, the origin-binding protein, and ICP8, are all required in concert for punctate structure assembly. Interestingly, while monitoring the ordered assembly of these proteins, we detected localization-associated antigenic changes in the viral ICP8 protein, as described below.
The localization of ICP8 to these intranuclear structures involves a series of sequential binding states between ICP8 and the host cell that can be biochemically defined by different fractionation characteristics and solubilization requirements such as detergent and/or DNase treatment (42). Several early observations suggested to us that ICP8 might undergo a conformational change during this localization and maturation process. First, ICP8 can exist in two distinct oxidative forms, which migrate as a doublet by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). In pulse-chase protein-labeling experiments, these oxidative species appeared to be related as precursor and product (42). Second, within cells infected with an ICP27 null mutant virus, immunofluorescence assays revealed a partial defect in ICP8 localization to viral replication structures and a corresponding difference in ICP8 reactivity with the conformation-specific 39S monoclonal antibody (MAb) (15). Finally, more recent biochemical studies on purified ICP8 have demonstrated an apparent conformational change in ICP8 upon binding to single-stranded DNA in vitro (18).
Here we present the results of immunocytochemical analysis of infected cells and biochemical analysis of ICP8 showing that multiple antibodies recognize antigenically distinct forms of ICP8. Using an intracellular approach which allowed us to follow ICP8 topology during the assembly process, we observed that one of these antigenic species only occurs coincident with ICP8 assembly at intranuclear replication structures.
MATERIALS AND METHODS
Cells and viruses.
All cell lines were grown and maintained in Dulbecco's modified Eagle's medium (Irvine Scientific, Santa Ana, Calif.) containing 10% heat-inactivated fetal calf serum (FCS), 2 mM l-glutamine, streptomycin sulfate, and penicillin G potassium. Experiments were performed with Vero African green monkey kidney cells (American Type Culture Collection, Manassas, Va.).
HSV type 1 (HSV-1) wild-type (WT) strain KOS1.1 (33), originally obtained from M. Levine, was titrated on Vero cells by using an overlay of medium 199 (GIBCO, Gaithersburg, Md.) containing 1% heat-inactivated calf serum and 0.1% human immune serum. The ICP8 mutant viruses constructed from KOS 1.1, n11 and n11SV, and the ICP8-expressing cell line S-2 on which these mutant viruses were propagated and titrated have been described previously (23). The ICP27 mutant virus d27, also constructed from KOS1.1, was grown and titrated on the ICP27-expressing V27 cell line (65).
HSV-1 WT strain KOS was obtained from Priscilla Schaffer. Replication protein mutant viruses hr114 (UL52−) (24), hr80 (UL8−) (11), hr99 (UL5−) (95), and hr94 (UL9−) (53) containing lacZ insertions into the indicated genes, as well as the complementing cell lines 2D6 (UL52), SL8 (UL8), L5 (UL5), and 2B11 (UL9), were provided by Sandra Weller. The UL42 deletion mutant virus, CgalΔ42, and the complementing cell line U9 (37) were provided by Paul Johnson. Don Coen provided both the HP66 polymerase mutant virus, which contains a 2.3-kbp deletion and lacZ insertion into the UL30 open reading frame, and the complementing cell line DP6 (56). KOS virus was used as the WT strain in all experiments which included the replication protein mutants, as these mutant viruses were constructed from this parental strain.
Infections.
For all experiments, viruses were diluted in cold phosphate-buffered saline containing 0.1% glucose and 1% heat-inactivated newborn-calf serum and were incubated with cells at a multiplicity of infection (MOI) of 20 PFU per cell. After 1 h at 37°C, the inoculum was removed, and cells were overlayed with medium 199 containing 1% newborn-calf serum. When indicated, sodium phosphonoacetate (PAA; Sigma Chemicals, St. Louis, Mo.) at a concentration of 400 μg/ml was included in the overlay media of infections to inhibit viral DNA replication (45, 54).
Antibodies.
Several antibodies were used for immunofluorescence staining and immunoprecipitation of ICP8 (Table 1). Paul Olivo provided the R219 polyclonal antiserum (61). MAbs LP-793 and LP-710 were provided by Lenore Pereira, and the 10-E3 MAb was obtained from Kathleen Shriver. The 3-83 polyclonal antiserum has been described previously (41). The 39S MAb (72) ascites fluid was generated in mice by the injection of hybridoma cells obtained from the American Type Culture Collection.
TABLE 1.
Antibodies and antisera
| Name | Type | Reference for antibody | Specificity | References for specificity |
|---|---|---|---|---|
| 39S | MAb | 72 | Native ICP8 | D. M. Knipe, unpublished data; W. Ruyechan, unpublished data |
| R219 | Polyclonal antiserum | 61 | Residues 15-221 | 61 |
| 10-E3 | MAb | 68 | Residues 1169-1196 | M. Gao and D. M. Knipe, unpublished data |
| LP-710 | MAb | L. Pereira, unpublished data | Residues 412-590 | M. Gao and D. M. Knipe, unpublished data |
| LP-793 | MAb | L. Pereira, unpublished data | Residues 933-1160 | M. Gao and D. M. Knipe, unpublished data |
| 3-83 | Polyclonal antiserum | 41 | Denatured and native ICP8 | 21, 41 |
Indirect immunofluorescence.
Cells were grown on glass coverslips as described above. At 6 h postinfection (hpi) or 48 h posttransfection, cells were fixed for 10 min in 3.7% formaldehyde, permeabilized for 2 min in −20°C acetone, and immunostained as described previously (64). The following primary antibodies were used at the indicated dilutions: 39S ascites (1:80), 3-83 (1:100), LP-793 (1:40), LP-710 (1:40), 10-E3 (1:40), and R219 (1:20). As the goal of these immunofluorescence assays was to detect whether these antibodies exhibited a stronger affinity for specific populations of ICP8, titration of antibodies was performed to determine the concentration at which these differences could be best observed (S. Uprichard and D. M. Knipe, unpublished results). To reduce nonspecific background, diluted R219 polyclonal antiserum was preabsorbed on fixed, permeabilized, uninfected Vero cells prior to immunofluorescence staining. The fluorochrome-conjugated secondary antibodies used in all experiments were fluorescein-conjugated goat anti-rabbit immunoglobulin G (1:200) and rhodamine-conjugated goat anti-mouse immunoglobulin G (1:100) (Cappel Laboratories). Cells were mounted on glass slides in glycerol-gelatin (Sigma) containing 1.3 μg of p-phenyldiamine per ml and examined by fluorescence and phase-contrast microscopy with a Zeiss photomicroscope (40).
Metabolic labeling and immunoprecipitation.
Cells were infected with virus as described above at an MOI of 20 PFU per cell. For pulse-chase experiments, at 4.5 hpi, cells were rinsed and incubated at 37°C for 10 min in methionine-free minimal essential medium (Met− MEM; ICN Biomedicals, Costa Mesa, Calif.). Met− MEM containing 100 μCi of [35S]methionine (ICN) per ml and 1% FCS was then added on ice, and cells were pulse-labeled for 3 min by submerging the flasks in a 37°C water bath (42). Incorporation was stopped on ice, labeling medium was removed, and 5 ml of 199 medium-1% FCS containing unlabeled methionine at a concentration of 200 μM was added to cultures for the chase periods indicated. After the chase period at 37°C, cells were harvested in ice-cold phosphate-buffered saline containing 100 μM phenylmethanesulfonyl fluoride (PMSF; Sigma), 2 μM Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK; Sigma), and 50 μM N-ethylmaleimide (NEM; Sigma). Cells were collected by centrifugation and frozen at −80°C. For other experiments, infected cells were labeled from 4.5 to 5.5 hpi in Met− MEM containing 50 μCi of [35S]methionine per ml, 10% 199, and 1% FCS immediately prior to harvest.
To generate cell lysates for immunoprecipitation, the cell pellets were resuspended in nonionic detergent IP buffer (10 mM Tris-HCl [pH 7.6], 50 mM NaCl, 0.5% NP-40, 100 μM PMSF, 2 μM TLCK, 50 μM NEM) at a concentration of 4.25 × 106 cells/ml and pulse-sonicated eight times at low power for 1 s. Because ICP8 contains a large number of cysteine residues and runs as a doublet of different oxidative species by SDS-PAGE (40), in the experiments reported here we included NEM in the immunoprecipitation buffer to alkylate free sulfhydryl groups and thereby trap any disulfide intermediates present at the time of harvest. Similar results were obtained in the absence of NEM (results not shown). In the indicated experiments, various buffers were added to aliquots of the lysate to adjust the salt and detergent concentrations. Antibodies were used in limiting quantities, enough to bind approximately 50% of the ICP8 present in the cells infected with WT virus, in an effort to avoid any potential antibody-induced changes in ICP8 structure (data not shown). After a 1-h incubation at 4°C with the indicated primary antibody, immune complexes were formed with fixed Staphylococcus aureus cells (CalBiochem, La Jolla, Calif.) for 40 min on ice. Bound complexes were collected and washed three times with 500 μl of wash buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 100 μM PMSF, 2 μM TLCK, 50 μM NEM), resuspended in SDS sample buffer, boiled, and resolved by SDS-PAGE with a 9.25% polyacrylamide gel. Following electrophoresis, gels were fixed, dried, and exposed to Kodak BioMax film for autoradiographic images and phosphorimager screens for quantitation. ICP8 bands on the phosphorimages were quantified by using Bio-Rad ImageQuant software. The percentage of ICP8 immunoprecipitated was calculated as the fraction of ICP8 in the immunoprecipitate relative to the amount of ICP8 in the lysate.
Plasmids.
Plasmid pSV8.3 contains the ICP8 open reading frame expressed from the simian virus 40 (SV40) early gene promoter (21, 22). The cytomegalovirus immediate-early promoter-driven constructs that express the viral helicase-primase genes, pCM-UL5, pCM-UL8, and pCM-UL52 (30), were provided by Diane Hayward.
Transfections.
Cells were grown on glass coverslips to 70% confluence. Transfections were performed using calcium phosphate precipitation (4). A total of 2 μg of DNA, which consisted of 0.5 μg of pSV8.3 plus either 1.5 μg of pUC19 carrier DNA or 0.5 μg of each of the three helicase-primase expression plasmids indicated above, was added to each coverslip. At 48 h posttransfection, cells were fixed, and immunofluorescence was performed.
RESULTS
Time-dependent conformational changes in ICP8.
To determine whether ICP8 undergoes any conformational changes during its maturational process, we examined the reactivity of ICP8 with several antibodies and antisera (Table 1). One of the antibodies used, the 39S MAb, has the properties of an ICP8-specific, conformation-dependent antibody because it recognizes ICP8 from infected cells (44) and purified native ICP8 (D. M. Knipe, unpublished data; W. Ruyechan unpublished data) but does not recognize denatured ICP8 by Western blot analysis (M. Gao, S. Uprichard, and D. M. Knipe, unpublished data) or by immunoprecipitation in the presence of 0.2% SDS (Table 2). In addition, a series of ICP8 in-frame linker insertion mutations indicated that small insertions throughout the protein can abolish the 39S epitope (E. Villarreal and D. M. Knipe, unpublished data) and several temperature-sensitive ICP8 proteins are not recognized by the 39S MAb at the nonpermissive temperature (D. M. Knipe, unpublished data). Denaturation or mutational changes likely alter the conformation of ICP8 so that the 39S epitope is no longer present in the molecule.
TABLE 2.
Immunoprecipitation of ICP8 with 39S MAb under different conditions
| Buffer NaCl concentration (mM) | % of ICP8 immunoprecipitateda |
|---|---|
| 50 | 31 |
| 150 | 31 |
| 500 | 33 |
| 1,000 | 32 |
| 50 (plus 0.2% SDS) | 5 |
Lysate was prepared from cells infected with hr80 mutant virus, and aliquots were adjusted to the buffer conditions listed. The percentage of ICP8 immunoprecipitated was calculated based on the total amount of ICP8 present in a duplicate aliquot as determined directly from an SDS-PAGE protein profile in which ICP8 is a prominent band.
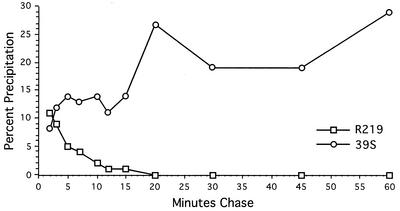
To determine whether this conformational epitope was present immediately following synthesis of ICP8 or required maturation of the protein, we performed [35S]methionine pulse-chase labeling of infected-cell proteins and assayed for the availability of specific ICP8 epitopes by immunoprecipitation with the 39S MAb or the N-terminal-specific R219 serum. Small amounts of the pulse-labeled ICP8 were precipitated by the 39S MAb immediately following synthesis (Fig. 1). However, we observed an increase in the amount of ICP8 recognized by the 39S MAb over the first 20 min of chase (Fig. 1). In contrast, R219 reactivity was highest just after ICP8 synthesis and was progressively lost during the subsequent 20-min chase period (Fig. 1). Thus, we detected a time-dependent formation of the 39S epitope and a corresponding decrease in the availability of the R219 epitope on ICP8. In similar experiments, the 3-83 polyclonal anti-ICP8 serum recognized the majority of ICP8 at all time points regardless of the length of the chase period (data not shown).
FIG. 1.
Reactivity of ICP8 with antibodies as a function of chase time. Vero cells were infected with WT KOS1.1 virus at an MOI of 20 and pulse-labeled with [35S]methionine for 3 min at 4.5 hpi. At the indicated times after labeling, infected cells were harvested, and ICP8 was immunoprecipitated from aliquots of each lysate, using the conformation-dependent MAb 39S or the polyclonal antiserum R219, which recognizes the N terminus of ICP8. Immune complexes were resolved by SDS-PAGE and quantified with a phosphorimager. The amounts immunoprecipitated are expressed as a percentage of the total ICP8 in each sample.
Cytoplasmic ICP8 is recognized by R219 but not by 39S.
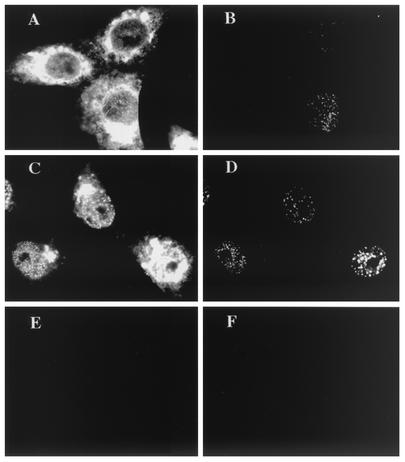
To determine whether these antigenic changes occurred upon entry of ICP8 into the cell nucleus, we assayed the R219 and 39S reactivities of cytoplasmic versus nuclear ICP8. ICP8 that remained in the cytoplasm was generated by infection of cells with the n11 ICP8 mutant virus, which encodes a truncated ICP8 in which the carboxy-terminal 28 residues encompassing the nuclear localization signal (NLS) of the protein have been deleted (22, 23). To examine a nuclear form of the n11 ICP8 protein, we used a second mutant virus, n11SV, which encodes the n11 ICP8 fused to the SV40 large T-antigen NLS (23).
Dual-label immunofluorescence of infected cells was performed at 6 hpi with the polyclonal R219 serum and 39S MAb. R219 staining detected large amounts of ICP8 in the cytoplasm of cells infected with the n11 virus and possibly small amounts of nuclear ICP8 in these cells (Fig. 2A). However, the 39S MAb did not recognize the cytoplasmic ICP8 in these cells but did recognize the small amount of nuclear punctate ICP8 in these cells (Fig. 2B). The n11 ICP8 fused to the SV40 large T-antigen NLS localized to small intranuclear structures and reacted with R219 (Fig. 2C) and 39S (Fig. 2D). Hence, the n11 polypeptide was able to form the 39S epitope, but localization of ICP8 into the nucleus was required for formation of the epitope and recognition by the 39S MAb. In contrast, R219 appeared to possess strong reactivity with cytoplasmic ICP8 (Fig. 2A and C), indicating that the N-terminal epitope(s) recognized by this polyclonal serum was accessible for binding primarily before ICP8 entered the nucleus. It is not known what type of interactions n11 ICP8 has with other viral replication proteins and hence to what degree normal viral replication complexes form; however, with this mutant, we do see R219 staining of ICP8 at intranuclear foci as well.
FIG. 2.
Reactivity of cytoplasmic ICP8. Vero cells were infected with ICP8 mutant virus n11 (A and B) or n11SV (C and D) or were mock infected (E and F). At 6 hpi, dual-label immunofluorescence was performed with ICP8 antibodies R219 (A, C, and E) and 39S (B, D, and F).
The 39S epitope is detected on ICP8 only at specific intranuclear sites.
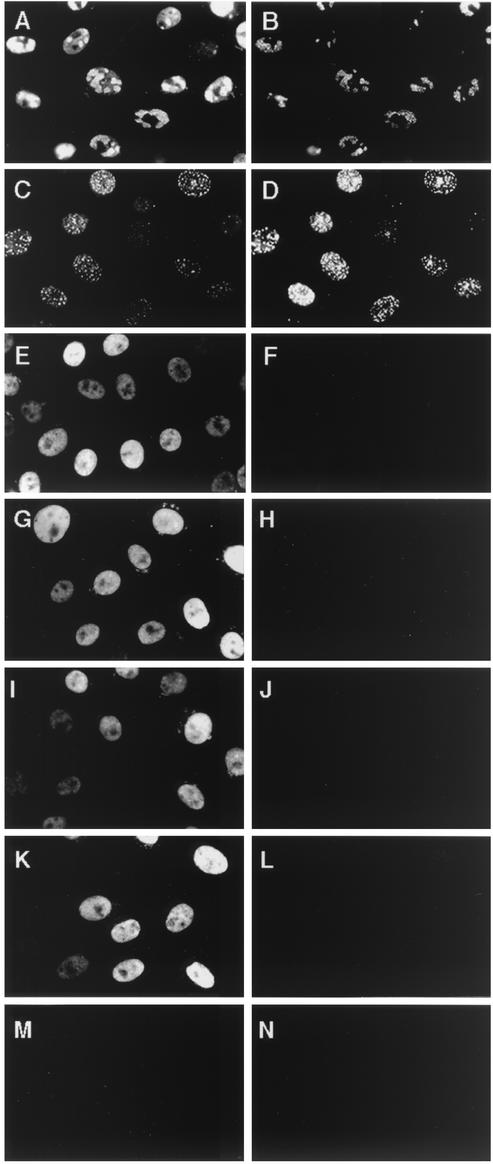
Because much of the n11 and n11SV that entered the nucleus localized to punctate prereplicative structures, it could not be determined whether nuclear entry was sufficient for 39S reactivity or whether the antigenic difference was associated with assembly of ICP8 at specific intranuclear sites. To make this distinction, we examined 39S reactivity in cells infected with mutant viruses in which ICP8 enters the nucleus but does not assemble at prereplicative sites. A defect in intranuclear localization of ICP8 was exhibited by four mutant viruses, each deficient for one component of the viral helicase-primase (hr114 [UL52−], hr80 [UL8−], and hr99 [UL5−]) or for the origin-binding protein (hr94 [UL9−]) (48, 51). Cells were infected with WT virus or one of these mutant viruses, and 39S reactivity was assayed by dual-label immunofluorescence in conjunction with the anti-ICP8 3-83 serum.
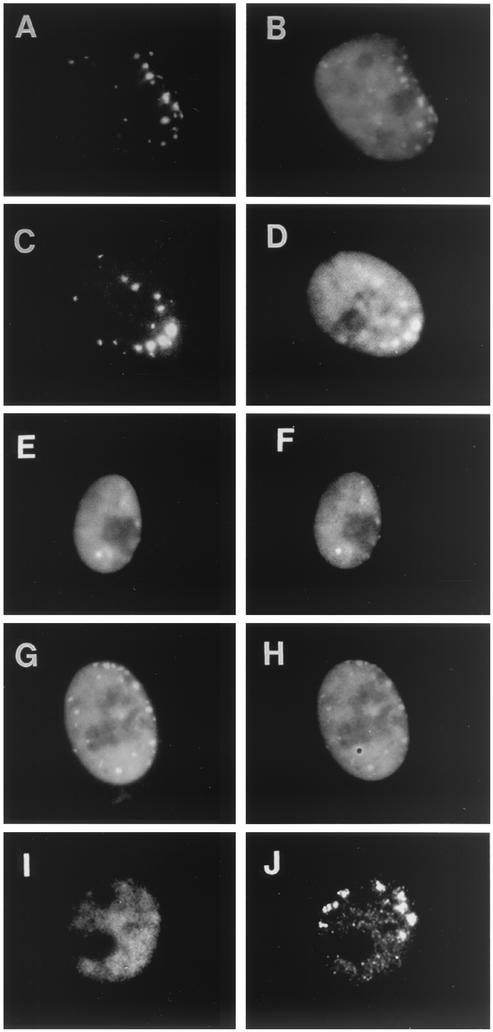
At 6 hpi, the majority of ICP8 in the WT-infected cells was in replication compartments and was detected by 3-83 and 39S antibodies (Fig. 3A and B, respectively). Likewise, when WT virus replication was blocked by incubation with PAA, both antibodies reacted with the ICP8 at punctate prereplicative sites (Fig. 3C and D). As has been seen previously (48, 51), in cells infected with the UL52 mutant virus, staining with the 3-83 antiserum showed ICP8 to be diffusely distributed throughout the nucleus (Fig. 3E). However, under these conditions little or no reactivity with the conformation-specific 39S antibody was detected (Fig. 3F). Similar results were obtained with the UL8 mutant virus (Fig. 3G and H), the UL5 mutant virus (Fig. 3I and J), and the UL9 mutant virus (Fig. 3K and L). Thus, ICP8 entry into the nucleus was not sufficient for 39S reactivity. Like 3-83, however, the R219 serum did bind to the diffuse ICP8 in these mutant virus-infected cells (S. Uprichard and D. M. Knipe, unpublished data).
FIG. 3.
Reactivity of diffuse ICP8 expressed in cells infected with ICP8 localization-defective viruses. Vero cells were infected with WT virus (A and B), WT plus PAA (C and D), UL52− virus (E and F), UL8− virus (G and H), UL5− virus (I and J), or UL9− virus (K and L) or were mock infected (M and N). At 6 hpi, dual-label ICP8 immunofluorescence was performed with 3-83 antiserum (left panels) or 39S MAb (right panels).
While neither component of the viral polymerase (UL30 or UL42) is required for ICP8 localization to prereplicative sites (8, 26, 64), we wanted to determine if the association of these proteins with ICP8 at replication structures was necessary for induction of the 39S epitope. Therefore, dual-label immunofluorescence with 39S and 3-83 antibodies was performed on cells infected with viruses containing null mutations in UL30 (HP66) or UL42 (CgalΔ42). As observed above (Fig. 3), ICP8 in WT-infected cells in the presence of PAA localized to punctate foci and was recognized by either 3-83 or 39S (Fig. 4A and B). As expected, in cells infected with the HP66 Pol− virus (Fig. 4C) or the CgalΔ42 UL42− virus (Fig. 4E), 3-83 antiserum staining showed ICP8 to be assembled at prereplicative sites regardless of the presence (Fig. 4) or absence (data not shown) of PAA. Strong 39S reactivity was detected in cells infected with HP66 (Fig. 4D) or CgalΔ42 (Fig. 4F). Therefore, neither component of the viral polymerase holoenzyme was required to induce 39S reactivity.
FIG. 4.
ICP8 39S reactivity in the absence of UL30 or UL42. Vero cells were infected with WT virus (A and B), UL30− virus (C and D), or UL42− virus (E and F) or were mock infected (G and H) in the presence of PAA. At 6 hpi, dual-label immunofluorescence was performed with the ICP8 antibody 3-83 (left panels) or 39S (right panels).
Availabilities of several ICP8 epitopes change coincident with ICP8 localization to replication structures.
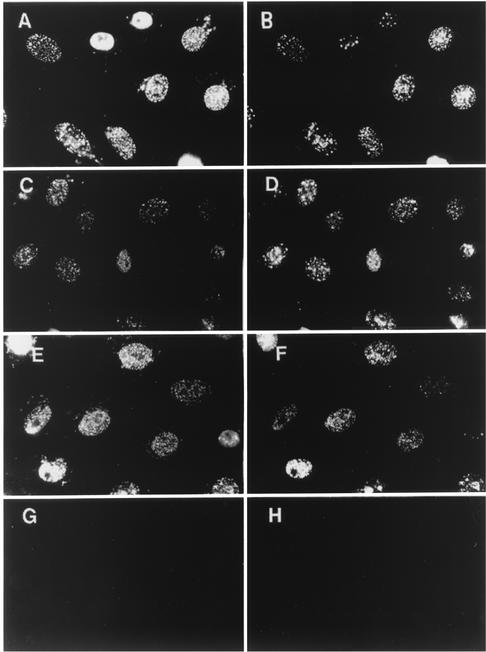
To test if other antigenic differences could be detected between diffuse ICP8 and ICP8 assembled at replication sites, we used additional ICP8 antibodies with defined epitopes (Table 1) to map the topology of ICP8 within infected cells. To ensure the availability of both diffuse and intranuclearly localized populations of ICP8 within individual nuclei, we infected cells with the ICP27 null mutant virus d27-1, which exhibits a partial defect in ICP8 localization (15), likely due to the underexpression of UL5, UL8, UL52, and UL9 (83). Dual-label immunofluorescence was performed to assess the reactivity characteristics of various MAbs compared with that of 3-83, while polyclonal R219 reactivity was assayed in parallel with the 39S MAb (Fig. 5). No staining of mock-infected cell nuclei was observed with any of these antibodies (results not shown).
FIG. 5.
ICP8 reactivity with various antibodies that recognized defined epitopes. Vero cells were infected with d27 virus at an MOI of 20. At 6 hpi, dual-label immunofluorescence was performed either with the polyclonal serum 3-83 (B, D, F, and H) in conjunction with ICP8 monoclonal antibody 39S (A), LP-710 (C), 10-E3 (E), or LP-793 (G) or with R219 serum (I) or 39S (J).
In cells infected with this ICP27 mutant virus, 3-83 polyclonal serum detected both diffuse and intranuclear localized ICP8 (Fig. 5B). Consistent with the previous observation of altered 39S reactivity in d27-infected cells (15), 39S MAb reacted only with ICP8 that accumulated in replication structures (Fig. 5A). Similar to the 39S MAb, LP-710 MAb reacted only with ICP8 in replication structures (Fig. 5C), although 3-83 staining showed the presence of large amounts of diffuse nuclear ICP8 in these cells (Fig. 5D). In contrast, MAbs 10-E3 (Fig. 5E) and LP-793 (Fig. 5G) reacted with both diffuse and intranuclearly localized ICP8, similar to what was seen with 3-83 serum (Fig. 5F and H). Although it was not clear from this analysis if 10-E3 and/or LP-793 exhibited any preferential reactivity for unassembled ICP8, in many cells R219 did appear to have higher affinity for diffuse nuclear ICP8 (Fig. 5I) than for the 39S-reactive ICP8 at replication structures (Fig. 5J). Yet it should be noted that R219 did react to some extent with the ICP8 at viral replication structures, especially at higher antibody concentrations (data not shown). Hence, this panel of ICP8 antibodies revealed several antigenic differences in WT ICP8 coincident with localization to intranuclear sites. These included the appearance of the 39S conformational epitope, exposure of the previously unavailable central residues recognized by LP-710, and at least a partial masking of the amino-terminal end of ICP8 that is recognized by R219.
Changes in antibody reactivity appear to be the result of intramolecular conformational changes.
While one might expect epitopes on ICP8 to become buried by other proteins upon assembly into replication complexes, we observed not only the partial disappearance (R219) but also the appearance (39S and LP-710) of ICP8-specific epitopes coincident with protein localization to these nuclear structures. Because of the number and varied nature of these antigenic changes, they might be due to conformational changes in ICP8 rather than a simple masking of epitopes due to intermolecular protein-protein interactions. To address the possibility that these antigenic changes might be due to masking of epitopes by intermolecular interactions between ICP8 and other proteins, we assayed ICP8 antibody reactivity under stringent buffer conditions that have been shown to solubilize ICP8 from other proteins and the nuclear matrix (40, 64).
Using radiolabeled UL8 mutant virus-infected cells as a source of diffusely distributed, unassembled ICP8, we determined whether we could “reveal” the 39S epitope prior to structure assembly by disrupting any potential protein complexes. To dissociate ICP8 from protein complexes with which it might be associated, aliquots of infected-cell lysate were sonicated and subjected to increasing salt concentrations, which has previously been shown to solubilize ICP8 (64). Immunoprecipitation of ICP8 after such treatment showed no significant increase in 39S reactivity (Table 2). In contrast, the inclusion of SDS decreased 39S immunoprecipitation of ICP8 (Table 2), consistent with the 39S epitope being eliminated by SDS denaturation. These results suggested that in these localization mutant virus-infected cells no transient intermolecular interactions were masking the 39S epitope of diffuse ICP8.
Similarly, ICP8 from radiolabeled cells infected with WT virus was subjected to the same manipulations to determine whether the R219 epitope would become more accessible upon disruption of replication protein complexes. As might be expected based on the previously determined time course of R219 reactivity (Fig. 1), R219 recognized very little ICP8 from cells infected with WT virus in which the majority of ICP8 is generally localized to viral replication structures (Table 3). Subjecting WT-infected cell lysates to high-salt treatment prior to immunoprecipitation did not expose the R219 epitope (Table 3). Infected lysates had to be incubated with SDS before significant R219 reactivity was detected (0.2% SDS) (Table 3). Hence, upon assembly into replication structures the R219 epitope(s) (N terminus residues 15 to 211) appeared to become buried within the ICP8 molecule such that the protein had to be denatured to reveal it.
TABLE 3.
Immunoprecipitation of ICP8 with R219 polyclonal antiserum under different conditions
| Buffer NaCl concentration (mM) | % of ICP8 immunoprecipitateda |
|---|---|
| 50 | 3 |
| 100 | 2 |
| 150 | 2 |
| 300 | 1 |
| 500 | 1 |
| 150 (plus 0.2% SDS) | 32 |
Lysate was prepared from cells infected with WT virus, and aliquots were adjusted to the buffer conditions listed. The percentage of ICP8 immunoprecipitated was calculated based on the total amount of ICP8 present in a duplicate aliquot as determined directly from an SDS-PAGE protein profile in which ICP8 is a prominent band.
Formation of the 39S epitope on ICP8 in transfected cells.
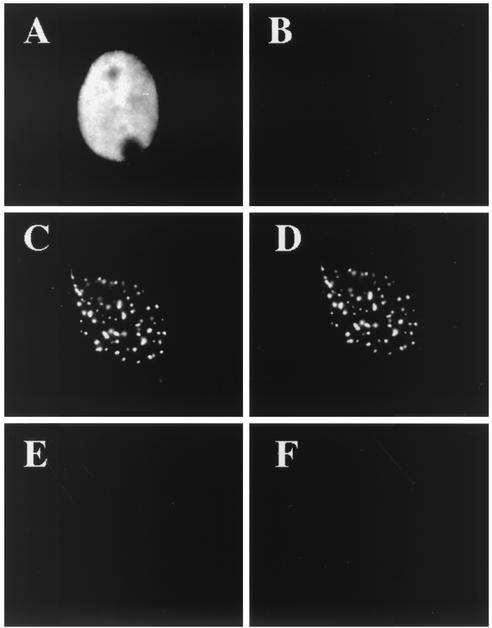
To determine which viral gene products were required to promote the formation of the 39S epitope, we examined the reactivity of ICP8 expressed in cells transfected with different combinations of viral genes. When ICP8 was expressed alone, nearly all of the transfected cells showed a diffuse nuclear pattern of ICP8, as observed with 3-83 (Fig. 6A), but little to no reactivity was detected with the 39S MAb (Fig. 6B). Consistent with the results of previous studies (52, 64), ICP8 did have some intrinsic ability to form the 39S epitope and localize to specific sites within the nucleus because a few of the 3-83-positive cells showed a 39S staining pattern that resembled punctate prereplicative sites or had 39S staining of ICP8 that was somewhat granular in appearance (S. Uprichard and D. M. Knipe, results not shown). As previously demonstrated (48, 52, 82), however, cotransfection of the UL5, UL8, and UL52 helicase-primase genes with the ICP8 gene resulted in nearly all of the cells showing ICP8 localized to intranuclear structures (Fig. 6C). The ICP8 at these punctate structures was clearly recognized by 39S (Fig. 6D). Hence, while UL5, UL8, and UL52 were not absolutely required for 39S reactivity, they did significantly enhance formation of the 39S epitope in ICP8 within the nucleus.
FIG. 6.
ICP8 localization and structure in transfected cells. Vero cells were transfected with plasmids expressing ICP8 (A and B); ICP8, UL5, UL8, and UL52(C and D); or negative control pUC19 DNA (E and F). At 48 h posttransfection, dual-label immunofluorescence was performed with 383 (left panels) or 39S (right panels).
Genetic analysis of ICP8 reveals a correlation between 39S reactivity and ICP8 DNA binding.
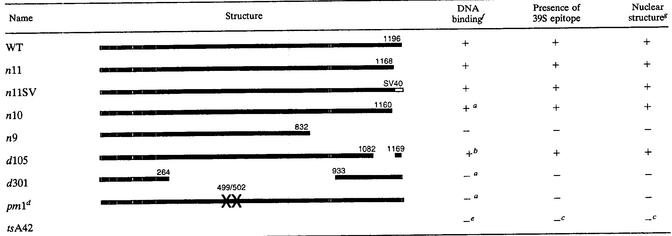
To investigate the possible functional significance of these antigenic changes in ICP8, we compared the 39S reactivity of multiple ICP8 mutant proteins with their DNA-binding phenotypes. ICP8 reactivity with the 39S MAb was tested by immunoprecipitation and immunofluorescence. The DNA-binding ability of these mutants had previously been determined by chromatography of infected-cell lysates through DNA cellulose columns (summarized in Table 4). Immunofluorescence assays performed on cells infected with various ICP8 mutant viruses revealed that only localization-competent ICP8 proteins were detected by 39S, further documenting the strict relationship between ICP8 39S reactivity and its intracellular localization (Table 4 and results not shown). Furthermore, in all cases, 39S reactivity correlated with the ability of each mutant ICP8 protein to be retained on DNA cellulose columns (Table 4). While one trivial explanation for this type of correlation could be that the 39S antibody might bind to the ICP8 DNA-binding domain, this seems unlikely because preincubation of WT ICP8 with the 39S antibody does not inhibit the binding of the protein to DNA (43). Some of the mutant ICP8 molecules that are not recognized by 39S, such as pm1, show reduced solubility (21), indicating conformational changes in the protein. Based on these results, we hypothesize that the formation of the 39S epitope may be part of a conformational change that allows ICP8 to bind to DNA or is caused by the binding of ICP8 to DNA.
TABLE 4.
Properties of mutant ICP8 proteins
Construction and DNA binding of n10, d301, and pm1 reported by Gao and Knipe (21a).
Reference 21b.
Analysis and references in Weller et al. (87a).
Complete analysis of pm1 reported in reference 21.
Leinbach et al. (45a).
+, greater than 75% of WT activity; −, less than 50% of WT activity.
Ability to localize to nuclear DNA replication structures.
DISCUSSION
The specificity of antibody recognition has proven to be a useful tool for detecting and mapping the tertiary structure of many proteins, including, for example, myosin (32, 81, 89), creatine kinase (59), p53 (20, 28, 93), influenza RNA polymerase (79), and hepatitis C virus E2 glycoprotein (14, 80). In this study, we found that several antibodies specific for the HSV ICP8 SSB can serve as immunochemical probes for the investigation of ICP8 topology during replication complex assembly. Using these antibodies, we observed time-dependent changes in ICP8 epitope availability and determined that these and other antigenic changes occur coincident with ICP8 localization to replication complexes within the nucleus. Although not specifically addressed here, in light of the fact that ICP8 assembles at two distinct types of prereplicative sites within infected cells (50, 82), it should be noted that these antigenic changes occur upon ICP8 assembly at both S phase-associated and S phase-independent viral structures (S. Uprichard and D. M. Knipe, results not shown).
A priori, it might be expected that formation of protein complexes at replication sites could mask some ICP8 epitopes. However, we detected a more complex change in reactivity that includes not only the reduced accessibility of the R219 epitope(s) but also the appearance of conformational (39S) and linear (LP-710) ICP8 epitopes. Hence, we currently favor the hypothesis that these varied antigenic changes are due to intramolecular changes in ICP8 conformation. The fact that disruption of ionic interactions in infected-cell lysates by high-salt treatment did not alter ICP8 antibody reactivity supports this hypothesis. However, additional analysis of ICP8 structure is needed to definitively define the nature of these antigenic changes. With the relatively recent progress in determining suitable crystallization conditions for ICP8 (55), it may be possible to produce high-resolution X-ray diffraction data to confirm and extend the observations reported here.
Relationship between ICP8 localization and changes in ICP8 antibody reactivity.
It is intriguing that we observed 39S- and LP-710-reactive ICP8 only at specific intranuclear sites, either prereplicative sites or replication compartments, but it is unclear whether localization causes or results from these changes in ICP8 conformation. The 39S form of ICP8 may be found only at viral replication structures because an ICP8 conformational change is needed for the targeting of ICP8 to the appropriate intranuclear sites. For example, because UL5, UL8, and UL52 are known to greatly enhance the localization of ICP8 to punctate intranuclear sites, it is possible that the 39S epitope is formed upon ICP8 complex formation with the helicase-primase proteins, resulting in the localization of the entire complex to potential replication sites. Alternatively, ICP8 may obtain the 39S conformation subsequent to arriving at the proper nuclear sites by interactions with previously deposited viral or cellular factors, possibly DNA.
Relationship between ICP8 DNA binding and changes in ICP8 antibody reactivity.
Another obvious issue to consider is whether and how these antigenic changes are related to ICP8 activity. Using sensitive biochemical assays, other studies have shown conformational changes in purified ICP8 upon binding to DNA (18, 70). Our genetic analysis showed an association of DNA binding with the presence of the 39S epitope. Thus, the conformational change we observed may be related to those observed biochemically. Because formation of the 39S epitope occurs in transfected cells, a complete viral genome or viral origin of replication is not needed. Therefore, one possible interpretation is that interaction with other viral proteins leads to a conformational change that forms the 39S epitope. If this were the case, it would raise the intriguing possibility that localization to prereplicative sites and/or interaction with other viral proteins leads to an activation of the DNA-binding activity of ICP8. Such localization-associated regulation of DNA binding could function to preferentially target ICP8 DNA binding to viral DNA. Arguing against this possibility, other researchers have used ICP8 expressed in the absence of other viral proteins and have observed apparently normal DNA-binding properties (27, 55). On the other hand, there are also many precedents of proteins that change conformation upon binding to DNA (reviewed in references 6, 25, 58, 73, and 86). Both of these possibilities are discussed in more detail below.
Conformational activation of ICP8 DNA binding: potential for redox regulation.
The existence of multiple oxidative isomers of ICP8 (42) raises the possibility that the conformation of ICP8 and its ability to bind DNA may be subject to redox regulation. Not only has redox regulation been proposed in the DNA-binding activity of several transcription factors, such as the bovine papillomavirus E2 protein (57), the cellular protooncogenes c-fos and c-jun (1, 60, 92), NF-κB (7, 78), Pax proteins (76), and the bacterial regulatory proteins OxyR and SoxR (reviewed in references 38, 63, and 74) but it also has been shown to regulate the DNA binding of the multifunction eukaryotic replication protein RPA (62, 94). In the case of OxyR, oxidation of Cys199 has been shown to induce a conformational change in the protein (74, 77). Likewise, modification of two cysteine residues modulates Pax-8 DNA-binding activity (10). More specifically, the zinc finger domains of RPA and a number of transcription factors have been shown to mediate the redox regulation of these proteins (reviewed in references 62 and 91).
Consistent with the importance of a cysteine residue(s) for ICP8 DNA binding, the alkylating agent NEM has been shown to inhibit ICP8 DNA binding in vitro (69). In addition, several aspects of ICP8 primary structure could predispose this protein to being redox sensitive. First, ICP8 is a zinc-binding protein with a putative C2H2 class zinc finger motif from residues 499 to 512 (27). Second, ICP8 contains the small basic sequence motif RCR (residues 245 to 247), which is homologous to the conserved redox factor 1 (Ref-1) responsive element present in many redox-sensitive transcription factors. Ubiquitous nuclear Ref-1 stimulates DNA binding of AP-1 (KCR), NF-κB (KICR), Myb (KQCR), ATF-2 (RCR), and other related factors (92). Finally, within the proposed ICP8 DNA-binding domain is the cysteine-containing sequence ACGPCP (residues 662 to 667), which is homologous to the active site of thioredoxin (WCGPCK) (reviewed in reference 31).
An interesting situation that arises if the antigenic forms of ICP8 described in this report do correspond with the different oxidative species of ICP8 previously observed is that both oxidative forms of ICP8 would exist in the nucleus. Hence, an exciting aspect of this potential redox regulation is that it would be a localization-specific oxidation of a protein within the nucleus, which would mean that a differential exists in the nuclear redox environment.
DNA binding could induce ICP8 conformational changes.
An alternative possibility is that ICP8 may undergo conformational alterations as a result of DNA binding. The reported quenching of ICP8 tryptophan fluorescence, changes in protease cleavage patterns, and changes in antibody quenching of fluorescein-5-maleimide-modified ICP8 in the presence of single-stranded DNA are consistent with a DNA-induced conformational change (18, 70). Changes in protein conformation upon DNA binding have been documented for a number of proteins (reviewed in reference 73), including the HSV polymerase (86), the VP16 interacting protein Oct-1 (58), and the cellular SSB RP-A (25). Similar to what has been found with the analogous cellular protein RP-A (6), such structural changes in ICP8 could serve to facilitate interactions with other proteins and thus promote replication complex assembly on DNA, or they could be the result of cooperative interactions between adjacent ICP8 molecules.
In conclusion, we have observed altered antibody reactivity of the HSV SSB ICP8 coincident with its assembly at viral replication structures, likely due to conformational changes in ICP8 before, during, or as a result of this process. These changes in ICP8 are associated with its interactions with the HSV helicase-primase protein complex and with the ability of ICP8 to bind to DNA. Further analysis of ICP8 structure to determine what specifically induces the 39S epitope should ultimately help clarify the relationship between ICP8 structure, DNA binding, and localization.
Acknowledgments
We thank Sandra Weller, Paul Johnson, and Don Coen for providing some of the mutant viruses used in this study. We also thank Paul Olivo, Lenore Pereira, and Kathy Shriver for supplying anti-ICP8 antibodies.
This research was supported by Public Health Service grant CA26345 from the National Cancer Institute. S.L.U. was supported by NIH training grant T21 AI07245.
REFERENCES
- 1.Abate, C., L. Patel, F. J. Rauscher III, and T. Curran. 1990. Redox regulation of fos and jun DNA-binding activity in vitro. Science 249:1157-1161. [DOI] [PubMed] [Google Scholar]
- 2.Arai, K., and A. Kornberg. 1981. Mechanism of dnaB protein action III. J. Biol. Chem. 256:5260-5266. [PubMed] [Google Scholar]
- 3.Arai, K., R. Low, J. Kobori, J. Shlomai, and A. Kornberg. 1981. Mechanism of dnaB protein action V. J. Biol. Chem. 256:5273-5280. [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. E. Struhl. 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 5.Bauer, C. E., S. Elsen, and T. H. Bird. 1999. Mechanisms for redox control of gene expression. Annu. Rev. Microbiol. 53:495-523. [DOI] [PubMed] [Google Scholar]
- 6.Blackwell, L. J., J. A. Borowiec, and I. A. Mastrangelo. 1996. Single-stranded-DNA binding alters human replication protein A structure and facilitates interaction with DNA-dependent protein kinase. Mol. Cell. Biol. 16:4798-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan, P., and L. A. O'Neill. 1995. Effects of oxidants and antioxidants on nuclear factor κB activation in three different cell lines: evidence against a universal hypothesis involving oxygen radicals. Biochim. Biophys. Acta 1260:167-175. [DOI] [PubMed] [Google Scholar]
- 8.Bush, M., D. R. Yager, M. Gao, K. Weisshart, A. I. Marcy, D. M. Coen, and D. M. Knipe. 1991. Correct intranuclear localization of herpes simplex virus DNA polymerase requires the viral ICP8 DNA-binding protein. J. Virol. 65:1082-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calder, J. M., E. C. Stow, and N. D. Stow. 1992. On the cellular localization of the components of the herpes simplex virus type 1 helicase-primase complex and the viral origin-binding protein. J. Gen. Virol. 73:531-538. [DOI] [PubMed] [Google Scholar]
- 10.Cao, X., F. Kambe, S. Ohmori, and H. Seo. 2002. Oxidoreductive modification of two cysteine residues in paired domain by Ref-1 regulates DNA-binding activity of Pax-8. Biochem. Biophys. Res. Commun. 297:288-293. [DOI] [PubMed] [Google Scholar]
- 11.Carmichael, E. P., and S. K. Weller. 1989. Herpes simplex virus type 1 DNA synthesis requires the product of the UL8 gene: isolation and characterization of an ICP6::lacZ insertion mutation. J. Virol. 63:591-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Challberg, M. D. 1986. A method for identifying the viral genes required for herpesvirus DNA replication. Proc. Natl. Acad. Sci. USA 83:9094-9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clairborne, A., H. Miller, D. Parsonage, and R. P. Ross. 1993. Protein-sulfenic acid stabilization and function in enzyme catalysis and gene regulation. FASEB J. 7:1484-1490. [DOI] [PubMed] [Google Scholar]
- 14.Clayton, R. F., A. Owsianka, J. Aitken, S. Graham, D. Bhella, and A. H. Patel. 2003. Analysis of antigenicity and topology of E2 glycoprotein present on recombinant hepatitis C virus-like particles. J. Virol. 76:7672-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curtin, K. D., and D. M. Knipe. 1993. Altered properties of the herpes simplex virus ICP8 DNA-binding protein in cells infected with ICP27 mutant viruses. Virology 196:1-14. [DOI] [PubMed] [Google Scholar]
- 16.de Bruyn Kops, A., and D. M. Knipe. 1988. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell 55:857-868. [DOI] [PubMed] [Google Scholar]
- 17.Dekker, J., P. N. Kanellopoulos, J. A. van Oosterhout, G. Stier, P. A. Tucker, and P. C. van der Vliet. 1998. ATP-independent DNA unwinding by the adenovirus single-stranded DNA binding protein requires a flexible DNA binding loop. J. Mol. Biol. 277:825-838. [DOI] [PubMed] [Google Scholar]
- 18.Dudas, K. C., S. K. Scouten, and W. T. Ruyechan. 2001. Conformational change in the herpes simplex single-strand binding protein induced by DNA. Biochem. Biophys. Res. Commun. 288:184-190. [DOI] [PubMed] [Google Scholar]
- 19.Frankel, A. D., and P. S. Kim. 1991. Modular structure of transcription factors: implication for gene regulation. Cell 65:717-719. [DOI] [PubMed] [Google Scholar]
- 20.Gannon, J. V., R. Greaves, R. Iggo, and D. P. Lane. 1990. Activating mutations in p53 produce a common conformational effect. A monoclonal antibody specific for the mutant form. EMBO J. 9:1595-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao, M., J. Bouchey, K. Curtin, and D. M. Knipe. 1988. Genetic identification of a portion of the herpes simplex virus ICP8 protein required for DNA-binding. Virology 163:319-329. [DOI] [PubMed] [Google Scholar]
- 21a.Gao, M., and D. M. Knipe. 1989. Genetic evidence for multiple nuclear functions of the herpes simplex virus ICP 8 DNA-binding protein. J. Virol. 63:5258-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21b.Gao, M., and D. M. Knipe. 1991. Potential role for herpes simplex virus ICP8 DNA replication protein in late gene expression. J. Virol. 65:2666-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao, M., and D. M. Knipe. 1992. Distal protein sequences can affect the function of a nuclear localization signal. Mol. Cell. Biol. 12:1330-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao, M., and D. M. Knipe. 1993. Intragenic complementation of herpes simplex virus ICP8 DNA-binding protein mutants. J. Virol. 67:876-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldstein, D. J., and S. K. Weller. 1988. An ICP6::lacZ insertional mutagen is used to demonstrate that the UL52 gene of herpes simplex virus type 1 is required for virus growth and DNA synthesis. J. Virol. 62:2970-2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomes, X. V., L. A. Henricksen, and M. S. Wold. 1996. Proteolytic mapping of human replication protein A: evidence for multiple structural domains and a conformational change upon interaction with single-stranded DNA. Biochemistry 35:5586-5595. [DOI] [PubMed] [Google Scholar]
- 26.Goodrich, L. D., P. A. Schaffer, D. I. Dorsky, C. S. Crumpacker, and D. S. Parris. 1990. Localization of the herpes simplex virus type 1 65-kilodalton DNA-binding protein and DNA polymerase in the presence and absence of viral DNA synthesis. J. Virol. 64:5738-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupte, S. S., J. W. Olson, and W. T. Ruyechan. 1991. The major herpes simplex virus type-1 DNA-binding protein is a zinc metalloprotein. J. Biol. Chem. 266:11413-11416. [PubMed] [Google Scholar]
- 28.Halazonetis, T. D., L. J. Davis, and A. N. Kandil. 1993. Wild-type p53 adopts a “mutant”-like conformation when bound to DNA. EMBO J. 12:1021-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halazonetis, T. D., and A. N. Kandil. 1993. Conformational shifts propagate from the oligomerization domain of p53 to its tetrameric DNA binding domain and restore DNA binding to select p53 mutants. EMBO J. 12:5057-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heilbronn, R., and H. zur Hausen. 1989. A subset of herpes simplex virus replication genes induces DNA amplification within the host cell genome. J. Virol. 63:3683-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmgren, A. 1985. Thioredoxin. Annu. Rev. Biochem. 54:237-271. [DOI] [PubMed] [Google Scholar]
- 32.Horowitz, A., K. M. Trybus, D. S. Bowman, and F. S. Fay. 1994. Antibodies probe for folded monomeric myosin in relaxed and contracted smooth muscle. J. Cell Biol. 126:1195-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hughes, R. G., Jr., and W. H. Munyon. 1975. Temperature-sensitive mutants of herpes simplex virus type 1 defective in lysis but not in transformation. J. Virol. 16:275-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hupp, T. R., D. W. Meek, C. A. Midgley, and P. D. Lane. 1992. Regulation of the specific DNA binding function of p53. Cell 71:875-886. [DOI] [PubMed] [Google Scholar]
- 35.Jezewska, M. J., and W. Bujalowski. 1996. Global conformational transitions in Escherichia coli primary replicative helicase DnaB induced by ATP, ADP, and single-stranded DNA binding. Multiple conformational states of the helicase hexamer. J. Biol. Chem. 271:4261-4265. [DOI] [PubMed] [Google Scholar]
- 36.Johnson, L. N., and M. O'Reilly. 1996. Control by phosphorylation. Curr. Opin. Struct. Biol. 6:762-769. [DOI] [PubMed] [Google Scholar]
- 37.Johnson, P. A., M. G. Best, T. Friedmann, and D. S. Parris. 1991. Isolation of a herpes simplex virus type 1 mutant deleted for the essential UL42 gene and characterization of its null phenotype. J. Virol. 65:700-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim, S. O., K. Merchant, R. Nudelman, W. F. J. Beyer, T. Keng, J. DeAngelo, A. Hausladen, and J. S. Stamler. 2002. OxyR: a molecular code for redox-related signaling. Cell 109:383-396. [DOI] [PubMed] [Google Scholar]
- 39.Knipe, D. M. 1989. The role of viral and cellular nuclear proteins in herpes simplex virus replication. Adv. Vir. Res. 37:85-123. [DOI] [PubMed] [Google Scholar]
- 40.Knipe, D. M., M. P. Quinlan, and A. E. Spang. 1982. Characterization of two conformational forms of the major DNA-binding protein encoded by herpes simplex virus 1. J. Virol. 44:736-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knipe, D. M., D. Senechek, S. A. Rice, and J. L. Smith. 1987. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J. Virol. 61:276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knipe, D. M., and A. E. Spang. 1982. Definition of a series of stages in the association of two herpesviral proteins with the cell nucleus. J. Virol. 43:314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee, C. K., and D. M. Knipe. 1985. An immunoassay for the study of DNA-binding activities of herpes simplex virus protein ICP8. J. Virol. 54:731-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, C. K., and D. M. Knipe. 1983. Thermolabile in vivo DNA-binding activity associated with a protein encoded by mutants of herpes simplex virus type 1. J. Virol. 46:909-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leinbach, S. S., J. M. Reno, L. F. Lee, A. F. Isbell, and J. A. Boezi. 1976. Mechanism of phosphonoacetate inhibition of herpesvirus-induced DNA polymerase. Biochemistry 15:425-430. [DOI] [PubMed] [Google Scholar]
- 45a.Leinbach, S. S., J. F. Capto, and T. K. Pickett. 1984. Deoxyribonucleoprotein complexes and DNA synthesis of herpes simplex virus type 1. Virology 137:287-296. [DOI] [PubMed] [Google Scholar]
- 46.Lin, K., P. K. Hwang, and R. J. Fletterick. 1997. Distinct phosphorylation signals converge at the catalytic center in glycogen phosphorylases. Structure 5:1511-1523. [DOI] [PubMed] [Google Scholar]
- 47.Lindsley, J. E., and J. C. Wang. 1993. Study of allosteric communication between promoters by immunotagging. Nature 361:749-750. [DOI] [PubMed] [Google Scholar]
- 48.Liptak, L., S. L. Uprichard, and D. M. Knipe. 1996. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J. Virol. 70:1759-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lowry, D. F., A. F. Roth, P. B. Rupert, F. W. Dahlquist, F. J. Moy, P. J. Domaille, and P. Matsumura. 1994. Signal transduction in chemotaxis. A propagating conformation change upon phosphorylation of CheY. J. Biol. Chem. 269:26358-26362. [PubMed] [Google Scholar]
- 50.Lukonis, C. J., J. Burkham, and S. K. Weller. 1997. Herpes simplex virus type 1 prereplicative sites are a heterogeneous population: only a subset are likely to be precursors to replication compartments. J. Virol. 71:4771-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lukonis, C. J., and S. K. Weller. 1996. Characterization of nuclear structures in cells infected with herpes simplex virus type 1 in the absence of viral DNA replication. J. Virol. 70:1751-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lukonis, C. J., and S. K. Weller. 1997. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J. Virol. 71:2390-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malik, A. K., R. Martinez, L. Muncy, E. P. Carmichael, and S. K. Weller. 1992. Genetic analysis of the herpes simplex virus type 1 UL9 gene: isolation of a LacZ insertion mutant and expression in eukaryotic cells. Virology 190:702-715. [DOI] [PubMed] [Google Scholar]
- 54.Mao, J. C. H., E. E. Robishaw, and L. R. Overby. 1975. Inhibition of DNA polymerase form herpes simplex virus-infected Wi-38 cells by phosphonoacetic acid. J. Virol. 15:1281-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mapelli, M., and P. A. Tucker. 1999. Crystallization and preliminary X-ray crystallographic studies on the herpes simplex virus 1 single-stranded DNA binding protein. J. Struct. Biol. 128:219-222. [DOI] [PubMed] [Google Scholar]
- 56.Marcy, A. I., D. R. Yager, and D. M. Coen. 1990. Isolation and characterization of herpes simplex virus mutants containing engineered mutations at the DNA polymerase locus. J. Virol. 64:2208-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McBride, A. A., R. D. Klausner, and P. M. Howley. 1992. Conserved cysteine residue in the DNA-binding domain of the papillomavirus type 1 E2 protein confers redox regulation of the DNA-binding activity in vitro. Proc. Natl. Acad. Sci. USA 89:7531-7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Misra, V., S. Walker, P. Yang, S. Hayes, and P. O'Hare. 1996. Conformational alteration of Oct-1 upon DNA binding dictates selectivity in the differential interactions with related transcriptional co-activators. Mol. Cell. Biol. 16:4404-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morris, G. E. 1989. Monoclonal antibody studies of creatine kinase. Biochem. J. 257:461-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okuno, H., A. Akahori, H. Sato, S. Xanthoudakis, T. Curran, and H. Iba. 1993. Escape from redox regulation enhances the transformation activity of Fos. Oncogene 8:695-701. [PubMed] [Google Scholar]
- 61.Olivo, P. D., N. J. Nelson, and M. D. Challberg. 1989. Herpes simplex virus type 1 gene products required for DNA replication: identification and overexpression. J. Virol. 63:196-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park, Y. W., J. Wilusz, and M. G. Katze. 1999. Regulation of eukaryotic protein synthesis: selective influenza viral mRNA translation is mediated by the cellular RNA-binding protein GRSF-1. Proc. Natl. Acad. Sci. USA 96:6694-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pomposiello, P. J., and B. Demple. 2001. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 19:109-114. [DOI] [PubMed] [Google Scholar]
- 64.Quinlan, M. P., L. B. Chen, and D. M. Knipe. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857-868. [DOI] [PubMed] [Google Scholar]
- 65.Rice, S. A., and D. M. Knipe. 1990. Genetic evidence for two distinct transactivation functions of the herpes simplex virus alpha protein ICP27. J. Virol. 64:1704-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roberts, S. G. E., and M. R. Green. 1994. Activator-induced conformational change in general transcription factor TFIIB. Nature 371:717-720. [DOI] [PubMed] [Google Scholar]
- 67.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2460. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 68.Rose, D. S., K. Shriver, D. S. Latchman, and N. B. LaThangue. 1986. A filamentous distribution for the herpes simplex virus type 2-encoded major DNA-binding protein. J. Gen. Virol. 67:1315-1325. [DOI] [PubMed] [Google Scholar]
- 69.Ruyechan, W. T. 1988. N-ethylmaleimide inhibition of the DNA-binding activity of the herpes simplex virus type 1 major DNA-binding protein. J. Virol. 62:810-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ruyechan, W. T., and J. W. Olson. 1992. Surface lysine and tyrosine residues are required for interaction of the major herpes simplex virus type 1 DNA-binding protein with single-stranded DNA. J. Virol. 66:6273-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sabapathy, K., M. Klemm, R. Jaenisch, and E. F. Wagner. 1997. Regulation of ES cell differentiation by functional and conformational modulation of p53. EMBO J. 16:6217-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Showalter, S. D., M. Zweig, and B. Hampar. 1981. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect. Immun. 34:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Spolar, R. S., and M. T. Record, Jr. 1994. Coupling of local folding to site-specific binding of proteins to DNA. Science 263:777-784. [DOI] [PubMed] [Google Scholar]
- 74.Storz, G., L. A. Tartaglia, and B. N. Ames. 1990. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science 248:189-194. [DOI] [PubMed] [Google Scholar]
- 75.Stow, N. D. 1992. Herpes simplex virus type 1 origin-dependent DNA replication in insect cells using recombinant baculoviruses. J. Gen. Virol. 73:313-321. [DOI] [PubMed] [Google Scholar]
- 76.Tell, G., A. Scaloni, L. Pellizzari, S. Formisano, C. Pucillo, and G. Damante. 1998. Redox potential controls the strucutre and DNA-binding of the paired domain. J. Biol. Chem. 273:25062-25072. [DOI] [PubMed] [Google Scholar]
- 77.Toledano, M. B., I. Kullik, F. Trinh, P. T. Baird, T. D. Schneider, and G. Storz. 1994. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism of differential promoter selection. Cell 78:897-909. [DOI] [PubMed] [Google Scholar]
- 78.Toledano, M. B., and W. J. Leonard. 1991. Modulation of the transcription factor NF-κB binding by oxidation-reduction in vitro. Proc. Natl. Acad. Sci. USA 88:4328-4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Toyoda, T., K. Masunaga, Y. Ohtsu, K. Hara, N. Hamada, T. Kashiwagi, and J. Iwahashi. 2000. Antibody-scanning and epitope-tagging methods; molecular mapping of proteins using antibodies. Curr. Protein Pept. Sci. 1:303-308. [DOI] [PubMed] [Google Scholar]
- 80.Triyatni, M., J. Vergalla, A. R. Davis, K. G. Hadlock, S. K. Foung, and T. J. Liang. 2002. Structural features of envelope proteins on hepatitis C virus-like particles as determined by anti-envelope monoclonal antibodies and CD81 binding. Virology 298:124-132. [DOI] [PubMed] [Google Scholar]
- 81.Trybus, K. M., and L. Henry. 1989. Monoclonal antibodies detect and stabilize conformational states of smooth muscle myosin. J. Cell Biol. 109:2879-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Uprichard, S. L., and D. M. Knipe. 1997. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology 229:113-125. [DOI] [PubMed] [Google Scholar]
- 83.Uprichard, S. L., and D. M. Knipe. 1996. Herpes simplex virus ICP27 mutant viruses exhibit reduced expression of specific DNA replication genes. J. Virol. 70:1969-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Veenstra, T. D., K. L. Johnson, A. J. Tomlinson, T. A. Craig, R. Kumar, and S. Naylor. 1998. Zinc-induced conformational changes in the DNA-binding domain of the vitamin D receptor determined by electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 9:8-14. [DOI] [PubMed] [Google Scholar]
- 85.Villemain, J. L., and D. P. Giedroc. 1993. Energetics of arginine-4 substitution mutants in the N-terminal cooperativity domain of T4 gene 32 protein. Biochemistry 32:11235-11246. [DOI] [PubMed] [Google Scholar]
- 86.Weisshart, K., A. A. Kuo, G. R. Painter, L. L. Wright, P. A. Furman, and D. M. Coen. 1993. Conformational changes induced in herpes simplex virus DNA polymerase upon DNA binding. Proc. Natl. Acad. Sci. USA 90:1028-1032. (Erratum, 90:4763.) [DOI] [PMC free article] [PubMed]
- 87.Weller, S. K. 1991. Genetic analysis of HSV-1 genes required for genome replication, p. 105-136. In E. K. Wagner (ed.), Herpesvirus transcription and its regulation. CRC Press, Boca Raton, Fla.
- 87a.Weller, S. K., K. J. Lee, D. Sabourin, and P. A. Schaffer. 1983. Genetic analysis of temperature-sensitive mutants which define the gene for the major herpes simplex virus type 1 DNA-binding protein. J. Virol. 45:354-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.White, A., X. Ding, J. C. van der Spek, J. R. Murphy, and D. Ringe. 1998. Structure of the metal-ion-activated diphtheria toxin repressor/tox operator complex. Nature 394:502-506. [DOI] [PubMed] [Google Scholar]
- 89.Winkelmann, D. A., S. Lowey, and J. L. Press. 1983. Monoclonal antibodies localize changes in myosin heavy chain isozymes during avian myogenesis. Cell 34:295-306. [DOI] [PubMed] [Google Scholar]
- 90.Wu, C. A., N. J. Nelson, D. J. McGeoch, and M. D. Challberg. 1988. Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J. Virol. 62:435-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu, X., N. H. Bishopric, D. J. Discher, B. J. Murphy, and K. A. Webster. 1996. Physical and functional sensitivity of zinc finger transcription factors to redox change. Mol. Cell. Biol. 16:1035-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xanthhoudakis, S., G. Miao, F. Wang, Y. C. Pan, and T. Curran. 1992. Redox activation of Fos-Jun DNA binding activity is mediated by a DNA repair enzyme. EMBO J. 11:3323-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yewdell, J. W., J. V. Gannon, and D. P. Lane. 1986. Monoclonal antibody analysis of p53 expression in normal and transformed cells. J. Virol. 59:444-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.You, J. S., M. Wang, and S. H. Lee. 2000. Functional characterization of zinc-finger motif in redox regulation of RPA-ssDNA interaction. Biochemistry 39:12953-12958. [DOI] [PubMed] [Google Scholar]
- 95.Zhu, L. A., and S. K. Weller. 1992. The UL5 gene of herpes simplex virus type 1: isolation of a lacZ insertion mutant and association of the UL5 gene product with other members of the helicase-primase complex. J. Virol. 66:458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]