Abstract
The antigenic diversity of human immunodeficiency virus type 1 (HIV-1) represents a significant challenge for vaccine design as well as the comprehensive assessment of HIV-1-specific immune responses in infected persons. In this study we assessed the impact of antigen variability on the characterization of HIV-1-specific T-cell responses by using an HIV-1 database to determine the sequence variability at each position in all expressed HIV-1 proteins and a comprehensive data set of CD8 T-cell responses to a reference strain of HIV-1 in infected persons. Gamma interferon Elispot analysis of HIV-1 clade B-specific T-cell responses to 504 overlapping peptides spanning the entire expressed HIV-1 genome derived from 57 infected subjects demonstrated that the average amino acid variability within a peptide (entropy) was inversely correlated to the measured frequency at which the peptide was recognized (P = 6 × 10−7). Subsequent studies in six persons to assess T-cell responses against p24 Gag, Tat, and Vpr peptides based on autologous virus sequences demonstrated that 29% (12 of 42) of targeted peptides were only detected with peptides representing the autologous virus strain compared to the HIV-1 clade B consensus sequence. The use of autologous peptides also allowed the detection of significantly stronger HIV-1-specific T-cell responses in the more variable regulatory and accessory HIV-1 proteins Tat and Vpr (P = 0.007). Taken together, these data indicate that accurate assessment of T-cell responses directed against the more variable regulatory and accessory HIV-1 proteins requires reagents based on autologous virus sequences. They also demonstrate that CD8 T-cell responses to the variable HIV-1 proteins are more common than previously reported.
An important first step in the development of an effective human immunodeficiency virus type 1 (HIV-1) vaccine is to understand the correlates of protective immunity in infected persons. An increasing body of evidence points to a central role of HIV-1-specific cytotoxic T lymphocytes (CTL) and T helper cells in controlling viral replication (4, 6, 9-11, 14, 23, 24, 28, 38, 41, 43). Recent advances in immunological methods, including Elispot assays and flow-based techniques, have allowed for the first time a more comprehensive assessment of the magnitude and breadth of T-cell responses directed against all expressed HIV-1 proteins in large numbers of infected individuals (3, 13, 34, 35, 50).
Initial data emerging from these studies demonstrate that HIV-1-specific T-cell responses are broadly directed against all HIV-1 proteins, with the strongest responses clustering in the more conserved regions of HIV-1 Gag, Pol, and Nef (3, 13, 34, 35). Despite the comprehensive assessment of the breadth and magnitude of virus-specific T-cell responses, the majority of these recent studies have failed to demonstrate a correlation between gamma interferon (IFN-γ) production of virus-specific T cells and viral load or CD4+ T-cell counts (3, 13, 34, 35), suggesting either a true lack of correlation or limitations in the currently used assays to identify the correlates of protective immunity in HIV-1 infection.
Studies dissecting the role of virus-specific CD8+ and CD4+ T-cell responses in HIV-1 infection are complicated by the HLA heterogeneity in many cohorts and the substantial genetic diversity within the viral population that results from high replication and mutation rates as well as from immunological selection pressure (7, 15, 19, 20, 25, 26, 31-33, 42, 46, 48, 49, 51). Viral strains from the same HIV-1 clade can differ by more than 5 to 25% of the amino acids, depending on the particular HIV-1 protein under consideration (49). This sequence variability has an important influence on the cellular immune responses generated in an infected individual, as each step in the recognition of T-cell epitopes and the generation of virus-specific T-cell responses has potential constraints imposed by sequence specificity.
Single amino acid changes within and flanking a CD8+ T-cell epitope impact the processing of the epitope by the intracytoplasmic immunoproteasome and peptidases in the endoplasmic reticulum (12, 17, 47). In addition, the binding of an 8- to 12-amino-acid epitope to the binding groove of HLA class I molecules and the recognition of the HLA-epitope complex by the T-cell receptor are highly specific (40, 44, 45). A number of studies have demonstrated that single amino acid substitutions within T-cell epitopes can reduce or abrogate binding to class I and/or recognition by CD8+ T cells (4, 6, 10, 15, 18, 20, 25, 26). Thus, the use of reagents based on sequences derived from laboratory prototype HIV-1 isolates or clade consensus sequences is likely to favor detection of responses in well-conserved areas of the genome, as T-cell responses directed against more variable regions may be missed due to sequence differences between the autologous virus sequence and the reagents used (51). The high degree of both intra- and interclade sequence variability of HIV-1 may therefore pose a major challenge for the accurate characterization of HIV-1-specific T-cell responses.
In this study we assessed the impact of viral sequence variability on the characterization of virus-specific T-cell responses against conserved and variable proteins of HIV-1. We demonstrate that overlapping peptides spanning relatively conserved regions of HIV-1 are significantly more frequently recognized by HIV-1-specific T cells than peptides spanning highly variable regions of the virus. Furthermore, the use of peptides based on the autologous virus allowed enhanced detection of T-cell responses directed against the variable regions of HIV-1 Tat and Vpr, indicating that these variable regions serve as targets for cellular immune responses. These data highlight the importance of assessing virus-specific immune responses directed against the autologous virus in HIV-1-infected individuals in order to characterize the actual breadth and magnitude of the cellular immune response.
MATERIALS AND METHODS
Subjects.
A data set of HIV-1-specific T-cell responses that had been generated by comprehensive screening of blood samples from 57 infected individuals at different stages of HIV-1 infection (3) was used to correlate peptide entropy and recognition by T cells. In addition, HIV-1-specific T-cell responses directed against peptides spanning HIV-1 clade B consensus sequences or spanning the autologous virus sequences of HIV-1 p24 Gag, Vpr, and Tat were compared in six individuals with symptomatic acute HIV-1 infection. The mean viral loads at the time of diagnosis of acute HIV-1 infection in these six individuals were 7.95 × 106 copies of HIV-1 RNA per ml of plasma (range, 0.25 × 106 to 30.7 × 106 copies of HIV-1 RNA), and the mean CD4+ T-cell count was 501 cells per μl (range, 365 to 667 cells). Five of the six individuals were treated at the time of acute HIV-1 infection with highly active antiretroviral therapy. Study subjects were recruited from the Massachusetts General Hospital, the Fenway Community Health Center and the Lemuel Shattuck Hospital in Boston. The study was approved by the institutional review boards, and all subjects gave written informed consent.
Sequencing of autologous virus.
Viral DNA was isolated from peripheral blood mononuclear cells (PBMC) (5 × 106 cells) as described previously (6). We determined the proviral template copy number by serial dilution of PBMC DNA in nested PCR amplifications. All PCR amplifications were done with procedural safeguards, including aliquoting of all reagents and physical separation of sample processing and post-PCR handling steps. Replicate control amplifications with no template were included in every PCR experiment to test for carryover contamination. Amplifications with 10 genome equivalents of 8E5 cell DNA were also performed as an internal control to monitor the efficiency of the PCR.
To reduce template resampling, we pooled multiple first-round PCR products and reamplified the pool in a second round for derivation and sequence analysis. The first-round products were used to amplify the desired regions. PCR cycling conditions were generally 94°C for 2 min, 35 to 50 cycles of 30 s at 94°C, 30 s at 56°C, and 2 min at 72°C, and a final extension of 68°C for 20 min. Purified PCR fragments were then either bulk sequenced or cloned (TOPO TA; Invitrogen, Carlsbad, Calif.) with plasmid DNA isolated by miniprep (QiaPrep Turbo Miniprep), as described elsewhere (6, 46). At least 10 clones were sequenced per study subject. All PCR products were sequenced bidirectionally on an ABI 3100 Prism automated sequencer. Sequencher (Gene Codes Corp., Ann Arbor, Mich.) and MacVector 4.1 (Oxford Molecular) software programs were used to edit and align sequences. Sequences were subjected to Blast and phylogenetic analysis to test for the presence of carryover contaminants from other samples in the laboratory (27, 29).
Synthetic HIV-1 peptides.
Peptides were synthesized on an automated peptide synthesizer (MBS 396; Advanced Chemtech, Louisville, Ky.) with fluorenylmethoxycarbonyl chemistry. Overlapping peptides (17- to 19-mers, overlapping by 10 amino acids) spanning the expressed 2001 HIV-1 clade B consensus sequence (http://hiv-web.lanl.gov/immunology/index.html) of p24 Gag, Vpr, and Tat as well as overlapping peptides corresponding to the autologous virus sequence of these HIV-1 proteins in the six individuals studied were synthesized. The overlapping autologous sequence peptides were of the same length and overlapped and spanned the same amino acids as the corresponding consensus sequence overlapping peptides.
Calculation of peptide variability.
A Shannon entropy score was calculated as described previously (51) for each position of the 504 overlapping peptides after alignment to the 2002 alignment of sequences published in the Los Alamos National Database (http://hiv-web.lanl.gov), and an average entropy score for all positions in each overlapping peptide was determined. Entropy at each amino acid position was calculated as ΣPaalogPaa, where Paa is the proportion of each amino acid in the respective position (51). Positions where the majority of the sequences had gaps were excluded from consideration. When only a minority of sequences had gaps, however, the position was included and the gaps were treated as separate symbols.
Elispot assay.
Frozen PBMC were plated on 96-well polyvinylidene difluoride-backed plates (MAIP S45; Millipore, Bedford, Mass.) that had been coated with 100 μl of anti-IFN-γ monoclonal antibody 1-D1k (0.5 μg/ml; Mabtech, Stockholm, Sweden) overnight at 4°C. Overlapping peptides spanning the expressed HIV-1 clade B consensus sequence of HIV-1 p24 Gag, Vpr, and Tat or overlapping peptides spanning the autologous virus sequence of these HIV-1 proteins were added directly to the wells at a final concentration of 10−5 M along with 100,000 cells. The plates were incubated at 37°C in 5% CO2 overnight (14 to 16 h) and then processed as described previously (3, 8). IFN-γ-producing cells were counted by direct visualization and are expressed as spot-forming cells (SFC) per 106 cells. The number of specific IFN-γ-secreting T cells was calculated by subtracting the number of spots counted in the negative control wells. For negative controls, 100,000 PBMC were incubated with R10 alone, without adding peptides. Responses of >50 SFC per 106 input cells and higher than 3 times the mean background activity were considered positive. The positive control consisted of incubation of 100,000 PBMC with phytohemagglutinin.
Flow cytometric detection of antigen-induced intracellular IFN-γ.
Intracellular cytokine staining assays were performed as described elsewhere with minor modifications (13, 22, 39). HIV-1 p24 Gag, Tat, and Vpr overlapping peptides were combined in two pools of overlapping peptides per protein (p24 Gag: pool 1, peptides 1 to 12; pool 2, peptides 13 to 25; Tat: pool 1, peptides 1 to 6; pool 2, peptides 7 to 12; Vpr: pool 1, peptides 1 to 5; pool 2, peptides 6 to 11). Briefly, 0.5 × 106 to 1.0 × 106 PBMC were incubated with pools of overlapping peptides (final concentration of each peptide in the assay was 2 μM) and 1 μg/ml each of the monoclonal antibodies anti-CD28 and anti-CD49d (Becton Dickinson) at 37°C and 5% CO2 for 1 h before the addition of 10 μg of brefeldin A (Sigma, St. Louis, Mo.) per ml. Following a further 5-h incubation, the cells were placed at 4°C overnight. After surface marker labeling with anti-CD8 and anti-CD4 (Becton Dickinson), cells were fixed and permeabilized with a Caltag fixation/permeabilization kit (Caltag, Burlingham, Calif.), and anti-IFN-γ monoclonal antibody (Becton Dickinson) was added. Cells were analyzed on a FACSCalibur flow cytometer (Becton Dickinson Immunocytometry Systems, San Jose, Calif.). Control conditions were established by the use of autologous PBMC which had not been stimulated with peptide but otherwise had been treated identically.
Statistical analysis.
Statistical analysis and graphic presentation were done with SigmaPlot 5.0 (SPSS Inc., Chicago, Ill.). Results are given as mean ± standard deviation or median with range. Statistical analyses of significance (P values) were done with a nonparametric Pearson's rank statistic and Wilcoxon rank sum tests with the GNU free software R project for statistical computing, version 1.4.1, 2002 (www.R-project.org).
RESULTS
Average entropy and fraction of conserved amino acids within a peptide correlated with frequency of recognition by HIV-1-specific T cells.
The sequence diversity of HIV-1 poses a challenge for vaccine design and may influence the comprehensive assessment of HIV-1-specific immune responses in infected persons, as the sequence of the infecting strain is rarely used to assess immune responses. To estimate the impact of sequence variability of HIV-1 on the characterization of virus-specific T-cell responses, we compared reported sequence variability with the frequency of responses against a set of overlapping peptides spanning all HIV-1 proteins. These frequencies were derived from a comprehensive characterization of HIV-1-specific T-cell responses against a set of 504 overlapping peptides spanning the entire expressed clade B sequence with PBMC from 57 infected subjects in an IFN-γ Elispot assay (3). T-cell responses were broadly directed against a median of 14 epitopic regions, ranging from 2 to 42 targeted regions per study subject. The median total magnitude of HIV-1-specific T-cell responses in the 57 study subjects was 4,245 SFC/106 PBMC, ranging from 280 to 25,860 SFC/106 PBMC (3). Of the 504 overlapping peptides tested, 315 peptides (63%) were targeted by HIV-1-specific T cells of at least 1 of the 57 study subjects (range, 1 to 27 study subjects; median, 2).
For each of the 504 overlapping peptides, an average entropy score was calculated based on the 2002 alignment of HIV-1 clade B sequences of the Los Alamos Database. Entropy in each amino acid position was calculated as described in Materials and Methods. The median entropy score per peptide ranged from 0 in a peptide spanning a highly conserved region within reverse transcriptase to 1.475 in a peptide spanning a highly variable region of gp120 Env, with a median entropy score of 0.181 for all 504 peptides. Comparison of entropy values to T-cell responses detected against each peptide demonstrated that the average entropy correlated negatively with the frequency of recognition (Spearman's rho = −0.22, P = 6 × 10−7). The set of completely nonreactive peptides had higher entropy values than did reactive peptides (Wilcoxon rank sum P = 5.7 × 10−4), and the negative correlation between entropy and reaction frequency persisted even when the peptides that were not targeted in at least one individual were excluded from the analysis (rho = −0.21, P = 2 × 10−4).
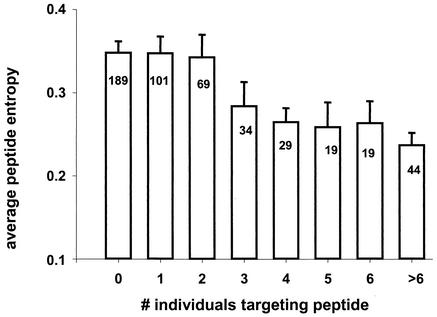
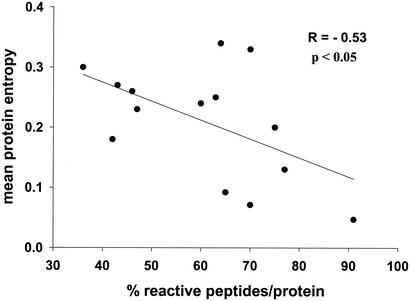
The relationship between the number of people that targeted a peptide and the entropy of the peptide is illustrated in Fig. 1; the mean entropy score gradually declined for peptides that were more frequently targeted. In a similar analysis, the fraction of invariant amino acid positions within a tested peptide increased with increased recognition of this peptide (data not shown; Spearman's rho = 0.22, P = 1.1 × 10−6). Overall, both peptide entropy and the fraction of invariant amino acid positions within a peptide tended to level off for peptides recognized by more than 5% of tested individuals (three or more individuals). At the protein level, the percentage of peptides within that protein that were targeted by at least one individual correlated inversely with the protein entropy (Fig. 2) and correlated positively with the fraction of invariant amino acid positions within the protein (data not shown). Taken together, these data suggest that overlapping peptides spanning more variable regions of HIV-1 are less frequently recognized by HIV-1-specific T-cell responses.
FIG. 1.
Influence of average peptide entropy on frequency of recognition. The average entropy score of an overlapping peptide was calculated based on the alignment of HIV-1 clade B sequences published in the Los Alamos National Database. The average entropy score is given for peptides targeted by none of the 57 tested individuals as well as for peptides targeted by one, two, three, four, five, six, and more than six individuals. The number of peptides targeted in each group is shown in the columns. A total of 504 overlapping peptides were tested.
FIG. 2.
Correlation between percentage of reactive peptides per protein and protein entropy. The mean protein entropy was calculated for p17 Gag, p24 Gag, p15 Gag, reverse transcriptase, integrase, protease, g41 Env, gp120 Env, Vpr, Vif, Vpu, Rev, Tat, and Nef. Mean protein entropy scores were correlated to the percentage of overlapping peptides per proteins that were targeted by HIV-1-specific T cells in the 57 study subjects. The greatest percentage of reactive peptides per protein was observed in p24 Gag; the least was observed in Vpu.
Detection of stronger HIV-1-specific CD8+ T-cell responses with the autologous virus sequence.
The above data suggest that immune responses to more variable regions of HIV-1 may be underestimated by current methods of detection that rely on single reference strains of HIV-1. We hypothesized that T-cell responses directed against peptides derived from autologous virus sequences would be stronger and more broadly directed than T-cell responses directed against peptides based on HIV-1 reference strain sequences or clade consensus sequences.
In order to assess HIV-1-specific T-cell responses directed against the autologous virus sequence, we determined autologous virus sequences encoding HIV-1 p24 Gag, Vpr, and Tat in six individuals within 2 weeks of presentation with acute HIV-1 infection. These three proteins were chosen for the present studies because previous data (51) have shown high sequence conservation (low average entropy) in HIV-1 p24 Gag, intermediate entropy in Vpr, and high average entropy in Tat, and they represented the spectrum of variability found in HIV-1. Persons with primary HIV-1 infection were chosen because virus is more homogeneous during acute infection (7), and isolates from later time points may already carry escape mutations that would hinder detection of early CTL responses (4, 37).
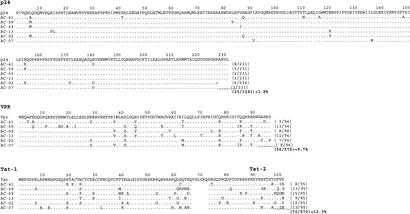
Alignment of the autologous virus sequences in these six study subjects was in line with these previous observations. Within HIV-1 p24 Gag, an average of 1.8 amino acids per 100 (1.8%) differed from the HIV-1 clade B consensus sequence (range, 0.87% to 3.5%), while an average of 9.7% (range, 7.3% to 13.5%; P < 0.001 compared to p24 Gag) and 12.3% (8.4% to 15.8%; P < 0.001 compared to p24 Gag) of amino acids differed from the HIV-1 clade B consensus sequence for Vpr and Tat, respectively (Fig. 3). The differences in sequence variability between HIV-1 Vpr and Tat did not reach statistical significance (P = 0.12).
FIG. 3.
Alignments of autologous virus sequences of p24 Gag, Vpr, and Tat from the study subjects to HIV-1 clade B 2001 consensus sequence. The amino acid sequence of the autologous virus determined in acute HIV-1 infection is shown for the six study subjects and aligned to the HIV-1 clade B consensus sequence of p24 Gag, Vpr, and Tat. The number of amino acids per protein that differed from the consensus sequence is given at the end of each sequence, and the average percent amino acid differences between autologous sequences and the clade B consensus sequence was calculated.
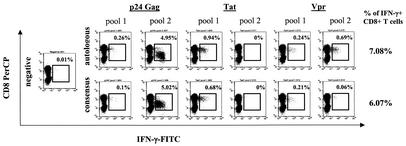
In order to generate overlapping peptides spanning the autologous virus sequence, an average of 26% (range, 8% to 52%) of the peptides spanning HIV-1 p24 Gag, 61% (range, 27% to 91%) of the peptides spanning HIV-1 Vpr, and 68% (range, 17% to 100%) of the peptides spanning HIV-1 Tat were different from the consensus sequence and had to be resynthesized. HIV-1-specific T-cell responses directed against pools of peptides spanning the consensus sequence of HIV-1 p24 Gag, Vpr, and Tat were compared to T-cell responses directed against pools containing the autologous variants of peptides spanning these proteins by intracellular IFN-γ staining analysis. Data are shown for individual AC-09 (Fig. 4), in whom 7.08% of CD8+ T cells produced IFN-γ after activation with pools of overlapping peptides spanning autologous sequences of HIV-1 p24 Gag, Vpr, and Tat, compared to 6.07% of CD8+ T cells activated by consensus sequence peptides.
FIG. 4.
Quantification of HIV-1-specific CD8+ T-cell responses directed against overlapping peptide pools derived from clade B consensus and autologous virus sequences with intracellular IFN-γ staining. The percentage of CD8+ T cells reactive to pools of overlapping peptides spanning either the clade B consensus sequence (upper row) or the autologous virus sequence (lower row) of p24 Gag (pools 1 and 2), Tat (pools 1 and 2), or Vpr (pools 1 and 2) was quantified by intracellular IFN-γ staining. The negative control with no peptides for stimulation but otherwise treated identically is shown. The magnitude of peptide-specific IFN-γ production by CD8+ T cells following stimulation is given after subtraction of background activity. CD8+ T-cell responses following stimulation with pools of peptides based on autologous sequences were higher (7.08%) than responses to consensus sequence peptides (6.07%).
HIV-1-specific CD8+ T-cell responses in the additional five study subjects were of lower magnitude for both autologous and consensus peptides compared to those of subject AC-09 but still showed stronger responses against peptides spanning autologous virus sequences compared to consensus sequences (Table 1). However, these differences did not reach statistical significance in the six individuals studied. No significant HIV-1-specific CD4+ T-cell responses were detectable in the six study subjects by intracellular IFN-γ staining on frozen samples (Fig. 4 and data not shown), and no peptide-specific IFN-γ production above background activity was observed in five HIV-1-negative control subjects with the same peptide pools. Taken together, these data indicate that HIV-1-specific CD8+ T-cell responses directed against the autologous virus may be underestimated with peptides based on HIV-1 clade B consensus sequences.
TABLE 1.
Percentage of CD8+ T cells producing IFN-γ in response to stimulation with pools of overlapping peptides spanning autologous or consensus sequences of p24 Gag, Vpr, and Tat using ICS
| Subject | % of CD8+ T cells producing IFN-γ
|
|||||
|---|---|---|---|---|---|---|
| p24 Gag
|
Vpr
|
Tat
|
||||
| Autologous | Consensus | Autologous | Consensus | Autologous | Consensus | |
| AC-09 | 5.21 | 5.12 | 0.93 | 0.27 | 0.94 | 0.68 |
| AC-41 | 0.36 | 0.32 | 0.42 | 0.31 | 0.19 | 0.23 |
| AC-13 | 0.16 | 0.04 | 0.06 | 0.05 | 0.02 | 0 |
| AC-59 | 0.03 | 0.02 | 0.01 | 0.01 | 0.11 | 0.05 |
| AC-02 | 0.03 | 0.01 | 0 | 0 | 0 | 0 |
| AC-07 | 0 | 0.01 | 0.01 | 0 | 0.09 | 0 |
| Mean ± SD | 0.97 ± 2.08 | 0.92 ± 2.06 | 0.24 ± 0.38 | 0.11 ± 0.14 | 0.23 ± 0.36 | 0.16 ± 0.27 |
Overlapping peptides spanning the autologous virus sequence allow the detection of HIV-1-specific T-cell responses missed by the use of peptides spanning HIV-1 consensus sequences.
We subsequently characterized the HIV-1-specific T-cell responses against HIV-1 p24 Gag, Vpr, and Tat at the single-peptide level in order to determine whether the above-described underestimation of responses was due to stronger responses directed against peptides spanning the autologous sequences, to the detection of additional responses with autologous sequences, or to a combination of both. In Fig. 5, the magnitude and breadth of HIV-1 p24 Gag-, Vpr-, and Tat-specific T-cell responses assessed by peptides spanning either the consensus sequence or the autologous sequence are compared.
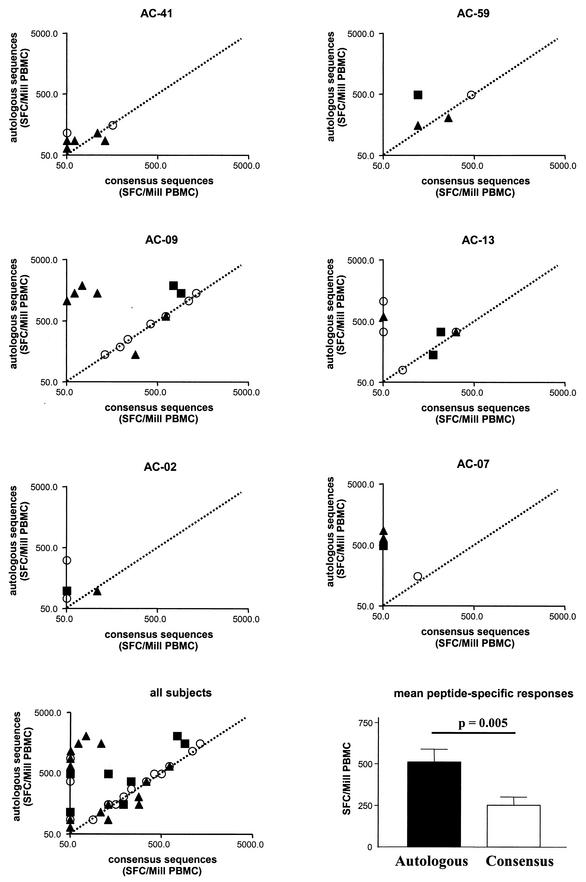
FIG.5.
Comparison of magnitude and breadth of HIV-1-specific T-cell responses determined with peptides spanning autologous virus sequences or HIV-1 clade B consensus sequences. For each of the six study subjects (top six panels) the magnitude of T-cell responses, given as spot-forming cells per million input PBMC (SFC/Mill PBMC), is shown for responses detected with overlapping peptides spanning the autologous virus (y axis) or the HIV-1 clade B consensus sequences (x axis). The dotted line indicates values equal for both conditions. In the bottom left panel, responses for all six individuals are combined. T-cell responses directed against HIV-1 p24 Gag are shown by open circles, responses directed against HIV-1 Vpr by black triangles, and responses directed HIV-1 Tat by black squares. In the lower right panel, the mean peptide-specific T-cell responses are compared between peptides spanning the autologous virus (black bar) and the HIV-1 clade B consensus sequences (white bar). Statistical significance was calculated with a two-tailed t test.
In all six individuals, stronger peptide-specific T-cell responses were observed with the peptides spanning the autologous sequences, with significantly higher mean peptide-specific T-cell responses directed against autologous sequence peptides (P = 0.005). In addition, a total of 12 of 42 (29%) peptide-specific responses in these six individuals were only detected with peptides spanning the autologous sequence, while all responses detected by consensus sequence peptides were also detected by the use of autologous sequence peptides. The underestimation of the magnitude of HIV-1-specific T-cell responses with consensus sequence peptides was most pronounced for the more variable proteins Vpr and Tat (mean magnitude of combined Vpr- and Tat-specific responses ± standard deviation: consensus peptides, 179 ± 221 SFC/106 PBMC; autologous peptides, 514 ± 522 SFC/106 PBMC, P = 0.007), while differences between the two peptide sets did not reach statistical significance for the more conserved protein p24 Gag (consensus peptides, 328 ± 396 SFC/106 PBMC; autologous peptides, 464 ± 443 SFC/106 PBMC, P = 0.34).
Remarkably, T-cell responses directed against autologous Tat and Vpr (514 ± 522 SFC/106 PBMC) were of equal or even higher magnitude than responses directed against autologous p24 Gag (464 ± 443 SFC/106 PBMC, P = 0.7), while mean p24 Gag-specific T-cell responses against consensus sequence peptides were higher than mean responses against Vpr and Tat (p24 Gag, 328 ± 396 SFC/106 PBMC; Vpr and Tat, 179 ± 221 SFC/106 PBMC; P = 0.16). Finally, a total of 4 out of 18 (22%) p24 Gag-specific T-cell responses were missed in the six study subjects with consensus sequence peptides, while 8 of 24 (33%) Vpr- and Tat-specific T-cell responses were missed. A number of these responses solely detected by autologous virus peptides represented immunodominant responses in the study subjects (Fig. 5).
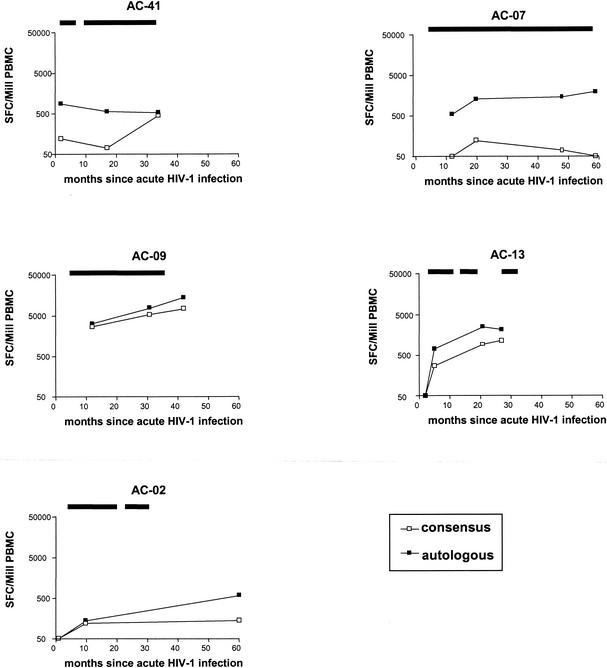
Longitudinal studies comparing the T-cell recognition of autologous and consensus sequence based peptides were performed in five of the six subjects. At all time points tested over a study period of up to 5 years, HIV-1-specific T-cell responses directed against the autologous virus were always stronger than responses directed against the consensus sequence peptides (Fig. 6). Taken together, these data demonstrate a significant underestimation of the magnitude and breadth of HIV-1-specific T-cell responses against the more variable regulatory and accessory HIV-1 proteins Vpr and Tat when peptides based on HIV-1 clade B consensus sequences are used.
FIG. 6.
Longitudinal assessment of HIV-1 p24 Gag-, Vpr-, and Tat-specific T-cell responses directed against peptides spanning autologous virus or clade B consensus sequences. The magnitude of HIV-1-specific T-cell responses directed against overlapping peptides spanning autologous virus or the HIV-1 clade B consensus sequences was determined longitudinally in five of the six study subjects. Magnitude of responses is given as spot-forming cells per million input PBMC (SFC/Mill PBMC). Total T-cell responses against peptides spanning autologous HIV-1 p24 Gag, Vpr, and Tat are shown as black symbols, while total responses against the corresponding clade B consensus sequence peptides are shown as open symbols. Periods of antiretroviral treatment are indicated by the black bars above each panel.
DISCUSSION
Recent advances in immunological methods for the first time have allowed more comprehensive assessment of the magnitude and breadth of HIV-1-specific T-cell responses in large numbers of infected individuals with the Elispot assays and flow-based techniques (3, 13, 34, 35, 50). A limitation of these studies is that the sets of overlapping peptides used for the assessment of responses are based on sequences of prototype laboratory HIV-1 isolates or clade consensus sequences. Here we show a negative correlation between the variability of a tested overlapping peptide and the frequency of recognition of this peptide with a comprehensive data set of HIV-1-specific T-cell responses in 57 HIV-1-infected individuals. These data suggest that peptides based on HIV-1 reference strains underestimate responses directed against the virus in more variable regions of the genome.
We subsequently tested this hypothesis in six individuals identified during acute HIV-1 infection by comparing virus-specific T-cell responses directed against overlapping peptides based on the HIV-1 clade B consensus sequence and autologous virus sequences of HIV-1 p24 Gag, Vpr, and Tat. The data demonstrate that HIV-1-specific T-cell responses directed against the autologous virus are significantly underestimated with peptides based on HIV-1 consensus sequences. This underestimation of T-cell responses was most pronounced within proteins of relatively high sequence variability (Vpr and Tat) and corresponded to some of the strongest peptide-specific responses detected.
The HIV-1 epidemic is characterized by high intraclade sequence diversity even within a defined geographic region, with regional epidemics clearly dominated by a single clade (49). This sequence diversity poses a significant challenge for both HIV-1 vaccine design and the comprehensive assessment of virus-specific immune responses in an infected subject, as the sequence of the infecting virus is rarely known. We have recently shown that HIV-1-specific optimal CD8+ T-cell epitopes are preferentially located within regions of low sequence diversity (51). Here we extend these initial studies with a comprehensive data set of HIV-1-specific T-cell responses generated in 57 HIV-1-infected individuals at different stages of disease and demonstrate that overlapping peptides spanning regions of low sequence variability (entropy) are recognized significantly more frequently than peptides spanning areas of high viral diversity.
Our data are in line with a number of recent comprehensive studies of HIV-1-specific T-cell responses with HIV-1 reference strain or clade consensus sequences that demonstrated that the relatively conserved HIV-1 regions of Gag, Pol, and Nef are frequently targeted by virus-specific T cells, while the variable proteins Vpu, Tat, Rev, and Env are less frequently targeted (1-3, 5, 13, 34, 35, 50). These data suggest that reagents based on HIV-1 reference strains or consensus sequences may have a limited potential to detect HIV-1-specific T-cell responses directed against more variable regions of the HIV-1 genome, although the consensus sequence may have more cross-reactive potential than single reference strains used in a heterologous setting (19, 49). Therefore, it is possible that the inability of recent studies to detect a negative association between HIV-1-specific T-cell responses and viral load or CD4 T-cell counts could be due to the use of reagents derived from laboratory prototype or clade consensus sequences, resulting in the lack of detection of responses directed against variable regions of the virus.
In order to further explore the basis for potential differences between T-cell responses directed against the autologous virus and responses detected by the use of HIV-1 clade B consensus sequences, we determined the autologous sequence of HIV-1 p24 Gag, Vpr, and Tat in six individuals with acute HIV-1 infection within 2 weeks of presentation with clinical symptoms. Individuals with acute HIV-1 infection were chosen for this study because viral sequences from later time points in an infected individual may have already escaped from key early CTL responses. In line with previous reports on HIV-1 sequence variability, the sequences of p24 Gag were highly conserved among the different individuals (and closely related to the HIV-1 p24 Gag consensus sequence), while HIV-1 Vpr and Tat showed significantly higher levels of sequence diversity. About 1 in 10 amino acids within Vpr and Tat differed between the autologous virus sequence and the HIV-1 clade B consensus sequence.
Since the length of an optimal CTL epitope ranges from 8 to 12 amino acids (16), every potential epitope within Vpr and Tat had a significant probability of containing at least one amino acid substitution. The presentation of a CD8+ T-cell epitope on major histocompatibility complex class I alleles and the recognition of the major histocompatibility complex-peptide complex by the T-cell receptor is a highly specific process, and single amino acid changes in an epitope can abolish presentation or recognition (4, 6, 10, 15, 18, 21, 37). Hence, these data highlight the potential limitations of peptides spanning HIV-1 clade consensus sequences to detect T-cell responses within these highly variable regions.
The comparison of the magnitude and breadth of HIV-1-specific T-cell responses directed against p24 Gag, Vpr, and Tat with peptides spanning autologous and consensus sequences confirmed these anticipated limitations of reagents based on consensus sequences to detect responses within variable regions. Twelve of 42 peptide-specific T-cell responses detected by the use of autologous virus sequences were missed by the sole use of consensus sequence peptides, and two-thirds of these peptides were located within Vpr and Tat. The fine mapping of the optimal CD8+ T-cell epitopes targeted by these T-cell responses is currently under way to determine the exact locations of the epitopes within these HIV-1 proteins. In addition, mean T-cell responses to peptides located within HIV-1 Vpr and Tat were of significantly higher magnitude when assessed with autologous virus sequences compared to consensus sequence peptides. In contrast, differences for p24 Gag did not reach statistical significance.
The detection of stronger and broader HIV-1-specific T-cell responses directed against the autologous peptide sequence determined during acute infection persisted over a study period of up to 5 years in the five individuals for whom longitudinal specimen were available. These findings are in contrast to recent data describing stronger “type-specific” CD8+ T-cell responses directed against HIVIIIB than directed against the autologous virus in progressive disease (30). These differences may be due to differences in the methods used, as overlapping synthetic peptides spanning the corresponding HIV-1 proteins were used in the present study, versus CD4+ T cells infected with virus in the study of Lee et al. (30). In addition, in our study autologous virus sequences were derived from time points during acute HIV-1 infection, whereas in the study by Lee et al., sequences were derived from time points during chronic progressive infection, when CTL escape might already be reflected in the autologous virus (4, 6, 15, 36, 37).
Taken together, our data demonstrate that the use of autologous peptides allows the detection of significantly stronger as well as broader T-cell responses directed against more variable proteins of the HIV-1 genome that would otherwise have been missed. We conclude that responses against the more variable regulatory and accessory HIV-1 proteins appear to be more common than previously reported. These data also demonstrate that the role of T-cell responses directed against the more variable regions of HIV-1 in the control of viral replication and their impact on viral escape from CTL-mediated immune pressure can best be assessed with reagents based on autologous sequences.
Acknowledgments
Marcus Altfeld and Marylyn Addo contributed equally to this work.
This study was supported by the Doris Duke Charitable Foundation (M.A., E.S.R., and B.D.W.), the National Institutes of Health (R01 AI50429, R37 AI128568, R01 AI30914, R01 AI44656, R01 AI40873, U01 AI41535, and U01 AI41531), the German Research Council (DFG) (M.M.A.), the American Foundation for AIDS Research (M.M.A.), Concerned Parents for AIDS Research (M.M.A.), the Partners/Fenway/Shattuck Center for AIDS Research (X.G.Y.), the University of Washington Center for AIDS Research (R.S. and J.I.M.), and the Howard Hughes Medical Institute (B.D.W.). M.A. is a recipient of a Doris Duke Clinical Scientist Development Award, and B.D.W. is a recipient of a Doris Duke Distinguished Clinical Scientist Award.
We thank Hong Zhao for help with sequencing.
REFERENCES
- 1.Addo, M. M., M. Altfeld, A. Rathod, M. Yu, X. G. Yu, P. J. Goulder, E. S. Rosenberg, and B. D. Walker. 2002. HIV-1 Vpu represents a minor target for cytotoxic T lymphocytes in HIV-1-infection. AIDS 16:1071-1073. [DOI] [PubMed] [Google Scholar]
- 2.Addo, M. M., M. Altfeld, E. S. Rosenberg, R. L. Eldridge, M. N. Philips, K. Habeeb, A. Khatri, C. Brander, G. K. Robbins, G. P. Mazzara, P. J. Goulder, and B. D. Walker. 2001. The HIV-1 regulatory proteins Tat and Rev are frequently targeted by cytotoxic T lymphocytes derived from HIV-1-infected individuals. Proc. Natl. Acad. Sci. USA 98:1781-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses but no correlation to viral load. J. Virol. 77:2081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 5.Altfeld, M., M. M. Addo, R. L. Eldridge, X. G. Yu, S. Thomas, A. Khatri, D. Strick, M. N. Phillips, G. B. Cohen, S. A. Islam, S. A. Kalams, C. Brander, P. J. R. Goulder, and B. D. Walker. 2001. Vpr is preferentially targeted by cytotoxic T lymphocytes during HIV-1 infection. J. Immunol. 167:2743. [DOI] [PubMed] [Google Scholar]
- 6.Altfeld, M., T. M. Allen, X. G. Yu, M. N. Johnston, D. Agrawal, B. T. Korber, D. C. Montefiori, D. H. O'Connor, B. T. Davis, P. K. Lee, E. L. Maier, J. Harlow, P. J. Goulder, C. Brander, E. S. Rosenberg, and B. D. Walker. 2002. HIV-1 superinfection despite broad CD8(+) T-cell responses containing replication of the primary virus. Nature 420:434-439. [DOI] [PubMed] [Google Scholar]
- 7.Altfeld, M., E. S. Rosenberg, R. Shankarappa, J. S. Mukherjee, F. M. Hecht, R. L. Eldridge, M. M. Addo, S. H. Poon, M. N. Phillips, G. K. Robbins, P. E. Sax, S. Boswell, J. O. Kahn, C. Brander, P. J. Goulder, J. A. Levy, J. I. Mullins, and B. D. Walker. 2001. Cellular immune responses and viral diversity in individuals treated during acute and early HIV-1 infection. J. Exp Med. 193:169-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altfeld, M. A., A. Trocha, R. L. Eldridge, E. S. Rosenberg, M. N. Phillips, M. M. Addo, R. P. Sekaly, S. A. Kalams, S. A. Burchett, K. McIntosh, B. D. Walker, and P. J. Goulder. 2000. Identification of dominant optimal HLA-B60- and HLA-B61-restricted cytotoxic T-lymphocyte (CTL) epitopes: rapid characterization of CTL responses by enzyme-linked immunospot assay. J. Virol. 74:8541-8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 10.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 11.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, and M. G. Lewis. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 12.Beekman, N. J., P. A. van Veelen, T. van Hall, A. Neisig, A. Sijts, M. Camps, P. M. Kloetzel, J. J. Neefjes, C. J. Melief, and F. Ossendorp. 2000. Abrogation of CTL epitope processing by single amino acid substitution flanking the C-terminal proteasome cleavage site. J. Immunol. 164:1898-1905. [DOI] [PubMed] [Google Scholar]
- 13.Betts, M. R., D. R. Ambrozak, D. C. Douek, S. Bonhoeffer, J. M. Brenchley, J. P. Casazza, R. A. Koup, and L. J. Picker. 2001. Analysis of total human immunodeficiency virus (HIV)-specific CD4+ and CD8+ T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 75:11983-11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 16.Brander, C., and P. Goulder. 2000. The evolving field of HIV CTL epitope mapping: new approaches for the identification of novel epitopes. In B. T. Korber, C. Brander, B. D. Walker, R. A. Koup, J. Moore, B. Haynes, and G. Meyer (ed.), HIV molecular database. Los Alamos National Laboratory, Los Alamos, N. Mex.
- 17.Chassin, D., M. Andrieu, W. Cohen, B. Culmann-Penciolelli, M. Ostankovitch, D. Hanau, and J. G. Guillet. 1999. Dendritic cells transfected with the nef genes of HIV-1 primary isolates specifically activate cytotoxic T lymphocytes from seropositive subjects. Eur. J. Immunol. 29:196-202. [DOI] [PubMed] [Google Scholar]
- 18.Evans, D. T., D. H. O'Connor, P. Jing, J. L. Dzuris, J. Sidney, J. da Silva, T. M. Allen, H. Horton, J. E. Venham, R. A. Rudersdorf, T. Vogel, C. D. Pauza, R. E. Bontrop, R. DeMars, A. Sette, A. L. Hughes, and D. I. Watkins. 1999. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 5:1270-1276. [DOI] [PubMed] [Google Scholar]
- 19.Gaschen, B., J. Taylor, K. Yusim, B. Foley, F. Gao, D. Lang, V. Novitsky, B. Haynes, B. H. Hahn, T. Bhattacharya, and B. Korber. 2002. Diversity considerations in HIV-1 vaccine selection. Science 296:2354-2360. [DOI] [PubMed] [Google Scholar]
- 20.Goulder, P. J., C. Brander, Y. Tang, C. Tremblay, R. A. Colbert, M. M. Addo, E. S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, M. Altfeld, S. He, M. Bunce, R. Funkhouser, S. I. Pelton, S. K. Burchett, K. McIntosh, B. T. Korber, and B. D. Walker. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412:334-338. [DOI] [PubMed] [Google Scholar]
- 21.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 22.Goulder, P. J., Y. Tang, C. Brander, M. R. Betts, M. Altfeld, K. Annamalai, A. Trocha, S. He, E. S. Rosenberg, G. Ogg, C. A. O'Callaghan, S. A. Kalams, R. E. McKinney, Jr., K. Mayer, R. A. Koup, S. I. Pelton, S. K. Burchett, K. McIntosh, and B. D. Walker. 2000. Functionally inert HIV-specific cytotoxic T lymphocytes do not play a major role in chronically infected adults and children. J. Exp. Med. 192:1819-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin, X., D. E. Bauer, S. E. Tuttleton, S. Lewin, A. Gettie, J. Blanchard, C. E. Irwin, J. T. Safrit, J. Mittler, L. Weinberger, L. G. Kostrikis, L. Zhang, A. S. Perelson, and D. D. Ho. 1999. Dramatic rise in plasma viremia after CD8(+) T-cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189:991-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalams, S. A., S. P. Buchbinder, E. S. Rosenberg, J. M. Billingsley, D. S. Colbert, N. G. Jones, A. K. Shea, A. K. Trocha, and B. D. Walker. 1999. Association between virus-specific cytotoxic T-lymphocyte and helper responses in human immunodeficiency virus type 1 infection. J. Virol. 73:6715-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klenerman, P., F. Lechner, M. Kantzanou, A. Ciurea, H. Hengartner, and R. Zinkernagel. 2000. Viral escape and the failure of cellular immune responses. Science 289:2003. [DOI] [PubMed] [Google Scholar]
- 26.Klenerman, P., Y. Wu, and R. Phillips. 2002. HIV: current opinion in escapology. Curr. Opin. Microbiol. 5:408. [DOI] [PubMed] [Google Scholar]
- 27.Korber, B. T., G. Learn, J. I. Mullins, B. H. Hahn, and S. Wolinsky. 1995. Protecting HIV databases. Nature 378:242-244. [DOI] [PubMed] [Google Scholar]
- 28.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Learn, G. H., Jr., B. T. Korber, B. Foley, B. H. Hahn, S. M. Wolinsky, and J. I. Mullins. 1996. Maintaining the integrity of human immunodeficiency virus sequence databases. J. Virol. 70:5720-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, S. K., Z. Xu, J. Lieberman, and P. Shankar. 2002. The functional CD8 T-cell response to HIV becomes type-specific in progressive disease. J. Clin. Investig. 110:1339-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, S. L., J. E. Mittler, D. C. Nickle, T. M. Mulvania, D. Shriner, A. G. Rodrigo, B. Kosloff, X. He, L. Corey, and J. I. Mullins. 2002. Selection for human immunodeficiency virus type 1 recombinants in a patient with rapid progression to AIDS. J. Virol. 76:10674-10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, S. L., T. Schacker, L. Musey, D. Shriner, M. J. McElrath, L. Corey, and J. I. Mullins. 1997. Divergent patterns of progression to AIDS after infection from the same source: human immunodeficiency virus type 1 evolution and antiviral responses. J. Virol. 71:4284-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439-1443. [DOI] [PubMed] [Google Scholar]
- 34.Novitsky, V., H. Cao, N. Rybak, P. Gilbert, M. F. McLane, S. Gaolekwe, T. Peter, I. Thior, T. Ndung'u, R. Marlink, T. H. Lee, and M. Essex. 2002. Magnitude and frequency of cytotoxic T-lymphocyte responses: identification of immunodominant regions of human immunodeficiency virus type 1 subtype C. J. Virol. 76:10155-10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novitsky, V., P. Gilbert, T. Peter, M. F. McLane, S. Gaolekwe, N. Rybak, I. Thior, T. Ndung'u, R. Marlink, T. H. Lee, and M. Essex. 2003. Association between virus-specific T-cell responses and plasma viral load in human immunodeficiency virus type 1 subtype C infection. J. Virol. 77:882-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novitsky, V., N. Rybak, M. F. McLane, P. Gilbert, P. Chigwedere, I. Klein, S. Gaolekwe, S. Y. Chang, T. Peter, I. Thior, T. Ndung'u, F. Vannberg, B. T. Foley, R. Marlink, T. H. Lee, and M. Essex. 2001. Identification of human immunodeficiency virus type 1 subtype C Gag-, Tat-, Rev-, and Nef-specific Elispot-based cytotoxic T-lymphocyte responses for AIDS vaccine design. J. Virol. 75:9210-9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, H. Horton, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8:493-499. [DOI] [PubMed] [Google Scholar]
- 38.Ogg, G. S., X. Jin, S. Bonhoeffer, P. R. Dunbar, M. A. Nowak, S. Monard, J. P. Segal, Y. Cao, S. L. Rowland-Jones, V. Cerundolo, A. Hurley, M. Markowitz, D. D. Ho, D. F. Nixon, and A. J. McMichael. 1998. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science 279:2103-2106. [DOI] [PubMed] [Google Scholar]
- 39.Pitcher, C. J., C. Quittner, D. M. Peterson, M. Connors, R. A. Koup, V. C. Maino, and L. J. Picker. 1999. HIV-1-specific CD4+ T cells are detectable in most individuals with active HIV-1 infection, but decline with prolonged viral suppression. Nat. Med. 5:518-525. [DOI] [PubMed] [Google Scholar]
- 40.Rammensee, H. G., T. Friede, and S. Stevanoviic. 1995. MHC ligands and peptide motifs: first listing. Immunogenetics 41:178-228. [DOI] [PubMed] [Google Scholar]
- 41.Rosenberg, E. S., M. Altfeld, S. H. Poon, M. N. Phillips, B. M. Wilkes, R. L. Eldridge, G. K. Robbins, R. T. D'Aquila, P. J. Goulder, and B. D. Walker. 2000. Immune control of HIV-1 after early treatment of acute infection. Nature 407:523-526. [DOI] [PubMed] [Google Scholar]
- 42.Russell, K. L., C. Carcamo, D. M. Watts, J. Sanchez, E. Gotuzzo, A. Euler, J. C. Blanco, A. Galeano, A. Alava, J. I. Mullins, K. K. Holmes, and J. K. Carr. 2000. Emerging genetic diversity of HIV-1 in South America. AIDS 14:1785-1791. [DOI] [PubMed] [Google Scholar]
- 43.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 44.Sette, A., and H. M. Grey. 1992. Chemistry of peptide interactions with MHC proteins. Curr. Opin. Immunol. 4:79-86. [DOI] [PubMed] [Google Scholar]
- 45.Sette, A., and G. T. Nepom. 2000. Antigen recognition. Curr. Opin. Immunol. 12:77-79. [DOI] [PubMed] [Google Scholar]
- 46.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sijts, A. J., S. Standera, R. E. Toes, T. Ruppert, N. J. Beekman, P. A. van Veelen, F. A. Ossendorp, C. J. Melief, and P. M. Kloetzel. 2000. MHC class I antigen processing of an adenovirus CTL epitope is linked to the levels of immunoproteasomes in infected cells. J. Immunol. 164:4500-4506. [DOI] [PubMed] [Google Scholar]
- 48.Walker, B. D., and P. J. Goulder. 2000. AIDS. Escape from the immune system. Nature 407:313-314. [DOI] [PubMed] [Google Scholar]
- 49.Walker, B. D., and B. T. Korber. 2001. Immune control of HIV: the obstacles of HLA and viral diversity. Nat. Immunol. 2:473-475. [DOI] [PubMed] [Google Scholar]
- 50.Yu, X. G., M. M. Addo, E. S. Rosenberg, W. R. Rodriguez, P. K. Lee, C. A. Fitzpatrick, M. N. Johnston, D. Strick, P. J. Goulder, B. D. Walker, and M. Altfeld. 2002. Consistent patterns in the development and immunodominance of human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T-cell responses following acute HIV-1 infection. J Virol. 76:8690-8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yusim, K., C. Kesmir, B. Gaschen, M. M. Addo, M. Altfeld, S. Brunak, A. Chigaev, V. Detours, and B. T. Korber. 2002. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J. Virol. 76:8757-8768. [DOI] [PMC free article] [PubMed] [Google Scholar]